Introduction
Esophageal squamous cell carcinoma (ESCC) is the
predominant histological subtype of esophageal carcinoma, and its
highest incidence and mortality rates have been observed in China
(1). Due to the high ability of
invasion, ESCC has a high mortality rate and extremely poor
prognosis, due to insidious symptomatology, late clinical
presentation and rapid progression (2). Despite the advancement of diagnostic
technologies and therapies, the prognosis for ESCC remains poor,
which is largely attributable to the high rates of extensive local
invasion and regional lymph node metastasis (3,4).
Consequently, improved preventive approaches and more effective
treatment modalities are required in order to improve the survival
rates of patients with ESCC.
Forkhead box protein M1 (FOXM1) is a member of the
FoxM family that consists of >50 proteins characterized by a
conserved 100 amino acid DNA binding domain. A previous study also
demonstrated that the dysregulation of FOXM1 is involved in a wide
range of human cancer subtypes, by functioning as classical
oncogenes and having an important function in the initiation,
promotion and progression of cancer (5). FOXM1 regulates the expression of a
number of targeted genes important to cell proliferation,
migration, apoptosis and tumor angiogenesis, suggesting it may have
a general function in tumor development and progression.
Overexpression of FOXM1 is linked with tumor development in various
types of cancer (6–9). Since FOXM1 suppression appears to
inhibit tumor progression, chemical compounds targeting FOXM1 may
serve as anticancer drugs in different types of tumor; however, the
expression of FOXM1 and its function in ESCCs remains unknown.
Materials and methods
Tissue specimens
A total of 78 fresh tumor tissues and paired
adjacent non-tumorous tissues were obtained from 78 patients (45
male and 33 female) with ESCC with a median age of 58 (age range
39–71) years who underwent surgery at the Department of Thoracic
Surgery, Provincial Hospital Affiliated to Shandong University
(Jinan, China) between July 2012 and June 2014. The aforementioned
patients were diagnosed with ESCC by a pathologist (Shandong
Provincial Hospital affiliated to Shandong University, Jinan,
China). The normal paired tissues were obtained from the distal
resection margins. All specimens were stored at −80°C until
analysis. In addition, the present study collected all the paraffin
blocks of the patients to perform immunohistochemical (IHC)
staining. No patients received radiotherapy or chemotherapy prior
to surgery. The present study was approved by the Research Ethics
Committee of Shandong University. Written informed consent was
obtained from all patients prior to enrollment in the present
study.
Cell culture
ESCC YES2, COLO-680N, EC9706 and KYSE450 cell lines
were provided by the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). All cell lines were grown in
RPMI-1640 supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 µg/µl
streptomycin and 100 µg/µl penicillin (pH 7.2–7.4) in a humidified
incubator containing 5% CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen tissues and cell
lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) for 30 min at room temperature. The quality of the RNA was
assessed by 1% denaturing agarose gel electrophoresis and
spectrophotometry. Complementary DNA (cDNA) was synthesized using a
SuperScript VILO cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The reaction was incubated in a MyCycler
Thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) for 10
min at 25°C, 60 min at 42°C and 5 min at 85°C. cDNA samples were
stored at −20°C prior to RT-PCR amplification. RT-qPCR was
performed using the SYBR Green reporter (Takara Biotechnology Co.,
Ltd., Dalian, China). GAPDH was used as an internal control for
each specific gene. A total of three independent experiments were
performed to analyze the relative gene expression level and each
sample was evaluated in triplicate. The following primers were
used: FOXM1 forward, 5′-GCGACAGGTTAAGGTTGAG-3′ and reverse,
5′-AGGTTGTGGCGGATGGAGT-3′; GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′.
Quantification was performed using the quantitative threshold cycle
(Cq) method and efficiency of the RT reaction (relative quantity,
2−ΔΔCq) (10).
