Introduction
Cutaneous squamous cell carcinoma (cSCC) is the
second most common keratinocyte cancer and accounts for ~30% of all
non-melanoma skin malignant tumors (1,2). The
5-year recurrence rate and metastasis of primary lesions is 8 and
5%, respectively. The fatality rate of cSCC metastasis is ~40%
(2). In general, long-term exposure
to ultraviolet light is the strongest risk factor, so the most
frequent sites of disease are the sun-exposed regions of head and
neck (3). Unless high-risk features
are present, the overwhelming majority of cSCC is curable with
operation, radiotherapy and/or chemotherapy (4). However, once invasion and metastasis
have occurred, the prognosis is poor.
Increasing evidence has demonstrated that
tumorigenesis, progression, invasion and metastasis of SCC involve
several signal transduction pathways and molecules, including
Notch, Integrin, tumor protein p53 (p53), epidermal growth factor
receptor (EGFR) (5). Recently,
researchers have demonstrated that autophagy might play a pivotal
role in the pathogenesis of cSCC (6,7). Autophagy
(term here used generally for macroautophagy) is a homeostatic
mechanism under starvation or stressful conditions, resulting in
the degradation of aggregated proteins and damaged organelles, and
subsequently buffering metabolic stress by recycling intracellular
components (8,9). The dysregulation of autophagy is closely
related with various pathological diseases, including
cardiomyopathy, muscular diseases, neurodegenerative disorders,
infection and cancer (10).
In cancer cells, autophagy demonstrates both
tumor-promoting and tumor-repressing properties, generally
depending on the cell type and context. During the initiation of
cancer, autophagy may mediate elimination of altered cytosolic
products, such as long-lived proteins or damaged organelles,
preventing cells from further DNA damage, genomic instability and
tumorigenesis (11,12). Once the cancer has formed, autophagy
contributes to tumor development by allowing tumor cells to survive
under starvation or hypoxia conditions (13). Autophagy has a ‘double swords’ role in
cSCC, but there is no definite time threshold for when autophagy
may promote SCC tumorigenesis or suppression, because the initial
time of pathogenesis is usually earlier than that of the clinical
diagnosis. Therefore, the relationship between autophagy and SCC
pathogenesis and progression is complex. Previous studies have,
however, indicated that autophagy-specific markers [high expression
of Beclin 1 and/or microtubule-associated protein 1 light chain 3
(LC3), and lower level of p62/sequestosome-1] may serve a key role
in controlling the tumorigenesis, progression and lymph-node
metastasis of cSCC and may be prognostic predictors of clinical
outcome (6,7,14).
Furthermore, in human squamous cell carcinoma of
head and neck (SCCHN), autophagy-related signaling pathways, such
as phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of
rapamycin (mTOR) or extracellular signal-regulated kinase (ERK) are
activated in some cases. Autophagy also alleviates apoptotic cell
death in SCCHN. Concomitant inhibition of AKT and autophagy
enhances efficient cisplatin-induced apoptosis in metastatic cSCC
(15–17). During invasion of cSCC,
epithelial-mesenchymal transition is paralleled by AKT activation
and AKT may serve as a treatment target to prohibit dissemination
of cSCC (18). However, contrary
evidence has suggested that activation of autophagic pathways is
associated with growth inhibition and senescence in metastatic cSCC
(19).
Notably, perineural invasion (PNI) is usually
defined as tumor cells invading into the perineural space,
well-recognized as one of the high-risk factors of cSCC. The
detection of PNI in cSCC indicates an aggressive tumor and predicts
a worse outcome, with higher rates of local recurrence, metastases
and poor survival (20). Clinical PNI
displays features of spread along central nerves, based on
clinical, radiological or histological evidence. Compared with
incidental PNI, clinical PNI is associated with a worse prognosis
(20) and requires a more aggressive
treatment approach. Furthermore, the 5-year overall survival rate
for clinical PNI in SCC ranges from 56–64% (21,22).
Although accumulating studies have explored the role
of autophagy-related genes in prognosis of this disease, most of
the previous study has been limited to analyzing one or two genes
in cell lines, tissue or animal models. At present, gene expression
analysis provides an efficient method for processing mass data. For
example, the finding of similar differentially expressed genes
(DEGs) in actinic keratosis (AK) and SCC by Padilla et al
(23) and Ra et al (24) confirmed that AK is a precursor lesion
of SCC and signified that both are closely related genetically. In
invasive SCC, gene analysis results suggested that IL-24
contributed to SCC invasion through enhancing focal expression of
matrix metalloproteinase (MMP) 7 (25). In addition, Ephrin type-B receptor 2
(EphB2) knockdown downregulated the gene expression of MMP1 and
MMP13, which are associated with biological functions such as cell
viability, migration and invasion of tumor cells (26). Warren et al (22) reported that alterations in the p53
pathway may be important in cSCC of head and neck (cSCCHN) with
clinical PNI, but not in cSCCHN without PNI (22). However, few analysis results of gene
expression signatures focusing on autophagy in cSCC invasion have
been documented. In addition, previous study from our research
group has demonstrated that RAB23 knockdown repressed cell
invasion, while RAB23 overexpression promoted cell invasion,
depending on the GTP-bound form of RAB23 (27). Therefore, whether RAB23 promotes cSCC
invasion may be directly associated with the autophagy process.
However, the association among RAB23 expression, autophagy and
tumor cells PNI in cSCCHN has not been defined.
In the present study, expression differences of
autophagy-related genes between cSCCHN and cSCCHN with clinical PNI
were explored. The original data series GSE86544 was downloaded
from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/). Then
autophagy-related gene expression profiles were screened within the
DEGs between cSCCHN and cSCCHN with clinical PNI. In addition, a
RAB23-related gene regulation network (http://autophagy-regulation.org/) was obtained. To
explore their biological functions and pathways, the
autophagy-related DEGs were further examined by Gene Ontology (GO)
and pathway enrichment analysis. Next, protein-protein interaction
(PPI) networks and sub-networks were constructed and analyzed.
Finally, the relationship among RAB23 expression, autophagy-related
key DEGs and tumor cells PNI was explored in cSCCHN. Collectively,
the present results demonstrated that a close correlation exists
among RAB23, autophagy and cSCCHN with clinical PNI and that
targeting these autophagy-related genes may be a promising
therapeutic strategy for cSCC.
Materials and methods
Microarray data
The microarray expression profile dataset GSE86544,
submitted by Warren et al (22) and based on the GEO Platform10588
Illumina HumanHT-12 V4.0 expression beadchip (Illumina, Inc., San
Diego, CA, USA), was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The dataset
contained 24 samples, including 9 cSCCHN, 7 cSCCHN with incidental
PNI and 8 cSCCHN with clinical PNI. In the present study, the
cSCCHN with clinical PNI or without PNI samples were analyzed by
bioinformatics. The dataset was downloaded from the GEO database on
February 2, 2017.
Data preprocessing and analysis of
DEGs
The original microarray data were transformed into
expression measures. Background correction, quartile data
normalization and probe synopsis were performed with GEO2R online,
using the Robust Multi-array Average (RMA) algorithm in the Raffy
package (https://www.bioconductor.org/packages/release/bioc/html/affy.html).
To obtain the P-value, multiple testing corrections were applied
with the Benjamini-Hochberg method. Only those genes exhibiting
log2 fold change (FC) >1.5 and adjusted P<0.05 were
considered to be DEGs. When multiple probes matched to the same
gene, the average expression value of probes was calculated.
Nonspecific probes were filtered.
Autophagy-related genes (https://www.ncbi.nlm.nih.gov/gene) were downloaded by
using the key words ‘autophagy’ and ‘Homo sapiens’. The 907
autophagy-related genes were obtained as a symbol gene list, and
was compared with the list of DEGs. The comparison resulted in a
list of 239 autophagy-related DEGs. The RAB23-related gene
regulation network (http://autophagy-regulation.org/) (28) was also searched, which is compatible
with Cytoscape software.
GO and Kyoto encyclopedia of genes and
genomes (KEGG) enrichment analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID 6.8; https://david.ncifcrf.gov/) consists of a set of
functional annotation tools, which have been developed for linking
functional terms with lists of genes by clustering algorithms
(29). To analyze the identified DEGs
at the functional level, GO enrichment analysis was performed to
annotate genes or gene products and identify characteristic
biological properties for genome or transcriptome data using the
DAVID online tool. Additionally, the KEGG pathway database was used
for systematic analysis of gene functions and related pathways.
P<0.05 was considered to indicate a statistically significant
difference.
Hierarchical clustering of DEGs and
data visualization
The series matrix file for GSE86544 was obtained
from GEO DataSets (downloaded on February 2nd, 2017). The Pearson's
correlation coefficient was performed using Cluster 3.0 with
average linkage in order to achieve the hierarchical heatmap
(30). Analysis results were
visualized employing the Java TreeView 1.1.6 version (31).
