Introduction
In recent years, gastric cancer incidence has
gradually decreased in certain countries; however, it remains one
of the main causes of cancer-associated mortality (1). Therefore, to improve patient prognosis,
it is urgent that more biomarkers for the diagnosis and treatment
of gastric cancer are identified. The development of gastric cancer
is a complex process that includes the inactivation of tumor
suppressor genes and the activation of oncogenes. p53, a
well-established tumor suppressor, has a critical role in the
anti-proliferative functions of cells. p53 mutations that lead to a
protein loss-of-function are the most common genetic changes in
cancer (2,3). Hence, it is important to find novel
target genes that regulate p53 expression directly or
indirectly.
Zinc finger (ZNF) proteins recognize various motifs
in RNAs and proteins. By interacting with DNA and proteins, ZNF
proteins can provide links between these molecule types (4). An increasing number of studies has
reported that ZNF proteins serve critical roles in human cancers
(5–15). For example, ZNF703 acts as an oncogene
in invasive gastric carcinoma, and its expression levels are
correlated with gastric carcinoma progression (16). The downregulation of ZNF306 expression
reduces tumorigenicity in colorectal cancer (17). It is notable that certain ZNF proteins
participate in tumorigenesis by influencing p53 activity. Through
preventing Mdm2-mediated p53 degradation, ZNF668 has been
considered an anti-oncogene in breast cancer (18). Similarly, ZNF307 inhibits p53 and p21
activity by enhancing EP300 and Mdm2 expression (19). In prostate cancer cells, ZNF280B
induces p53 nuclear export, leading to subsequent proteasomal
degradation (20). Therefore, ZNF280B
serves pro-growth and pro-survival functions in prostate cancer
cells.
In the present study, the expression of ZNF280B in
gastric cancer and its association with clinicopathological
parameters were explored. Furthermore, the biological role of
ZNF280B in growth of gastric cancer cells is investigated in
vitro and in vivo.
Materials and methods
Patients and samples
A total of 60 gastric cancer specimens obtained from
the Department of Pathology at The First Affiliated Hospital of
Henan University of Science and Technology (Lyuoyang, China) from
resections performed between July 2000 and March 2002 were
examined. The study included 36 male and 24 female patients, with a
mean age of 58.5 years (range, 45–72 years). Two experienced
pathologists participated in the study and were blinded to the
clinical information. Clinicopathological parameters for the
patients are included in Table I, and
the tissue samples were classified using a risk score TNM staging
system (21). Informed consent was
obtained on the collection of samples from each patient. The study
protocol was approved by the Medical Ethics Committee of the First
Affiliated Hospital and the College of Clinical Medicine of Henan
University of Science and Technology.
 | Table I.Association between ZNF280B expression
and the clinicopathological features of gastric cancer
patients. |
Table I.
Association between ZNF280B expression
and the clinicopathological features of gastric cancer
patients.
|
|
| ZNF280B
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Negative | Positive | P-value |
|---|
| Sex |
|
|
| 0.787 |
| Male | 36 | 14 | 22 |
|
|
Female | 24 | 8 | 16 |
|
| Age, years |
|
|
| 0.591 |
| ≤60 | 38 | 15 | 23 |
|
|
>60 | 22 | 7 | 15 |
|
| Tumor size, cm |
|
|
| 0.017a |
| ≥5 | 26 | 5 | 21 |
|
|
<5 | 34 | 17 | 17 |
|
| Tumor
differentiation |
|
|
| 0.573 |
|
Well/moderate | 40 | 16 | 24 |
|
| Poor | 20 | 6 | 14 |
|
| Tumor-node-metastasis
stage |
|
|
|
<0.001a |
| I/II | 41 | 21 | 20 |
|
|
III/IV | 19 | 1 | 18 |
|
Immunohistochemistry
Staining for ZNF280B was performed using
formalin-fixed, paraffin-embedded serial sections. Sections (4-µm
thick) were cut from the paraffin blocks and deparaffinized by
routine techniques. The slides were microwaved in citrate buffer
for 4 min for antigen retrieval. ZNF280B was detected with a rabbit
polyclonal antibody (dilution, 1:150; cat. no., AP17865a; Abgent,
Inc., San Diego, CA, USA). The antibody was incubated at 37°C for 3
h. The staining was detected with a biotinylated goat-anti rabbit
secondary antibody, incubated at 37°C for 2 h (dilution, 1:50; cat.
no., ZF-0311; OriGene Technologies, Inc., Rockville, Beijing,
China), avidin-biotin complexes and diaminobenzidine (both 5 mg/ml;
both Maxim Biomedical, Inc., Rockville, MD, USA), both incubated at
37°C for 5 min. Sections were then counterstained with hematoxylin
at 37°C for 5 min. The expression of ZNF280B was scored according
to the positive percentage and staining intensity of the stained
tumor cells. The percentage was scored as 0 (0–25%), 1 (26–50%), 2
(51–75%) and 3 (>75%). The staining intensity was scored as 0
(no staining), 1 (weakly stained), 2 (moderately stained) and 3
(strongly stained). If the product of multiplication between
staining intensity and the percentage of positive cells was ≥2, it
was considered immunoreaction positive (+). The goat anti-human
monoclonal Ki-67 staining was achieved by a 2 h primary antibody
incubation at 37°C (dilution, 1: 100; cat. no., D3B5; Gene Tech
Biotechnology Co., Ltd., Shanghai, P.R. China) and a
biotinylated-conjugated rabbit anti-goat immunoglobulin G secondary
antibody incubation at 37°C for 2 h (dilution, 1:50; cat. no.,
ZF-0314; OriGene Technologies, Inc., Rockville, Beijing, China),
and was scored according to the percentage of positively stained
gastric cancer cells at high magnification (×200).