Western blot analysis
Total protein was extracted using a lysis buffer and
protease inhibitor (Beyotime Institute of Biotechnology, Haimen,
China). Equivalent protein amounts (30 µg) were denatured in an SDS
sample buffer, and then separated by 10% SDS-PAGE (EMD Millipore,
Billerica, MA, USA) and transferred onto a polyvinylidene
difluoride membrane. Following blocking with 5% skimmed milk in
Tris-buffered saline with 0.1% Tween-20 (pH 7.6) at room
temperature for 1 h, the membranes were incubated with primary
antibodies at room temperature for 1 h against FOXM1 (1:1,000; cat.
no. ab83097; Abcam, Cambridge, UK) and subsequently with a mouse
anti-rabbit IgG-horseradish peroxidase (HRP) secondary antibody at
room temperature for 1 h (1:5,000; cat. no. BA1003; Boster
Biological Technology, Pleasanton, CA, USA). An enhanced
chemiluminescent chromogenic substrate was used to visualize the
bands and the intensity of the bands was quantified by densitometry
(Quantity One software; Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
IHC analysis
Primary ESCC tissues and adjacent normal tissues
obtained following surgery were subjected to IHC analysis. In
brief, formalin-fixed and paraffin-embedded specimens from the
Department of Pathology were sectioned at a thickness of 3–4 µm for
IHC. The samples were deparaffinized with xylene, and rehydrated
through a series of decreasing concentrations of ethanol (100, 95,
90, 80 and 70%) to water. A high-temperature antigen retrieval
method was performed using a citrate buffer solution (Maixin Bio,
Fujian, China), and the slides were immersed in 100 µl of 3%
hydrogen peroxide for 10 min at 37°C to block endogenous peroxidase
activity. Subsequent to washing with PBS, the sections were
incubated with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 30 min, followed by incubation with
anti-FOXM1 monoclonal antibody (1:100; cat. no. sc-376471; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight.
Following washing with PBS, the sections were incubated with a
Rabbit Anti-Mouse IgG-HRP secondary antibody (1:5,000; cat. no.
ab6728; Abcam) for 30 min at 37°C. The slides were stained with
diaminobenzidine (as the color reagent) for 5 min at 37°C and
hematoxylin (as a counterstain for the nuclei) for 2 min at
37°C.
PBS was used as a negative control for the staining
reactions. All slides were dehydrated in increasing concentrations
of ethanol and xylene. Finally, all sections were mounted with
neutral gum. The FOXM1 staining index was ranked according to the
percentage of FOXM1-positive cells, which were marked by yellow
particles observed in the tumor cytoplasm/cell plasma membrane. The
expression level of FOXM1 was classified into five groups according
to the proportion of positively staining cells: 0, absent; 1,
1–25%; 2, 26–50%; 3, 51–75%; 4, ≥76%. The staining intensity was
categorized as follows: 0, negative; 1, weak; 2, moderate and 3,
strong. The proportion and intensity scores were then multiplied to
obtain a total score. If the product of multiplication between
staining intensity and the percentage of positive cells was ≤4, it
was defined low expression, whereas overall scores >4 were
defined as high expression. The Olympus CKX41SF (Olympus
Corporation, Tokyo, Japan) light microscope was used at a
magnification of ×200.
Small interfering RNA (siRNA)
transfection
For the siRNA-knockdown experiment, double-stranded
RNA duplexes that targeted the human FOXM1 gene
(5′-GGUCCUGGACACAAUGAAUTT-3′) and the negative control (NC;
5′-UAACAAUGAGAGCACGGCTT-3′) were purchased from Invitrogen (Thermo
Fisher Scientific, Inc.). Cells were transfected with Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol.
MTT method
ESCC cells (5×103 cells/well) were seeded
into a 96-well plate with 200 µl in each well. Following culturing
for 24, 48 and 72 h at 37°C, freshly made-up 20 µl MTT was added,
and further incubated at 37°C for 4 h. The culture medium was
replaced with 150 µl/well dimethylsulfoxide, and the absorbance at
a wavelength of 490 nm was measured using 2104 EnVision®
Multilabel reader (cat. no. 2104–0010; PerkinElmer, Inc., Waltham,
MA, USA). Survival rate of tumor cells (%)=experimental group-A
value/control group A-value ×100.