PPI network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING; version 10.0) covers 5,214,234 proteins from 1,133
organisms (32). In the present
study, to evaluate the interactive relationships among DEGs, the
239 DEGs were mapped to the STRING database, and only
experimentally validated interactions (combined score >0.4) were
selected as significant.
PPI networks were constructed using the Cytoscape
3.3.0 software (http://www.cytoscape.org/). The degree of connectivity
is evident by the number of edges linked to a given node. The nodes
with a high degree are regarded as the key genes that possess vital
biological functions. Then, the degree of connectivity was analyzed
to obtain the key proteins in the PPI network. The plug-in
Molecular Complex Detection (MCODE) was used to screen the modules
of the PPI network (MCODE scores >30 and number of nodes >4)
in Cytoscape.
Statistical analysis
The relationship between RAB23 and the key genes was
analyzed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of DEGs
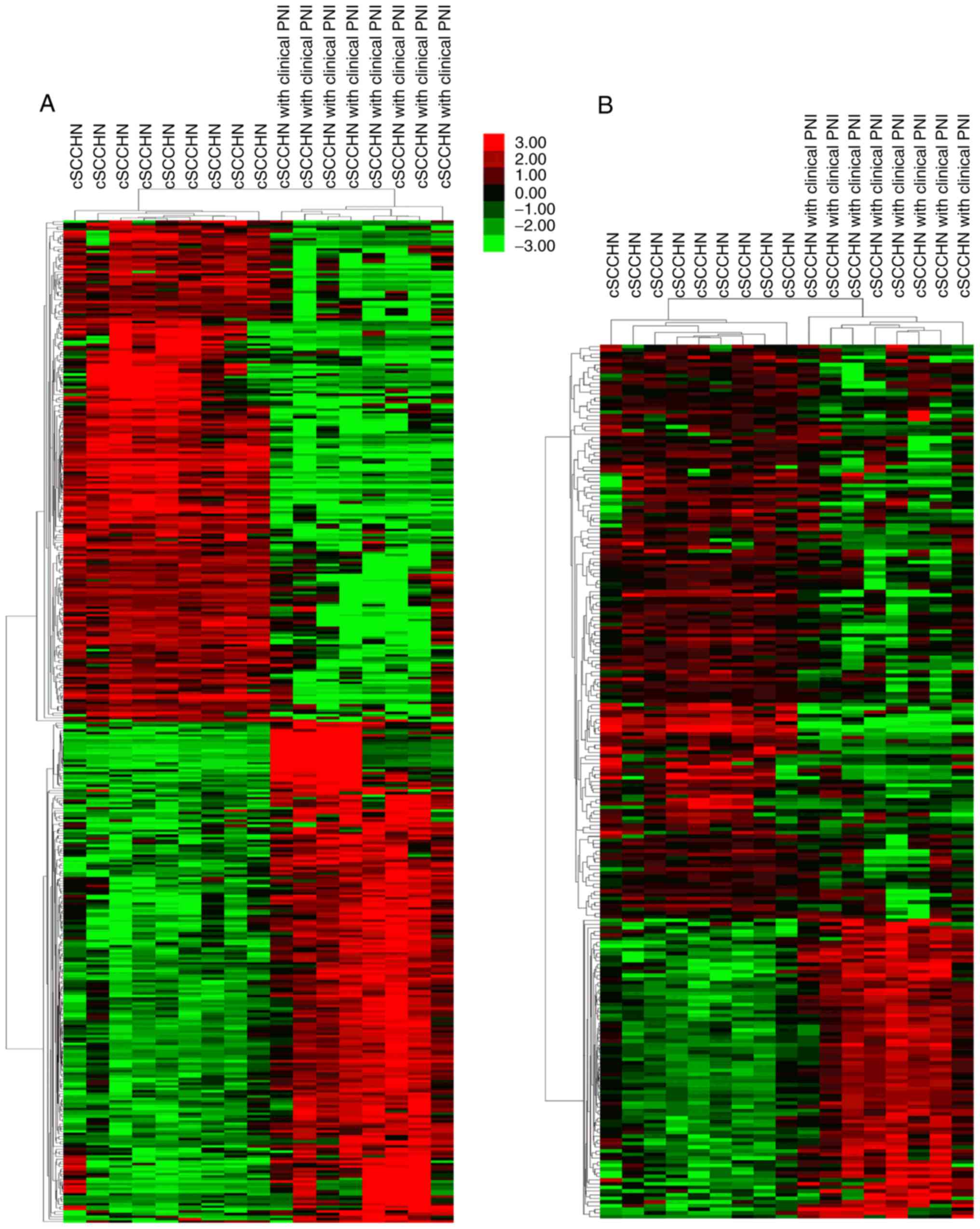
Following data processing, a total of 2,858 DEGs
were identified in cSCCHN with clinical PNI, compared with cSCCHN
samples without PNI involvement. These DEGs contained 921
upregulated and 1,937 downregulated genes, and the top 200 up and
downregulated genes were clustered (Fig.
1A). In comparison to DEGs, 239 autophagy-related genes were
screened and were also clustered employing the Series Matrix Files
(Fig. 1B).
GO and pathway enrichment
analysis
The 239 autophagy-related DEGs were uploaded to the
online DAVID tool in order to identify over-represented GO
categories and KEGG pathways. The results from GO analysis
demonstrated that upregulated DEGs mainly focused in biological
processes (BP), including tumor necrosis factor-mediated signaling
pathway, autophagy, positive regulation of MAP kinase activity, and
ERK1 and ERK2 cascade. The downregulated DEGs were significantly
enriched in autophagy, signal transduction, macroautophagy and
positive regulation of gene expression (Table I).
 | Table I.GO functional enrichment analysis for
the most significantly up and downregulated DEGs of autophagy. |
Table I.
GO functional enrichment analysis for
the most significantly up and downregulated DEGs of autophagy.
| Category | Fold Term/gene
function | Gene count | Genes | P-value | enrichment | FDR |
|---|
| A, Upregulated
autophagy-related DEGs |
|---|
|
|---|
| GOTERM_BP | GO:0033209/tumor
necrosis factor-mediated signaling pathway | 7 | TNFRSF10B, TNFSF4,
TNFSF11, KRT8, TNFSF13, CD40, AIM2 | 1.33E-05 | 13.28 | 0.021023 |
| GOTERM_BP |
GO:0006914/autophagy | 7 | EVA1A, ATG4C,
TRIM17, CHMP4A, EPM2A, ABL1, RAB33B | 2.63E-05 | 11.78 | 0.041524 |
| GOTERM_BP | GO:0045893/positive
regulation of transcription, DNA-templated | 11 | ATF4, CDKN2A,
HIF1A, PSEN1, AGT, SPI1, IGF1, KAT5, DDIT3, ARHGEF11, MT3 | 8.33E-05 | 4.78 | 0.13156 |
| GOTERM_BP | GO:0043406/positive
regulation of MAP kinase activity | 5 | FLT1, TNFSF11,
PSEN1, ERBB2, CD40 | 1.32E-04 | 18.97 | 0.207954 |
| GOTERM_BP | GO:0070371/ERK1 and
ERK2 cascade | 4 | TNFSF11, AGT, IGF1,
MT3 | 1.56E-04 | 37.32 | 0.24571 |
| GOTERM_CC |
GO:0005829/cytosol | 30 | ARFGAP1, MCL1,
XIAP, CHMP4A, TNFSF13, CDKN1A, ARHGAP33, HIF1A, DACT1, ATG4C,
MAPK8, ABL1, KPNA2, ABL2 | 2.45E-05 | 2.17 | 0.029494 |
| GOTERM_CC |
GO:0005615/extracellular space | 16 | SOGA1, FLT1,
TNFSF4, CSF1, CXCL2, IGF1, TNFSF13, CD40, TNFSF9, GRP, TNFSF11,
CTGF, AGT, SERPINA1, GOLM1, MT3 | 3.33E-04 | 2.85 | 0.400461 |
| GOTERM_CC |
GO:0048471/perinuclear region of
cytoplasm | 10 | HCRT, CDKN1A, CTGF,
CSF1, ERBB2, KAT5, ABL1, SRCAP, PRKCD, MT3 | 9.89E-04 | 3.86 | 1.185127 |
| GOTERM_CC |
GO:0005654/nucleoplasm | 22 | STX5, RAD51B, XIAP,
OLR1, MCL1, ARID5A, NR4A1, TNFSF13, KAT5, HIST2H4A, PRKCD, DDIT3,
ATF4, CDKN1A, TCF20, DACT1, HIF1A, CDKN2A, KRT8, MAPK8, ABL1,
KPNA2 | 0.003401 | 1.895 | 4.02117 |