Clonogenic assays
Clonogenic assay was performed in order to examine
the effect of FOXM1-siRNA on cell growth in ESCC cells. A total of
4×105 KYSE450 cells were plated in a 6-well plate.
Following a 24-h transfection at 37°C, the cells were trypsinized
and 1,000 single viable cells were plated in three 6-well plates.
The cells were then incubated for 14 days at 37°C in 5%
CO2/5% O2/90% N2. Colonies were
stained with 0.1% crystal violet for 10 min at 37°C, washed with
water and 10 random fields were counted manually using an Olympus
light microscope (CKX41SF; Olympus Corporation, Tokyo, Japan) at a
magnification of ×200. The colonies containing ≥100 cells were
scored. The surviving fraction in FOXM1 siRNA-transfected cells was
normalized to untreated control cells with respect to clonogenic
efficiency.
Cell migration and invasion
assays
The migratory and invasive activity of the FOXM1 or
control siRNA-transfected cells was evaluated using the Transwell
chambers equipped with a pore size of 8 µm (Corning Incorporated,
Corning, NY, USA), according to the manufacturer's protocol.
Following a 24-h transfection at 37°C, 2×104 ESCC
cells/well were resuspended in serum-free Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) and seeded
into the Transwell inserts either uncoated (for migration assay) or
coated (for invasion assay) with growth factor-reduced Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA), whereas the lower chambers
were filled with 500 µl Dulbecco's modified Eagle's medium
supplemented with 10% FBS. Following a 24-h incubation at 37°C, the
cells on the upper side of the insert filter were completely
removed by wiping with a cotton swab, and the cells that had
invaded were fixed in 90% methanol at 4°C for 10 min and stained
with 0.1% crystal violet for 10 min at 37°C. The cells were counted
manually under an inverted microscope in five random fields. Each
experiment was repeated in triplicate.
Statistical methods
Statistical analysis was performed using SPSS
version 17.0 software (SPSS, Inc., Chicago, IL, USA). Data were
presented as the mean ± standard deviation and three independent
experiments were analyzed. Data were compared using standard
one-way analysis of variance methodology for repeated evaluation,
followed by the Student-Newman-Keuls test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression level of FOXM1 in ESCC
tissues compared with paired para-cancerous tissues
The expression level of FOXM1 in 78 pairs of ESCC
cancerous and para-cancerous tissue samples was analyzed by
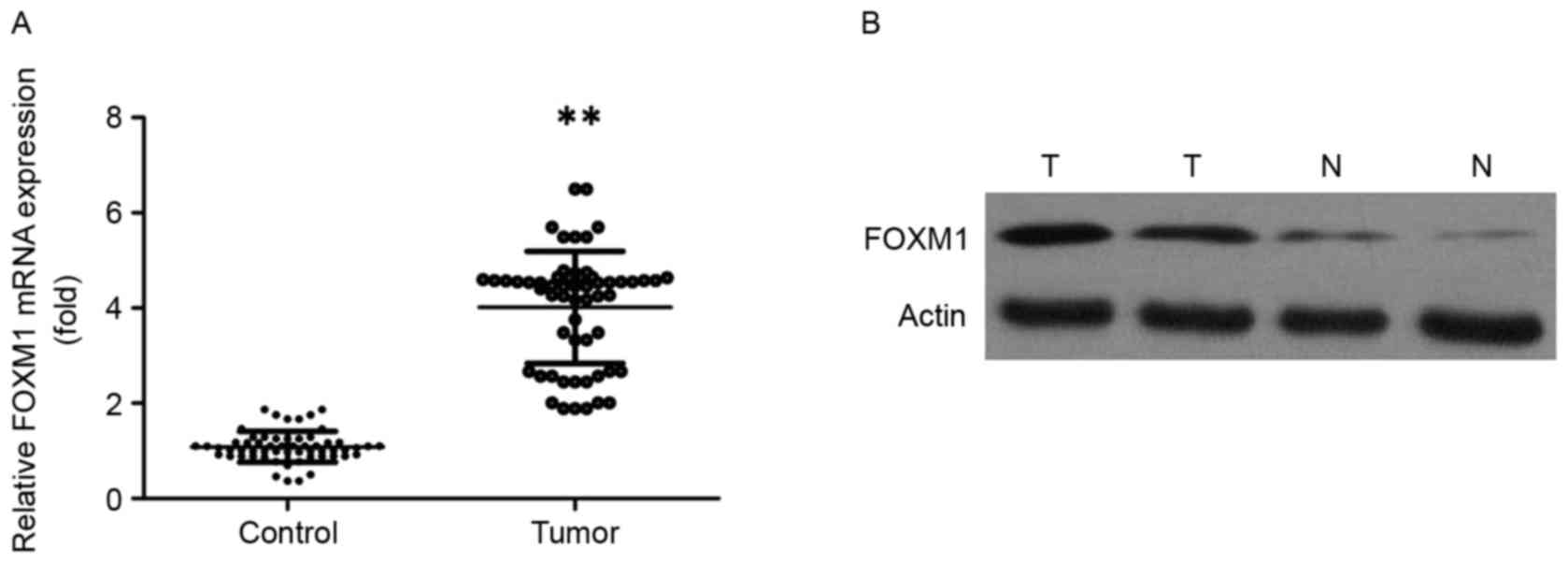
RT-qPCR, western blotting and IHC. The expression level of FOXM1
mRNA was significantly increased in ESCC tissues compared with in
normal paired tissues (Fig. 1A;
P<0.01). The results of western blotting also revealed that
FOXM1 protein levels were increased in tumor tissues compared with
the expression level in paired distal normal tissues (Fig. 1B). A total of 78 pairs of
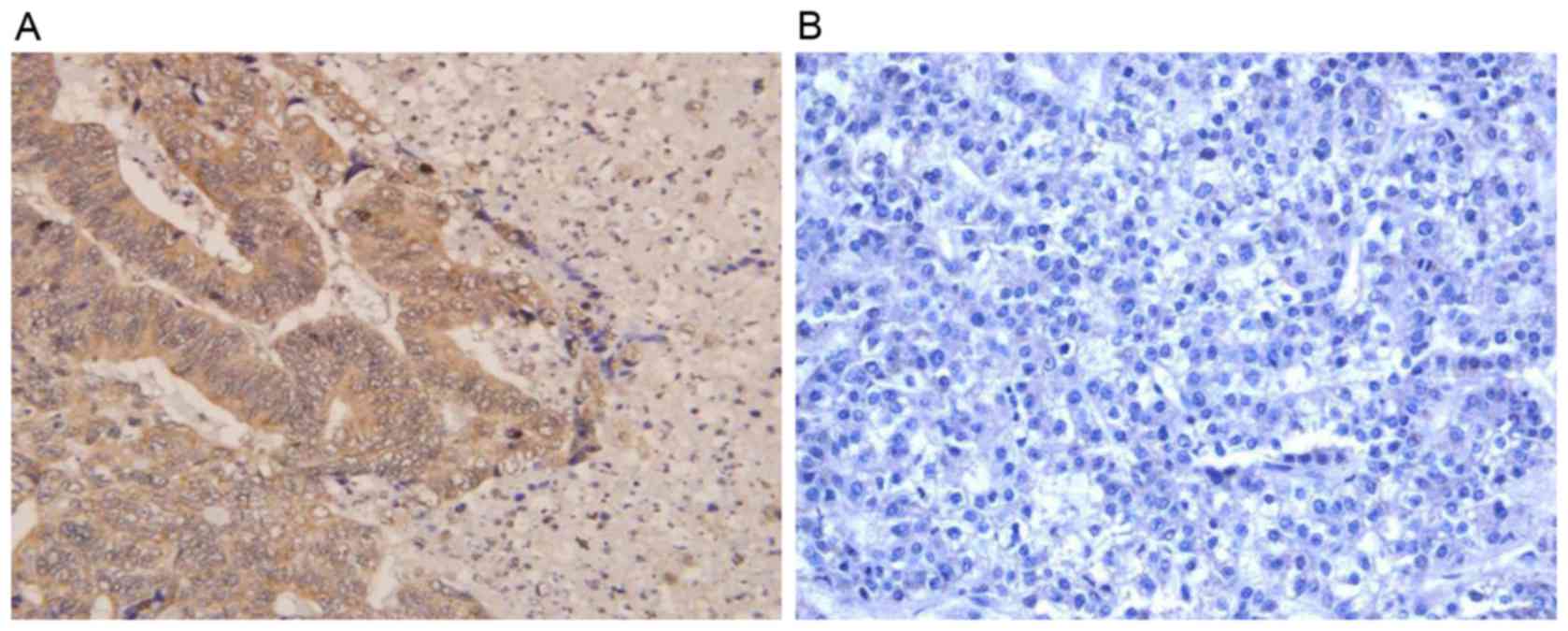
paraffin-embedded ESCC and adjacent non-cancerous tissues were
analyzed by IHC. The positive expression level of FOXM1 in ESCC
tissues (59/78, 75.64%) was significantly increased compared with
the adjacent non-cancerous tissues (30/78, 38.46%; P<0.01). As
presented in Fig. 2, FOXM1-positive
staining was predominantly observed in the nucleus and partially
occurred in the cytoplasm.