| GOTERM_CC |
GO:0005622/intracellular | 13 | STX5, SOCS3,
TRIM17, PRKCD, RAB33B, ARHGEF11, TNFSF11, TNFRSF10B, PSEN1, TGM2,
RAB23, MAPK8, MT3 | 0.007892 | 2.34 | 9.105383 |
| GOTERM_MF | GO:0005515/protein
binding | 54 | RAD51B, RAB23,
HIF1A, ARHGAP33, TAGLN, PSEN1, CLDN2, MAPK8, ARFGAP1, KPNA2, ERBB2,
CDKN1A, ATF4, TXNDC16 | 3.95E-05 | 1.46 | 0.050049 |
| GOTERM_MF | GO:0032813/tumor
necrosis factor receptor super family binding | 3 | TNFSF4, TNFSF11,
TNFSF9 | 1.68E-04 | 142.65 | 0.212711 |
| GOTERM_MF | GO:0005164/tumor
necrosis factor receptor binding | 4 | TNFSF4, TNFSF11,
TNFSF13, TNFSF9 | 2.31E-04 | 32.79 | 0.292052 |
| GOTERM_MF | GO:0005125/cytokine
activity | 5 | TNFSF4, TNFSF11,
CSF1, TNFSF13, TNFSF9 | 0.006134 | 6.76 | 7.495498 |
| GOTERM_MF | GO:0042826/histone
deacetylase binding | 4 | DACT1, HIF1A,
MAPK8, KPNA2 | 0.008748 | 9.32 | 10.52961 |
|
|
Category | Term/gene
function | Gene
count | Genes | P-value | Fold
enrichment | FDR |
|
| B, Downregulated
autophagy-related DEGs |
|
| GOTERM_BP |
GO:0006914/autophagy | 15 | TM9SF1, S100A8,
TOLLIP, TSG101, TFEB, FOXO1, SIRT2, TMEM208, SRPX, RB1CC1, ATG4A,
WDR24, MTOR, VPS28, HAP1 | 6.47E-12 | 13.24 | 1.07E-08 |
| GOTERM_BP | GO:0007165/signal
transduction | 27 | FGF7, PGF, TOLLIP,
BTRC, PPARG, ARHGAP19, KIT, TNFSF12, CXCL12, TNFRSF1A, CD34, TXN,
SMPD1, MST1R, MTOR, ARAP2 | 4.26E-06 | 2.73 | 0.007014 |
| GOTERM_BP |
GO:0006687/glycosphingolipid metabolic
process | 6 | GBA3, ARSF, SMPD1,
PSAPL1, KIT, GBA | 3.75E-05 | 15.66 | 0.061725 |
| GOTERM_BP |
GO:0016236/macroautophagy | 7 | CLN3, RB1CC1,
PRKAA1, MTOR, WIPI2, PIK3R4, ACBD5 | 4.42E-05 | 10.82 | 0.072771 |
| GOTERM_BP | GO:0010628/positive
regulation of gene expression | 11 | WNT10A, TNF, LAMP3,
CD34, PRKAA1, KIT, MTOR, MYC, GBA, TLR9, MYCN | 7.87E-05 | 4.93 | 0.12943 |
| GOTERM_CC |
GO:0005776/autophagosome | 8 | CLN3, ATG9B, SRPX,
HTT, WIPI2, PIK3R4, USP33, HAP1 | 8.14E-07 | 15.39 | 0.001036 |
| GOTERM_CC |
GO:0005615/extracellular space | 27 | TNF, S100A8,
SERPINA12, PGF, PSAPL1, TNFSF12, KIT, CXCL12, CCL27, KRT81, ARG1,
TNFRSF1A | 2.65E-05 | 2.47 | 0.033709 |
| GOTERM_CC |
GO:0005764/lysosome | 10 | CLN3, LAMP2, CD34,
AKR1B10, SMPD1, LRBA, PSAPL1, MTOR, HAP1, TLR9 | 9.26E-05 | 5.45 | 0.117814 |
| GOTERM_CC |
GO:0005829/cytosol | 44 | S100A8, TOLLIP,
BTRC, COPZ1, PPARG, ARHGAP19, PKMYT1, FOXO1, AURKA, BCL2L1, WIPI2,
MAPK3, PKLR, TXN, MTOR, VPS28 | 6.86E-04 | 1.63 | 0.869748 |
| GOTERM_CC |
GO:0016020/membrane | 32 | TNF, PGF, LRBA,
PKMYT1, BCL2L1, KIT, TNFSF12, CLN3, RAB32, LAMP2, LAMP3, CD209,
ARL8B, MTOR, DNAJB6, LRPPRC | 0.001292 | 1.79 | 1.631639 |
| GOTERM_MF |
GO:0043130/ubiquitin binding | 6 | ADRM1, TSG101,
TOLLIP, VPS28, USP33, SIRT2 | 4.90E-04 | 9.14 | 0.659317 |
| GOTERM_MF | GO:0004672/protein
kinase activity | 11 | FASTKD1, MAPK13,
PRKCI, PKMYT1, PRKAA1, AURKA, MAPK10, MTOR, FASTKD5, PIK3R4,
EPHA1 | 8.77E-04 | 3.64 | 1.176364 |
| GOTERM_MF | GO:0005515/protein
binding | 91 | RAD51D, FOXO1,
AURKA, TNFSF12, KRT80, APOE, PCBP1, FBXO27, BCL2L1, PACRGL, MYC,
TNF, BCL2L1, MTOR, VPS28 | 0.003005 | 1.23 | 3.980066 |
| GOTERM_MF |
GO:0048487/β-tubulin binding | 4 | HTT, ARL8B, SIRT2,
LRPPRC | 0.003329 | 13.20 | 4.40004 |
| GOTERM_MF | GO:0004707/MAP
kinase activity | 3 | MAPK13, MAPK3,
MAPK10 | 0.005902 | 25.47 | 7.677845 |
By KEGG analysis, the upregulated DEGs of autophagy
mainly focused on cytokine-cytokine receptor interaction, pathways
in cancer, ErbB signaling pathway, hypoxia inducible factor (HIF)-1
signaling pathway and transcriptional misregulation in cancer. By
contrast, the downregulated DEGs of autophagy distributed in
insulin resistance, pathways in cancer, forkhead box O (FoxO)
signaling pathway and insulin signaling pathway (Table II).
 | Table II.KEGG pathway enrichment analysis for
the most significant autophagy-related DEGs. |
Table II.
KEGG pathway enrichment analysis for
the most significant autophagy-related DEGs.