Expression level of FOXM1 in ESCC cell
lines
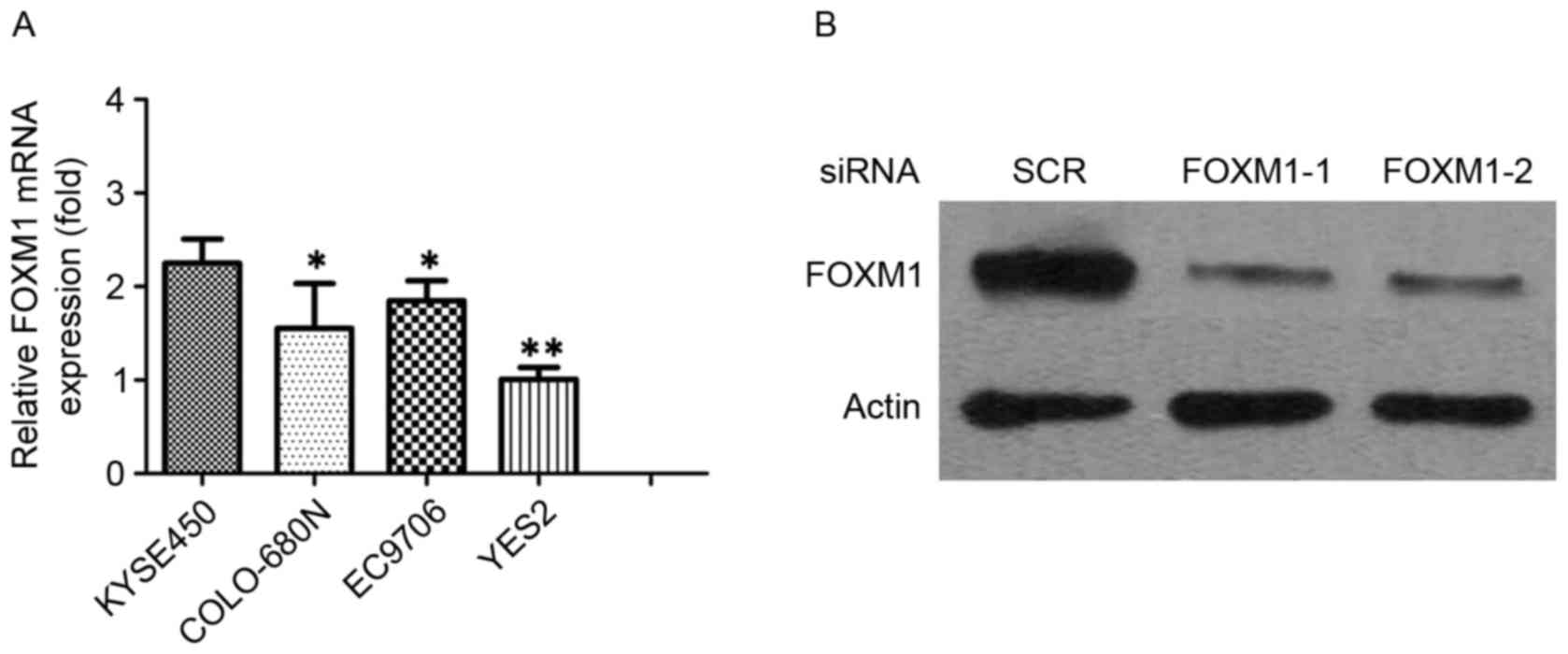
The present study evaluated FOXM1 expression levels
in four ESCC cell lines. FOXM1 was expressed in ESCC cell lines,
and KYSE450 expressed higher levels of endogenous FOXM1 expression
level when compared with YES2, COLO-680N and EC9706 cell lines
(Fig. 3A). It was reported that FOXM1
may have a function in cell proliferation regulation in various
types of human cancer. In order to investigate the function of
FOXM1 gene in human ESCC cells, KYSE450 cells were transiently
transfected with FOXM1 siRNA and NC siRNA. The expression levels of
FOXM1 were determined by RT-PCR and western blotting (Fig. 3B).
Knockdown of FOXM1 inhibited
proliferation of ESCC cells
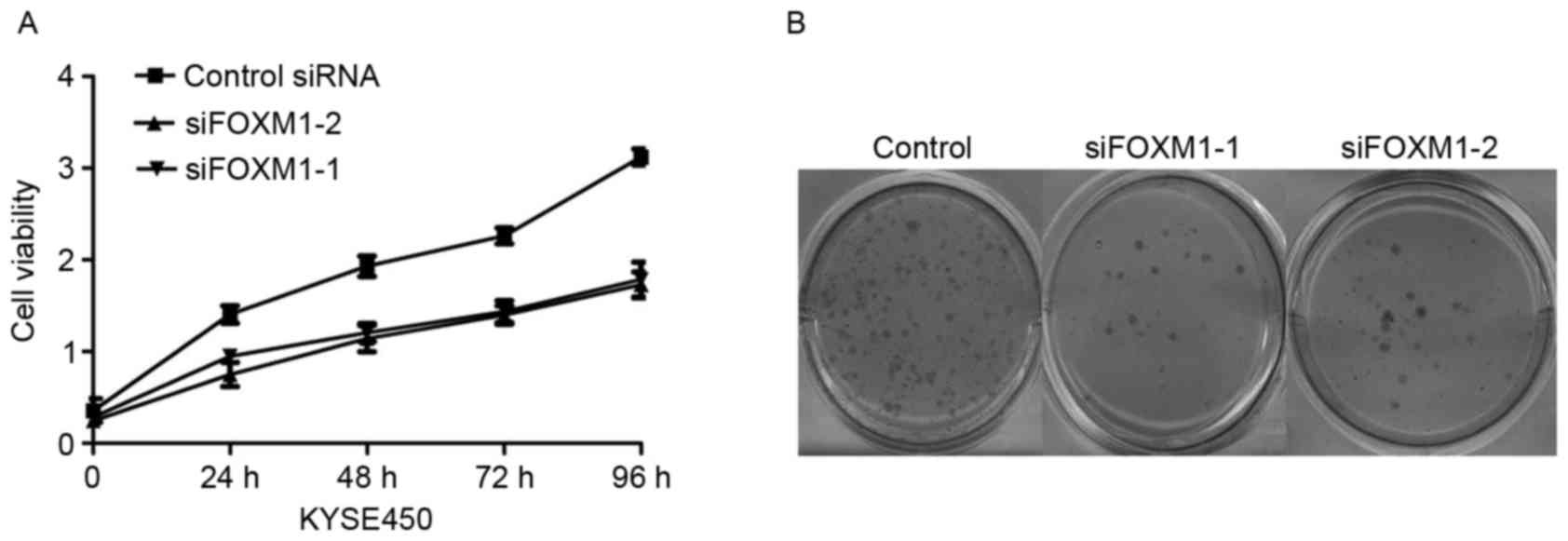
The present study further investigated the effects
of FOXM1 knockdown on cell proliferation in ESCC cells. Knockdown
of FOXM1 reduced the ESCC cell growth rate by MTT assay (Fig. 4A). These phenomena were further
confirmed by a colony formation assay (Fig. 4B). Furthermore, the results
demonstrated that the numbers of colonies in FOXM1-depleted KYSE450
cells were reduced compared with control-siRNA and untreated
cells.