| A, Upregulated
autophagy-related DEGs |
|---|
|
|---|
| Pathway ID | Name | Count | Genes | P-value | Fold
enrichment | FDR |
|---|
| hsa04060 | Cytokine-cytokine
receptor interaction | 9 | TNFSF4, FLT1,
TNFSF11, CXCR4, CSF1, TNFSF13, CD40, BMPR1B, TNFSF9 | 1.17E-04 | 5.75 | 0.136364 |
| hsa05200 | Pathways in
cancer | 11 | CDKN1A, CDKN2A,
HIF1A, XIAP, CXCR4, ERBB2, SPI1, IGF1, MAPK8, ABL1, ARHGEF11 | 2.03E-04 | 4.12 | 0.237542 |
| hsa04012 | ErbB signaling
pathway | 5 | CDKN1A, ERBB2,
MAPK8, ABL1, ABL2 | 0.002557 | 8.45 | 2.95308 |
| hsa04066 | HIF-1 signaling
pathway | 5 | CDKN1A, HIF1A,
FLT1, ERBB2, IGF1 | 0.003935 | 7.50 | 4.510872 |
| hsa05202 | Transcriptional
misregulation in cancer | 6 | CDKN1A, FLT1, SPI1,
IGF1, CD40, DDIT3 | 0.004902 | 5.25 | 5.590958 |
|
| B, Downregulated
autophagy-related DEGs |
|
| Pathway
ID | Name | Count | Genes | P-value | Fold
enrichment | FDR |
|
| hsa04931 | Insulin
resistance | 8 | TNFRSF1A, TNF,
FOXO1, PRKAA1, MAPK10, MTOR, PCK2, PPARGC1B | 2.33E-04 | 6.31 | 0.285418 |
| hsa05200 | Pathways in
cancer | 14 | WNT10A, WNT10B,
FGF7, PGF, PPARG, FOXO1, SMAD2, BCL2L1, KIT, MAPK10, CXCL12, MAPK3,
MTOR, MYC | 5.15E-04 | 3.04 | 0.629603 |
| hsa04068 | FoxO signaling
pathway | 8 | MAPK13, MAPK3,
FBXO25, FOXO1, PRKAA1, SMAD2, MAPK10, PCK2 | 8.63E-04 | 5.09 | 1.052464 |
| hsa04910 | Insulin signaling
pathway | 8 | MAPK3, PKLR, PRKCI,
FOXO1, PRKAA1, MAPK10, MTOR, PCK2 | 0.001026 | 4.95 | 1.250842 |
| hsa05142 | Chagas disease
(American trypanosomiasis) | 7 | TNFRSF1A, TNF,
MAPK13, MAPK3, SMAD2, MAPK10, TLR9 | 0.001228 | 5.74 | 1.49469 |
Finally, the intersection of genes from GO and KEGG
pathway analysis was obtained, including as the upregulated genes
HIF1A and mitogen-activated protein kinase 8 (MAPK8), and the
downregulated genes mammalian target of rapamycin (mTOR) and B-cell
lymphoma 2 like 1 (BCL2L1). These genes were considered to be key
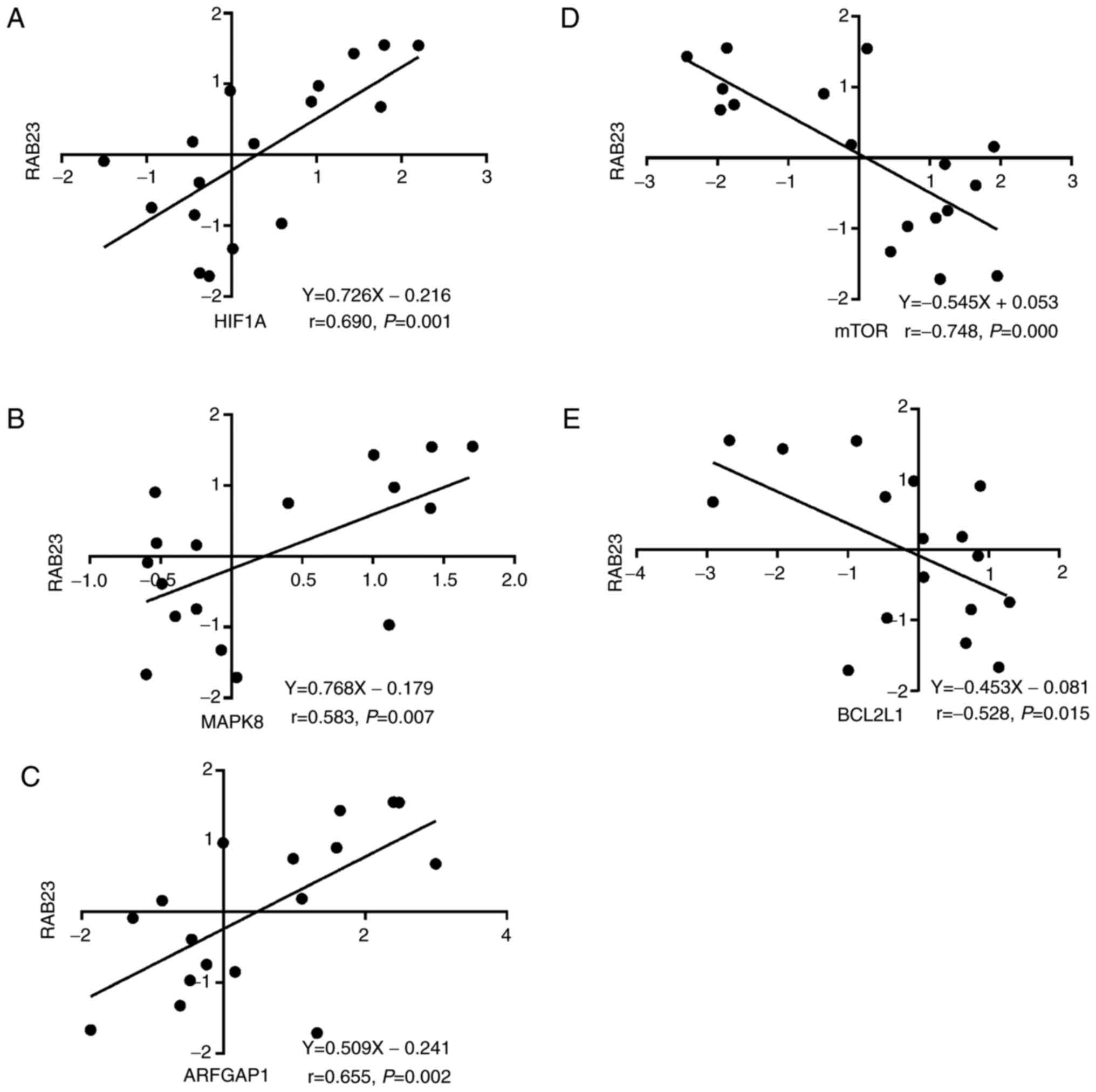
genes in cSCCHN with clinical PIN. In addition, as RAB23 gene
expression was positively correlated with HIF1A (P=0.001, r=0.690;
Fig. 2A), MAPK8 (P=0.007, r=0.583;
Fig. 2B) and ADP ribosylation factor
GTPase activating protein 1 (ARFGAP1; P=0.000, r=0.655; Fig. 2C), but negatively associated with mTOR
(P=0.002, r=−0.748; Fig. 2D) and
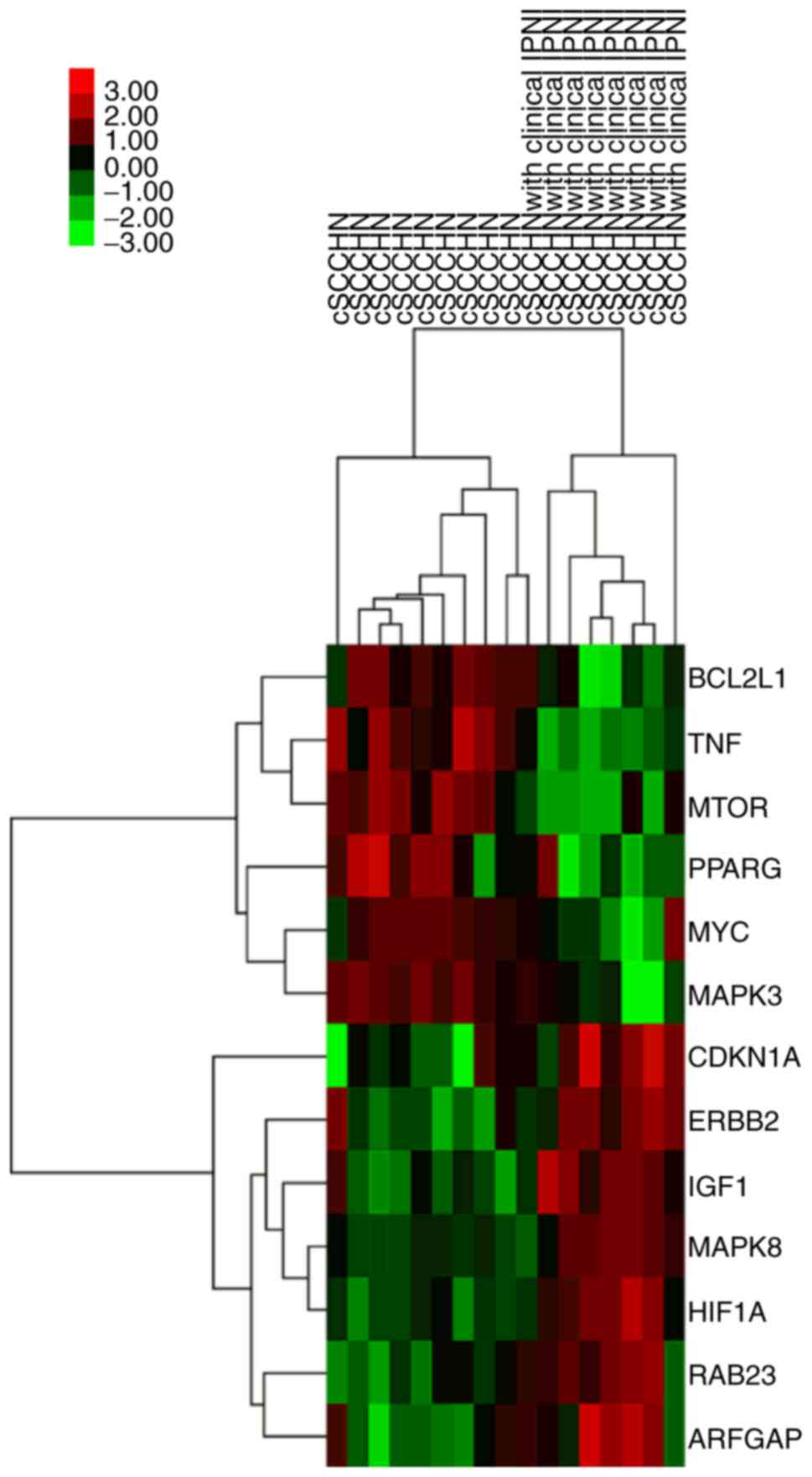
BCL2L1 (P=0.015, r=−0.528; Fig. 2E).
Hierarchical clustering analysis revealed consistent results
(Fig. 3).
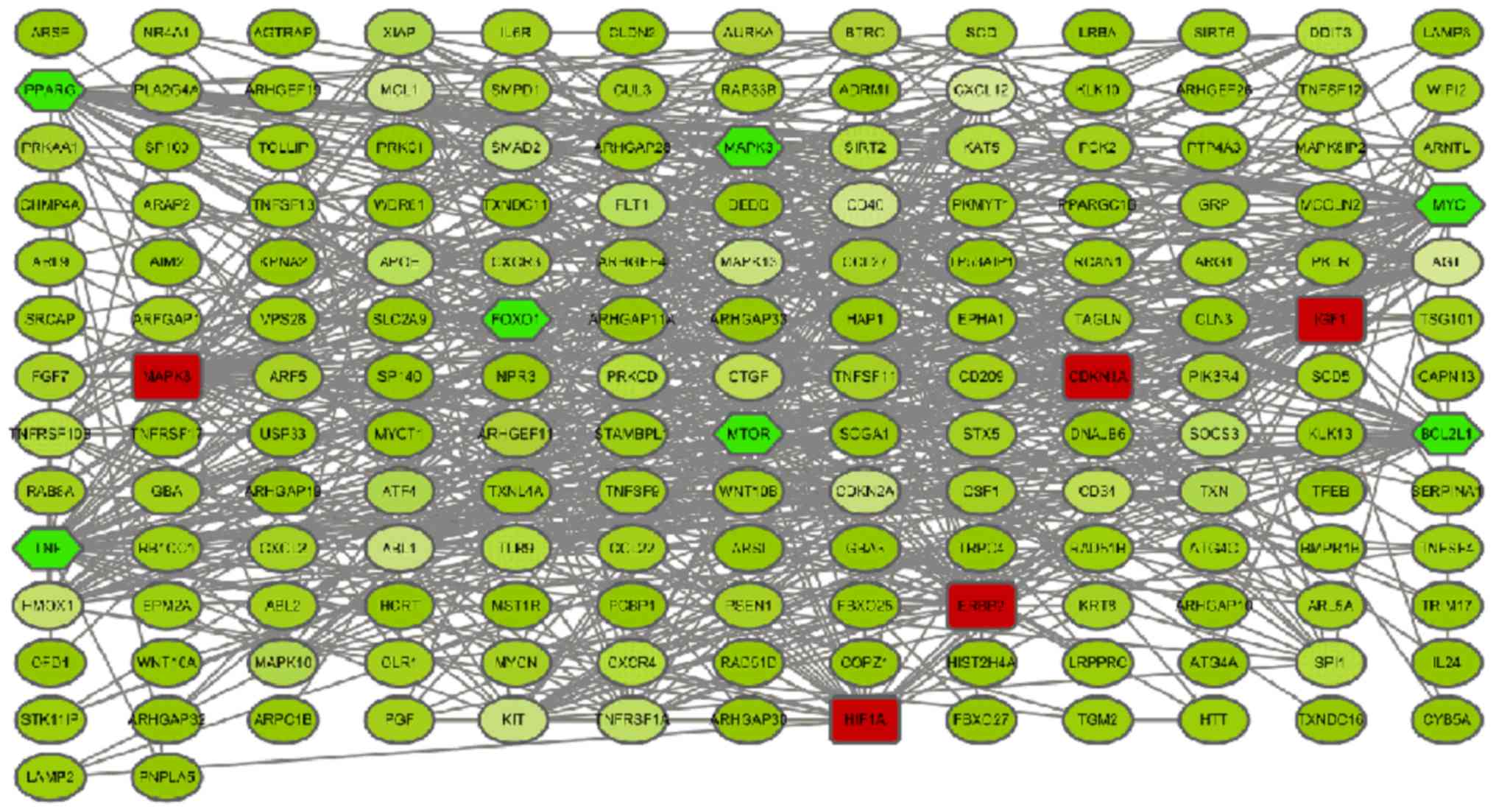
PPI network and module screening
Based on the information of the 239
autophagy-related genes contained in the STRING database, 171 nodes
and 669 edges formed (Fig. 4). A
total of 11 nodes were regarded as key proteins (degree ≥30) in the
PPI network. This included upregulated genes, such as MAPK8 with
highest node degree 41, followed by erb-b2 receptor tyrosine kinase
2 (ERBB2) (31), and HIF1A (30), and downregulated genes, such as tumor
necrosis factor (TNF) with highest node degree 44, followed by
MYC(42), BCL2L1 (36), mTOR (34), and peroxisome proliferator activated
receptor γ (PPARγ) (32). Modules of
PPI network were screened and subnetworks were constructed
(Fig. 5). In addition, a
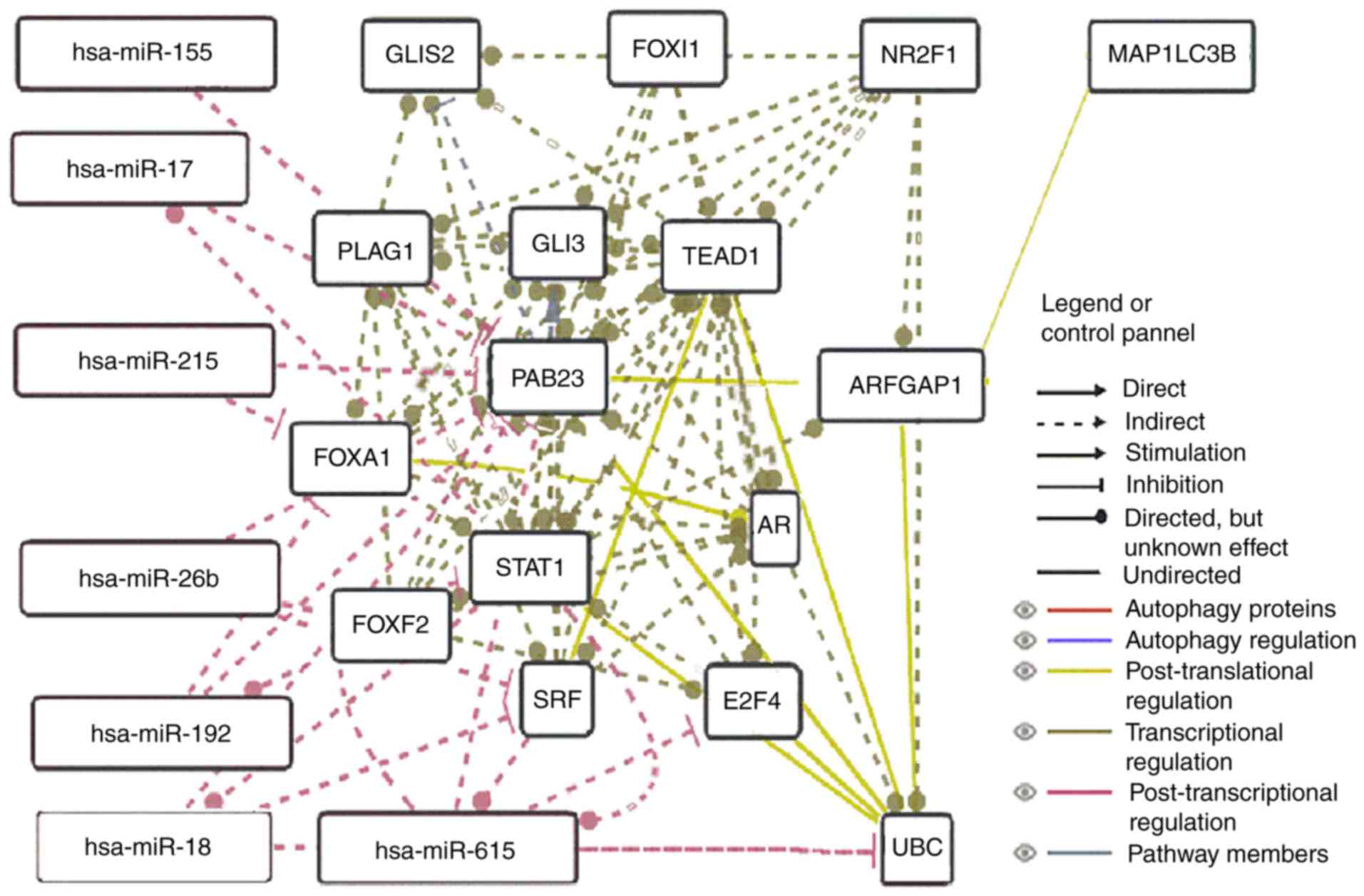
RAB23-related network predicted that there was a close association
among RAB23, UBC (ubiquitin C) and ARFGAP1 based on
post-translational regulation, suggesting a potential regulatory
role of RAB23 in ARFGAP1 (Fig.