Effect of altered FOXM1 expression
level on KYSE450 colon cancer cell migration and invasion in
vitro
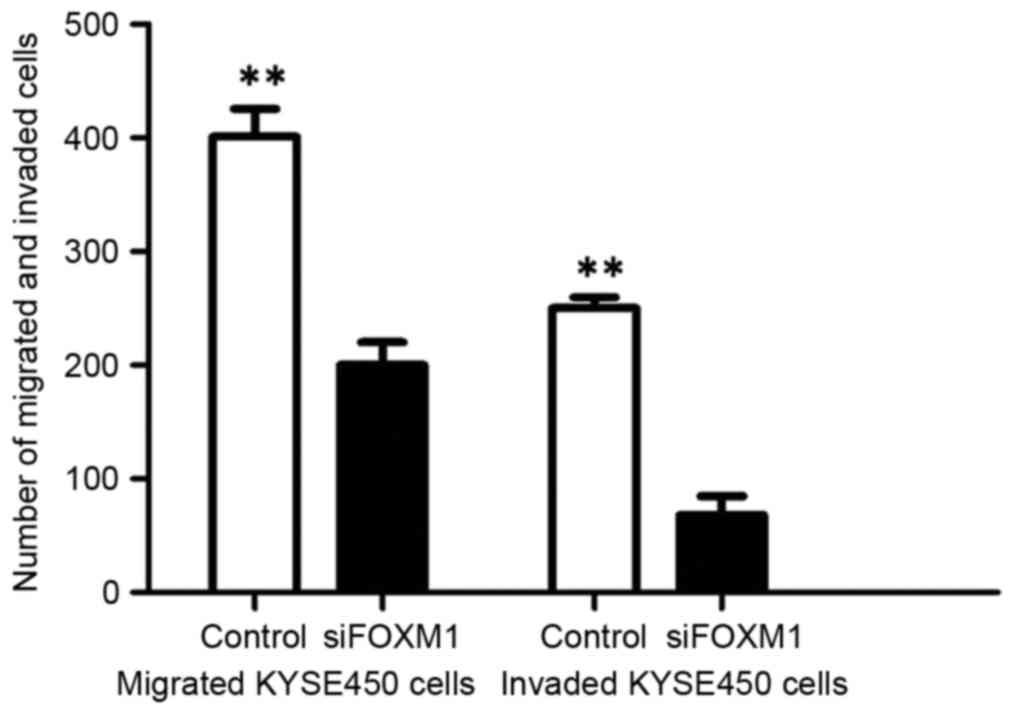
In order to determine whether attenuated FOXM1
expression level inhibited the migration and invasion of ESCC
cells, the present study analyzed the effect of FOXM1 on ESCC cell
migration by Transwell assay. Transwell invasion assay revealed
that FOXM1 siRNA-transfected cells revealed a low level of
penetration through the Matrigel-coated membrane compared with the
control cells (Fig. 5).
Discussion
The majority of patients with ESCC are diagnosed
with advanced stage disease and have poor long-term survival rates.
The efficacy of current regimens has reached a plateau and further
intensification of cytotoxic agents or radiation dose escalation
has been demonstrated to be associated with significant adverse
side effects. Despite advances in multimodality therapy, the
prognosis of ESCC remains poor and the overall 5-year survival is
<15%. Similar to other types of cancer, the development of ESCC
is believed to be a multiple-step process induced by the
accumulation of activation of oncogenes (11). At present, the exact cellular and
molecular mechanisms leading to ESCC have not been systematically
evaluated. Consequently, the requirement for the development of
effective targeted therapies aimed at treating specific mechanisms
underlying carcinogenesis are required in order to improve
survival.