6).
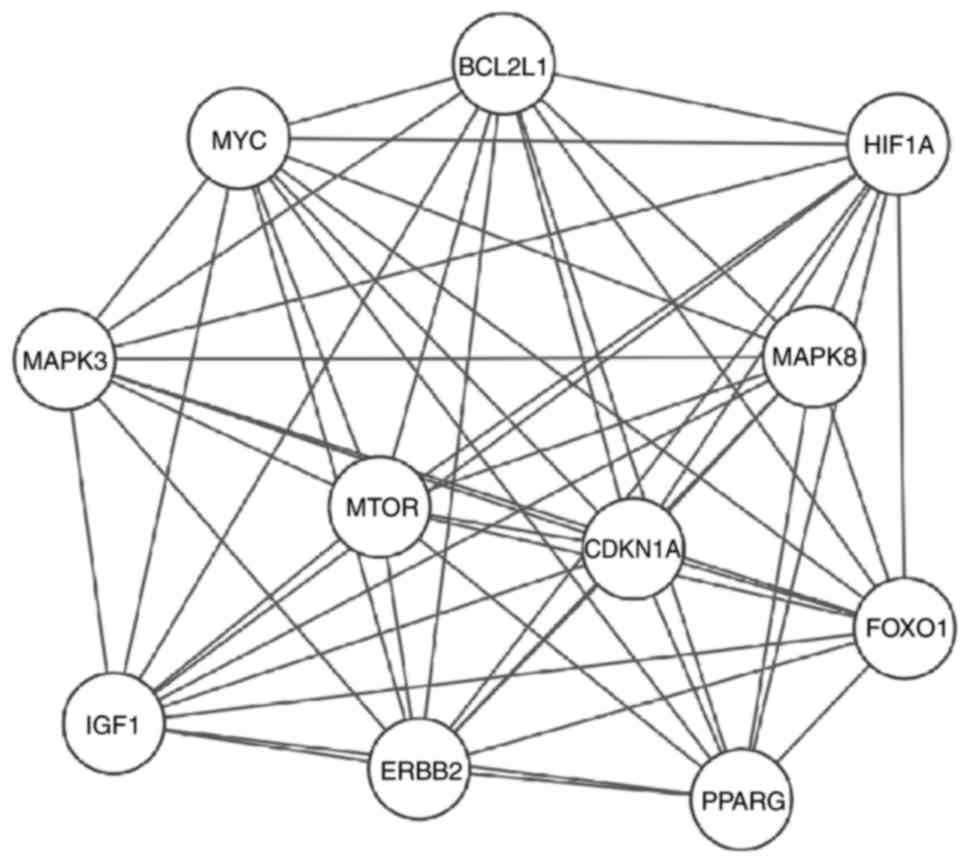 | Figure 5.A sub-network of key proteins was
constructed (degree ≥30). The upregulated proteins include MAPK8,
ERBB2, HIF1A, IGF1 and CDKN1A. The downregulated proteins include
MYC, BCL2L1, mTOR, PPARG and FOXO1. MAPK8, mitogen-activated
protein kinase 8; ERBB2, erb-b2 receptor tyrosine kinase 2; HIF,
hypoxia-inducible factor; IGF, insulin growth factor; CDKN1A,
cyclin dependent kinase inhibitor 1A; MYC, MYC proto-oncogene bHLH
transcription factor; BCL2L1, B-cell lymphoma 2 like 1; mTOR,
mammalian target of rapamycin; PPARG, peroxisome proliferator
activated receptor γ; FOXO1, Forkhead box O1. |
Discussion
Cutaneous squamous cell carcinoma (cSCC) accounts
for the second most common non-melanoma deriving skin cancer
worldwide. Although most of cases can be successfully treated with
surgery, there is a subset of lesions with invasion and metastasis,
resulting in severe morbidity and mortality (1,2,33). Histopathologically, lesions with
thickness >2 mm, poor differentiation, invasion of the
subcutaneous tissue or structures like vascular and lymphatic, or
involvement of nerves (>0.1 mm in diameter) predict poor
prognosis in patients with cSCC (22,34,35).
To date, although many gene expression data of SCCHN
or cSCC have been analyzed by bioinformatics methods, few reports
regarding the role of autophagy-related genes in cSCC have been
recorded (23,36,37).
In view of the variability of cSCC biological
behavior, early identification of high-risk factors for PNI in cSCC
is very important. However, to date, there has not been a gene
signature, especially for autophagy-related genes, that can
efficiently evaluate the degree of PNI in order to provide improved
early intervention treatment.
In the present study, gene expression profile data
were downloaded from the GEO database and analyzed for DEGs in
cSCCHN using bioinformatics analysis, focusing on autophagy-related
genes. A total of 2,858 DEGs were identified between cSCCHN with
clinical PNI and cSCCHN samples, containing 921 upregulated and
1,937 downregulated genes. Of these, 239 autophagy-related DEGs
(157 upregulated and 82 downregulated) were screened and clustered,
employing series matrix files.
Increasing evidence has demonstrated that
co-expressed genes, with similar expression profiles, frequently
participate in parallel biological process. To better understand
the interactions among the autophagy-related DEGs, GO and KEGG
pathway analysis were performed. The GO term analysis indicated
that upregulated DEGs were mainly involved in TNF-mediated
signaling pathway, autophagy, positive regulation of transcription
or MAP kinase activity and ERK1/2 cascade. Downregulated DEGs were
involved in autophagy, signal transduction, glycosphingolipid
metabolic process, macroautophagy and positive regulation of gene
expression. This is consistent with the knowledge that defective
function of TNF-mediated signaling pathway and autophagy is partly
responsible for tumor progression and invasion.
Furthermore, by KEGG pathway analysis, the
upregulated autophagy-related DEGs were enriched in
cytokine-cytokine receptor interaction, pathways in cancer, ErbB
signaling pathway, HIF-1 signaling pathway and transcriptional
misregulation in cancer. The enriched KEGG pathways of the
downregulated DEGs consisted of insulin resistance, pathways in
cancer, FoxO signaling pathway and insulin signaling pathway.
Previous studies have demonstrated that autophagy-related up and
downregulated genes in human breast cancer can predict the survival
rate and prognosis of breast cancer patients (38). Therefore, the key autophagy-related
DEGs identified in the present study, namely HIF1A, MAPK8, mTOR,
BCL2L1 and RAB23, may be involved in cSCCHN with clinical PNI.
Based on the PPI network, the top degree key proteins were
obtained. Among the DEGs, HIF1A and ERBB2 can be activated by
corresponding ligands or external stimuli signals, further
triggering signal transduction by downstream core member mTOR, or
pathways such as IKKB-mTOR, PI3K-AKT-mTOR or MEK-ERK1/2-mTOR, and
finally downregulate mTOR. Autophagy has the potential to promote
either cell survival or cell death, depending on the context of
different cancers, clinicopathological stages and post-irradiation
status (39). Notably, mTOR is the
first key autophagy-related gene and repressed mTOR may activate
the autophagy process (40). At
present, mTOR inhibitors, such as everolimus or temsirolimus,
inhibit cell proliferation of head and neck cancer cell lines in
vitro; however, in some phase I/II studies, only a partial
response was observed and the curative effect was variable.
Therefore, targeting both PI3K and mTOR concurrently is
hypothesized to be more efficient. Two studies have demonstrated
that NVP-BEZ235 (both a PI3K and mTOR kinase inhibitor) can induce
autophagy in cells, and combination treatment with autophagy
inhibitors, NVP-BEZ235 and radiation increased cell death (41,42). The
autophagy-related genes MAPK8, ERBB2 and HIF1A were found
upregulated in tumor cells suggesting a constitutive activation of
the autophagy machinery in cSCCHN with clinical PIN. Although TNF
was identified as one of the key genes which exhibit the highest
degree of connectivity, it was not included in the PPI subnetwork.
Other post-transcription regulation mechanisms may therefore be
involved, and this will need to be further verified in future
studies.
The second key gene HIF1A, functions to buffer
stressful response to hypoxia by activating transcription of
multiple genes and to maintain cellular and systemic homeostasis.
Therefore, HIF1A serves a key role in embryonic vascularization,
angiogenesis in tumor and pathophysiology process of ischemic
diseases. Hypoxia can activate autophagy in cancer cells and
further induce clearance of p62 protein, suggesting a role for p62
in the regulation of cell survival responses (43,44). In
the oral squamous cell carcinoma cell line Tca8113, the HIF1A
inhibitor PX-478 downregulated the expression levels of LC3-II/I
and inhibited the autophagy process (45). Low oxygen levels in SCCHN promote more
invasive phenotypes (46) and
correlate with poor local control after radiation. Blocking EGFR,
and hence PI3K-AKT signaling, led to improved oxygenation.
Constitutively active AKT favors HIF-1-dependent gene
transcription, which modulates angiogenesis, pH, and glucose
metabolism. In addition, other studies have highlighted the role of
transcription factors p53, E2F and HIF1A in regulating the
transcription of autophagy-related genes, including
autophagy-related 5 (ATG5), BCL2-interacting protein 3 (BNIP3),
Unc-51 like autophagy activating kinase 1 (ULK1), LC3 and DNA
damage-regulated autophagy modulator (DRAM), subsequently
activating autophagy in cancer cells in response to hypoxia or
chemotherapy (47,48). Furthermore, HIF1A appears to regulate
the expression of MYC in a direct way and is regarded as the key
node in the interactive network.
The third key gene MAPK8, also known as c-Jun
N-terminal kinase (JNK), participates in a wide variety of
biological processes, such as proliferation, differentiation and
transcription regulation. Various cell stimuli can activate MAPK8,
which subsequently targets specific transcription factors, and
finally mediates immediate-early gene expression. In addition,
MAPK8 is involved in apoptosis induced by UV radiation, which is
considered to be associated with the signal pathway of cytochrome
c-mediated cell death. Furthermore, MAPK8 pathway is indispensable
for TNF superfamily 10 (TNFSF)-induced autophagy. Blocking MAPK8
with the inhibitor SP600125 effectively reduced degradation of
BCL2L1 and expression of the autophagy-suppressing
BCL2L1-BECN1complex. Knockdown of TNF receptor associated factor 2
(TRAF2) or receptor interacting serine/threonine kinase 1 (RIPK1)
by small interfering RNA effectively repressed TNFSF10-induced
MAPK8 activation and autophagy levels (49).
RAB-like 3 (RABl3), as a member of the Rab subfamily
of small GTPases, participates in controlling cell proliferation
and vesicular trafficking. Recently, Zhang et al (50) demonstrated that knockdown of RABl3 in
lung cancer cells significantly increased cell death with autophagy
induction, as demonstrated by an elevated level of LC3-II.
Interestingly, RABl3 knockdown was also associated with enhanced
activation of MAPK8/9/10, except for MAPK11/12/13/14. Treatment
withSP600125 (a MAPK8/9/10-specific inhibitor) significantly
abolished RABl3 knockdown-induced LC3-II levels and autophagic cell
death (50). Similar to RABl3, RAB23
is likely involved in the autophagy process in cSCCHN with clinical
PNI: Nozawa et al (51)
reported that RAB9A and RAB23 were novel partners of autophagy
regulation against group A streptococcus infection. However, the
autophagosome-like structure or autophagosome lacked direct
evidence and needs to be additionally confirmed. Following
treatment with 30 mJ/cm2 UVB, wild-type RAB23 promotes
the expression of LC3-II and Beclin1, while knockdown of RAB23
decreases the levels of LC3-II and Beclin 1 (52). Consistent with this result, the
results of fluorescence microscopy demonstrated that wild-type
RAB23 largely increases the number of autophagosomes (52). In addition, RAB23 promotes cell
proliferation, migration and invasion in human astrocytoma,
possibly through upregulating Rac1 activity (53). The present data indicated ARFGAP1 may
be an effector of RAB23, which is associated with the autophagic
marker LC3B. Overall, RAB23 is likely to indirectly promote
autophagy.
As previously discussed, the anti-apoptotic proteins
of the BCL-2 family negatively regulate the assembly of phagophore
membrane. BCL-2 and BCL2L1 were proved to inhibit autophagy by
binding Beclin 1 at the BH3 domain site (54,55).
BCL2L1 protein is located at the outer mitochondrial membrane and
functions to regulate the opening of the outer mitochondrial
membrane channel, a kind of voltage-dependent anion channel (VDAC).
VDAC further regulates the mitochondrial membrane potential and
controls the production of reactive oxygen species (ROS) and
release of cytochrome c, both of which effectively induce cell
apoptosis (56). Knocking down
myeloid cell leukemia sequence-1 (Mcl-1) sensitizes oral SCC cells
to ABT-737 (a BH3 mimetic), which binds to BCL2L1 but not Mcl-1
(57).
Dysregulation of autophagy contributes to the
progression of cancer. Recently, based on gene expression profiles,
Gu et al (38) reported the
predictive value of autophagy for assessing prognosis of breast
cancer. The authors identified a set of eight autophagy-related
genes, including BCL2, GAPDH and vascular endothelial growth factor
A, which were closely related with overall survival in breast
cancer (38). Further analysis
demonstrated that the prognostic value of the autophagy signature
was independent of known clinical prognostic factors, such as ERBB2
status, lymph node status and p53 mutation status. The results from
the present study demonstrated that a close correlation may also
exist between autophagy and cSCC outcome, and that
autophagy-related genes are promising treatment target for skin
cancer.
In short, a total of 239 DEGs were identified
between cSCCHN with clinical PNI and cSCCHN without neural
involvement. Genes such as MAPK8, HIF1A, mTOR, BCL2L1 and RAB23 may
be potential therapeutic target genes in cSCCHN. Furthermore, RAB23
may promote autophagy in cSCC through ARFGAP1 in an indirect
way.
As far as clinical significance of these target
genes is concerned, several pathway inhibitors have exhibited
antitumor activity in vitro or in preclinical models, but
this has not always provided meaningful benefits to patients with
SCCHN. Therefore, agents or drugs that target multiple receptors
alone or in combination with other medicines will likely provide
the most efficient therapeutic effect for patients.
The present study has several limitations, such as
small sample size and lack of experimental verification, possibly
generating false positive results. Therefore, further experimental
studies with larger sample sizes will be needed in the future to
confirm the results.
Acknowledgements
The present study was funded by the National Natural
Sciences Foundation of China (grant nos. 31371412, 81572680 and
81673043).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rogers HW, Weinstock MA, Harris AR,
Hinckley MR, Feldman SR, Fleischer AB, Fleischer AB and Coldiron
BM: Incidence estimate of non-melanoma skin cancer in the United
States, 2006. Arch Dermatol. 146:283–287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eisemann N, Waldmann A, Geller AC,
Weinstock MA, Volkmer B, Greinert R, Breitbart EW and Katalinic A:
Non-melanoma skin cancer incidence and impact of skin cancer
screening on incidence. J Invest Dermatol. 134:43–50. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Narayanan DL, Saladi RN and Fox JL:
Ultraviolet radiation and skin cancer. Int J Dermatol. 49:978–986.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Veness MJ: Defining patients with
high-risk cutaneous squamous cell carcinoma. Australas J Dermatol.
47:28–33. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Echarri MJ, Lopez-Martin A and Hitt R:
Targeted therapy in locally advanced and Recurrent/metastatic head
and neck squamous cell carcinoma (LA-R/M HNSCC). Cancers (Basel).
8:E272016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yoshihara N, Takagi A, Ueno T and Ikeda S:
Inverse correlation between microtubule-associated protein
1A/1B-light chain 3 and p62/sequestosome-1 expression in the
progression of cutaneous squamous cell carcinoma. J Dermatol.
41:311–315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okura R and Nakamura M: Overexpression of
autophagy-related beclin-1 in cutaneous squamous cell carcinoma
with lymph-node metastasis. Eur J Dermatol. 21:1002–1003.
2011.PubMed/NCBI
|
|
8
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rabinowitz JD and White E: Autophagy and
metabolism. Science. 330:1344–1348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin S and White E: Role of autophagy in
cancer: Management of metabolic stress. Autophagy. 3:28–31. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lorin S, Hamai A, Mehrpour M and Codogno
P: Autophagy regulation and its role in cancer. Semin Cancer Biol.
23:361–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu
YC and Hung CH: Prognostic significance of p62/SQSTM1 subcellular
localization and LC3B in oral squamous cell carcinoma. Br J Cancer.
111:944–954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Claerhout S, Verschooten L, Van Kelst S,
De Vos R, Proby C, Agostinis P and Garmyn M: Concomitant inhibition
of AKT and autophagy is required for efficient cisplatin-induced
apoptosis of metastatic skin carcinoma. Int J Cancer.