Previous evidence has revealed that elevated
expression levels or activity of FOXM1 is associated with the
development and progression of numerous types of cancer (12). FOXM1 is a proliferation-associated
transcription factor with important functions in cell
proliferation, differentiation and apoptosis. The present study
demonstrated that the expression levels of FOXM1 were significantly
increased in ESCC tissues compared with in adjacent normal tissues,
at the transcriptional and translational levels. The results of the
present study were inconsistent with a previous study, which
revealed overexpression of FOXM1 in various types of human cancer.
Numerous previous studies have demonstrated that a reduction in
FOXM1 expression level resulted in inhibition of proliferation in
cancer cells and a marked decrease in tumor growth. The present
study used siRNA to knockdown FOXM1 expression in the ESCC KYSE450
cell line. The KYSE450 cell line was selected due to its high
abundance of FOXM1. Depletion of endogenous FOXM1 significantly
inhibited the viability of KYSE450 cells. These results are
consistent with the function of FOXM1 in cell proliferation.
Knockdown of FOXM1 suppressed cell migration of ESCC in
vitro. In the invasion assay and migration assays, FOXM1
siRNA-transfected cells revealed a low level of penetration through
the Matrigel-coated membrane compared with the control cells,
suggesting important functions for FOXM1 in human ESCC
tumorigenesis and distant metastasis. The results demonstrated that
FOXM1 has the potential to be a novel therapeutic target in
ESCC.
In summary, the results of the present study
suggested that FOXM1 mRNA and protein were clearly expressed to a
higher degree in ESCC tissues compared with paired para-cancerous
tissues. Downregulation of FOXM1 suppressed proliferation of ESCC
cells. In conclusion, FOXM1 may act as a novel target for ESCC
therapy.
References
|
1
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chai J and Jamal MM: Esophageal
malignancy: A growing concern. World J Gastroenterol. 18:6521–6526.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tejani MA and Burtness BA: Multi-modality
therapy for cancer of the esophagus and GE junction. Curr Treat
Options Oncol. 13:390–402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Raychaudhuri P and Park HJ: FoxM1: A
master regulator of tumor metastasis. Cancer Res. 71:4329–4333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun H, Teng M, Liu J, Jin D, Wu J, Yan D,
Fan J, Qin X, Tang H and Peng Z: FOXM1 expression predicts the
prognosis in hepatocellular carcinoma patients after orthotopic
liver transplantation combined with the Milan criteria. Cancer
Lett. 306:214–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu J, Deshmukh H, Payton JE, Dunham C,
Scheithauer BW, Tihan T, Prayson RA, Guha A, Bridge JA, Ferner RE,
et al: Array-based comparative genomic hybridization identifies
CDK4 and FOXM1 alterations as independent predictors of survival in
malignant peripheral nerve sheath tumor. Clin Cancer Res.
17:1924–1934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balli D, Zhang Y, Snyder J, Kalinichenko
VV and Kalin TV: Endothelial cell-specific deletion of
transcription factor FoxM1 increases urethane-induced lung
carcinogenesis. Cancer Res. 71:40–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calvisi DF, Pinna F, Ladu S, Pellegrino R,
Simile MM, Frau M, De Miglio MR, Tomasi ML, Sanna V, Muroni MR, et
al: Forkhead box M1B is a determinant of rat susceptibility to
hepatocarcinogenesis and sustains ERK activity in human HCC. Gut.
58:679–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Laoukili J, Stahl M and Medema RH: FoxM1:
At the crossroads of ageing and cancer. Biochim Biophys Acta.
1775:92–102. 2007.PubMed/NCBI
|
|
12
|
Ahmed M, Uddin S, Hussain AR, Alyan A,
Jehan Z, Al-Dayel F, Al-Nuaim A, Al-Sobhi S, Amin T, Bavi P and
Al-Kuraya KS: FoxM1 and its association with matrix
metalloproteinases (MMP) signaling pathway in papillary thyroid
carcinoma. J Clin Endocrinol Metab. 97:E1–E13. 2012. View Article : Google Scholar : PubMed/NCBI
|