127:2790–2803. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wright TJ, McKee C, Birch-Machin MA, Ellis
R, Armstrong JL and Lovat PE: Increasing the therapeutic efficacy
of docetaxel for cutaneous squamous cell carcinoma through the
combined inhibition of phosphatidylinositol 3-kinase/AKT signalling
and autophagy. Clin Exp Dermatol. 38:421–423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan TF, Bu LL, Wang WM, Ma SR, Liu JF,
Deng WW, Mao L, Yu GT, Huang CF, Liu B, et al: Tumor growth
suppression by inhibiting both autophagy and STAT3 signaling in
HNSCC. Oncotarget. 6:43581–43593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrette K, Van Kelst S, Wouters J,
Marasigan V, Fieuws S, Agostinis P, van den Oord J and Garmyn M:
Epithelial-mesenchymal transition during invasion of cutaneous
squamous cell carcinoma is paralleled by AKT activation. Br J
Dermatol. 171:1014–1021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choi SR, Chung BY, Kim SW, Kim CD, Yun WJ,
Lee MW, Choi JH and Chang SE: Activation of autophagic pathways is
related to growth inhibition and senescence in cutaneous squamous
cell carcinoma. Exp Dermatol. 23:718–724. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balamucki CJ, Mancuso AA, Amdur RJ, Kirwan
JM, Morris CG, Flowers FP, Stoer CB, Cognetta AB and Mendenhall WM:
Skin carcinoma of the head and neck with perineural invasion. Am J
Otolaryngol. 33:447–454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Warren TA, Panizza B, Porceddu SV, Gandhi
M, Patel P, Wood M, Nagle CM and Redmond M: Outcomes after surgery
and postoperative radiotherapy for perineural spread of head and
neck cutaneous squamous cell carcinoma. Head Neck. 38:824–831.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Warren TA, Broit N, Simmons JL, Pierce CJ,
Chawla S, Lambie DL, Quagliotto G, Brown IS, Parsons PG, Panizza BJ
and Boyle GM: Expression profiling of cutaneous squamous cell
carcinoma with perineural invasion implicates the p53 pathway in
the process. Sci Rep. 6:340812016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Padilla RS, Sebastian S, Jiang Z, Nindl I
and Larson R: Gene expression patterns of normal human skin,
actinic keratosis, and squamous cell carcinoma: A spectrum of
disease progression. Arch Dermatol. 146:288–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ra SH, Li X and Binder S: Molecular
discrimination of cutaneous squamous cell carcinoma from actinic
keratosis and normal skin. Mod Pathol. 24:963–973. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mitsui H, Suárez-Farinas M, Gulati N, Shah
KR, Cannizzaro MV, Coats I, Felsen D, Krueger JG and Carucci JA:
Gene expression profiling of the leading edge of cutaneous squamous
cell carcinoma: IL-24-driven MMP-7. J Invest Dermatol.
134:1418–1427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Farshchian M, Nissinen L, Siljamäki E,
Riihilä P, Toriseva M, Kivisaari A, Ala-Aho R, Kallajoki M,
Veräjänkorva E, Honkanen HK, et al: EphB2 promotes progression of
cutaneous squamous cell carcinoma. J Invest Dermatol.
135:1882–1892. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jian Q, Miao Y, Tang L, Huang M, Yang Y,
Ba W, Liu Y, Chi S and Li C: Rab23 promotes squamous cell carcinoma
cell migration and invasion via integrin β1/Rac1 pathway.
Oncotarget. 7:5342–5352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Turei D, Földvari-Nagy L, Fazekas D, Módos
D, Kubisch J, Kadlecsik T, Demeter A, Lenti K, Csermely P, Vellai T
and Korcsmáros T: Autophagy regulatory network-a systems-level
bioinformatics resource for studying the mechanism and regulation
of autophagy. Autophagy. 11:155–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
de Hoon MJ, Imoto S, Nolan J and Miyano S:
Open source clustering software. Bioinformatics. 20:1453–1454.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saldanha AJ: Java Treeview-extensible
visualization of microarray data. Bioinformatics. 20:3246–3248.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Parikh SA, Patel VA and Ratner D: Advances
in the management of cutaneous squamous cell carcinoma. F1000Prime
Rep. 6:702014. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miller SJ: Defining, treating, and
studying very high-risk cutaneous squamous cell carcinomas. Arch
Dermatol. 146:1292–1295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ross AS, Whalen FM, Elenitsas R, Xu X,
Troxel AB and Schmults CD: Diameter of involved nerves predicts
outcomes in cutaneous squamous cell carcinoma with perineural
invasion: an investigator-blinded retrospective cohort study.
Dermatol Surg. 35:1859–1866. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nindl I, Dang C, Forschner T, Kuban RJ,
Meyer T, Sterry W and Stockfleth E: Identification of
differentially expressed genes in cutaneous squamous cell carcinoma
by microarray expression profiling. Mol Cancer. 5:302006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rothenberg SM and Ellisen LW: The
molecular pathogenesis of head and neck squamous cell carcinoma. J
Clin Invest. 122:1951–1957. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gu Y, Li P, Peng F, Zhang M, Zhang Y,
Liang H, Zhao W, Qi L, Wang H, Wang C and Guo Z: Autophagy-related
prognostic signature for breast cancer. Mol Carcinog. 55:292–299.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim KW, Mutter RW, Cao C, Albert JM,
Freeman M, Hallahan DE and Lu B: Autophagy for cancer therapy
through inhibition of pro-apoptotic proteins and mammalian target
of rapamycin signaling. J Biol Chem. 281:36883–36890. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
O Farrell F, Rusten TE and Stenmark H:
Phosphoinositide 3-kinases as accelerators and brakes of autophagy.
FEBS J. 280:6322–6337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cerniglia GJ, Karar J, Tyagi S,
Christofidou-Solomidou M, Rengan R, Koumenis C and Maity A:
Inhibition of autophagy as a strategy to augment radiosensitization
by the dual phosphatidylinositol 3-kinase/mammalian target of
rapamycin inhibitor NVP-BEZ235. Mol Pharmacol. 82:1230–1240. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim KW, Myers CJ, Jung DK and Lu B:
NVP-BEZ-235 enhances radiosensitization via blockade of the
PI3K/mTOR pathway in cisplatin-resistant non-small cell lung
carcinoma. Genes Cancer. 5:293–302. 2014.PubMed/NCBI
|
|
43
|
Pursiheimo JP, Rantanen K, Heikkinen PT,
Johansen T and Jaakkola PM: Hypoxia-activated autophagy accelerates
degradation of SQSTM1/p62. Oncogene. 28:334–344. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X and Fan Z: The epidermal growth
factor receptor antibody cetuximab induces autophagy in cancer
cells by downregulating HIF-1alpha and Bcl-2 and activating the
beclin 1/hVps34 complex. Cancer Res. 70:5942–5952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li YN, Hu JA and Wang HM: Inhibition of
HIF-1α affects autophagy mediated glycosylation in oral squamous
cell carcinoma cells. Dis Markers. 2015:2394792015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Harris AL: Hypoxia - a key regulatory
factor in tumour growth. Nat Rev Cancer. 2:38–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pouysségur J, Dayan F and Mazure NM:
Hypoxia signalling in cancer and approaches to enforce tumour
regression. Nature. 441:437–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bohensky J, Shapiro IM, Leshinsky S,
Terkhorn SP, Adams CS and Srinivas V: HIF-1 regulation of
chondrocyte apoptosis: Induction of the autophagic pathway.
Autophagy. 3:207–214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He W, Wang Q, Xu J, Xu X, Padilla MT, Ren
G, Gou X and Lin Y: Attenuation of TNFSF10/TRAIL-induced apoptosis
by an autophagic survival pathway involving TRAF2- and
RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 8:1811–1821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang W, Sun J and Luo J: High expression
of Rab-like 3 (Rabl3) is associated with poor survival of patients
with non-small cell lung cancer via repression of
MAPK8/9/10-mediated autophagy. Med Sci Monit. 22:1582–1588. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nozawa T, Aikawa C, Goda A, Maruyama F,
Hamada S and Nakagawa I: The small GTPases Rab9A and Rab23 function
at distinct steps in autophagy during Group A Streptococcus
infection. Cell Microbiol. 14:1149–1165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zheng LQ, Chi SM and Li CX: Rab23′s
genetic structure, function and related diseases: A review. Biosci
Rep. 37:BSR201604102017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang M, Dong Q and Wang Y: Rab23 is
overexpressed in human astrocytoma and promotes cell migration and
invasion through regulation of Rac1. Tumour Biol. 37:11049–11055.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Maiuri MC, Le Toumelin G, Criollo A, Rain
JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K,
Tavernarakis N, et al: Functional and physical interaction between
Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 26:2527–2539.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Erlich S, Mizrachy L, Segev O, Lindenboim
L, Zmira O, Adi-Harel S, Hirsch JA, Stein R and Pinkas-Kramarski R:
Differential interactions between Beclin 1 and Bcl-2 family
members. Autophagy. 3:561–368. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yuan J, Zhang Y, Sheng Y, Fu X, Cheng H
and Zhou R: MYBL2 guides autophagy suppressor VDAC2 in the
developing ovary to inhibit autophagy through a complex of
VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 11:1081–1098. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Maji S, Samal SK, Pattanaik L, Panda S,
Quinn BA, Das SK, Sarkar D, Pellecchia M, Fisher PB and Dash R:
Mcl-1 is an important therapeutic target for oral squamous cell
carcinomas. Oncotarget. 6:16623–16637. 2015. View Article : Google Scholar : PubMed/NCBI
|