Cell lines and cell culture
Gastric cancer cell lines (SGC-7901, BGC-823 and
MGC-803) were obtained from the Department of Pathology, Guangdong
Medical University (Zhanjiang, China). MGC-803 cells were stably
transfected with pcDNA3.1-ZNF280B and pcDNA3.1 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Cells were positively selected
using G418 (500 µg/ml) for 4 weeks. Surviving colonies were
isolated and expanded. The cells stably transfected with
pcDNA3.1-ZNF280B and pcDNA3.1 were designated as MGC-803/ZNF280B
and negative control (NC) respectively. All cells were maintained
in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 µg/ml penicillin and 100 µg/ml
streptomycin at 37°C with 5% CO2.
Western blotting
Subsequent to washing in ice-cold PBS, cells were
lysed in lysis buffer [1% Triton X-100, 50 mM Tris-HCl (pH 7.5),
0.1% SDS, 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM
PMSF, 5 µl/ml leupeptin and 5 µl/ml aprotinin]. The BCA Protein
Assay Reagent kit (Pierce; Thermo Fisher Scientific, Inc.) was used
to evaluate protein concentrations. Total protein (80 µg) was
boiled for 8 min prior to loading onto a 10% polyacrylamide gel and
transferred to a polyvinylidene fluoride membrane. The membrane was
incubated with 5% non-fat dry milk overnight at 37°C. The membrane
was incubated with the ZNF280B primary antibody (dilution, 1:200),
followed by an HRP-conjugated secondary antibody (dilution,
1:2,000; cat. no., ZF-0311; Zhongshan Biology Co., Ltd., Foshan,
China). Lastly, detected proteins were visualized with an enhanced
chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.).
GAPDH was used as internal/loading control (dilution, 1:400; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA; catalog no.,
365062).
MTT and colony formation assay
An MTT assay was used to detect the proliferation
ability of MGC-803, NC and MGC-803/ZNF280B cells. The cells were
seeded in 96-well plates at a density of 1×104
cells/well. Following a 24-h incubation, 50 µl MTT (5 mg/ml) was
added to the medium. At 4 h, the medium was discarded, 150 µl
dimethyl sulfoxide was added into each well, and it was incubated
with rocking for 10 min. Finally, the absorbance of each well was
read at a wavelength of 492 nm. All experiments were performed in
triplicate.
Subsequent to incubation for 24 h, cells from each
group (MGC-803, NC and MGC-803/ZNF280B) were added to a six-well
plate at 200 cells per well for the colony formation assay. Then,
the tumor cells were incubated at 37°C and the medium was changed
every 5 days for 2 weeks. Finally, the tumor cells were fixed by
methanol, stained with trypan blue at 37°C for 20 min, and colonies
were counted using a microscope at low magnification (×100)
(Olympus Corporation, Tokyo, Japan). All experiments were performed
in triplicate.
Xenograft studies
A total of 9 female BALB/c nude mice (4–5 weeks of
age, 15–20 g) were purchased from the Shanghai Laboratory Animal
Center of the Chinese Academy of Sciences (Shanghai, China) and
kept in the Animal Center of Clinical Medicine of Henan University
of Science and Technology. The temperature was maintained at
25–27°C and 40–60% relative humidity in a 12-h light/12-h dark
cycle. Nude mice received sterilized water and food ad
libitum. Mice received humane care, and all animal experiments
were performed according to protocols approved by the Medical
Ethics Committee of Henan University of Science and Technology. For
the xenograft assay, nude mice (~8 weeks old) were anesthetized
with sodium pentobarbital (50 mg/kg) in a sterile environment.
Then, 2×106 MGC-803, NC and MGC-803/ZNF280B cells in 50
µl of PBS were subcutaneously injected into the mice (3 mice per
group). The mice were sacrificed by cervical dislocation on day 25.
Maximum tumor diameter exceeding 2 cm was deemed the humane
endpoint of the study. Subsequently, the tumors were collected and
the primary tumor weight was measured.
Statistical analysis
Statistical analyses were performed using SPSS 16.0
for Windows (SPSS, Inc., Chicago, IL, USA). The association between
ZNF280B and clinicopathological parameters was assessed using
Fisher's exact test. Multi-group comparisons were made with a
one-way analysis of variance with a Student-Newman-Keuls post hoc
test. P<0.05 was considered to indicate a statistically
significant difference. All results are expressed as the mean ±
standard deviation.
Results
ZNF280B expression is associated with
the clinicopathological factors of patients with gastric
cancer
To investigate the expression status of ZNF280B in
gastric cancer, ZNF280B immunohistochemical staining was performed
on 60 gastric cancer specimens. The results indicated that ZNF280B
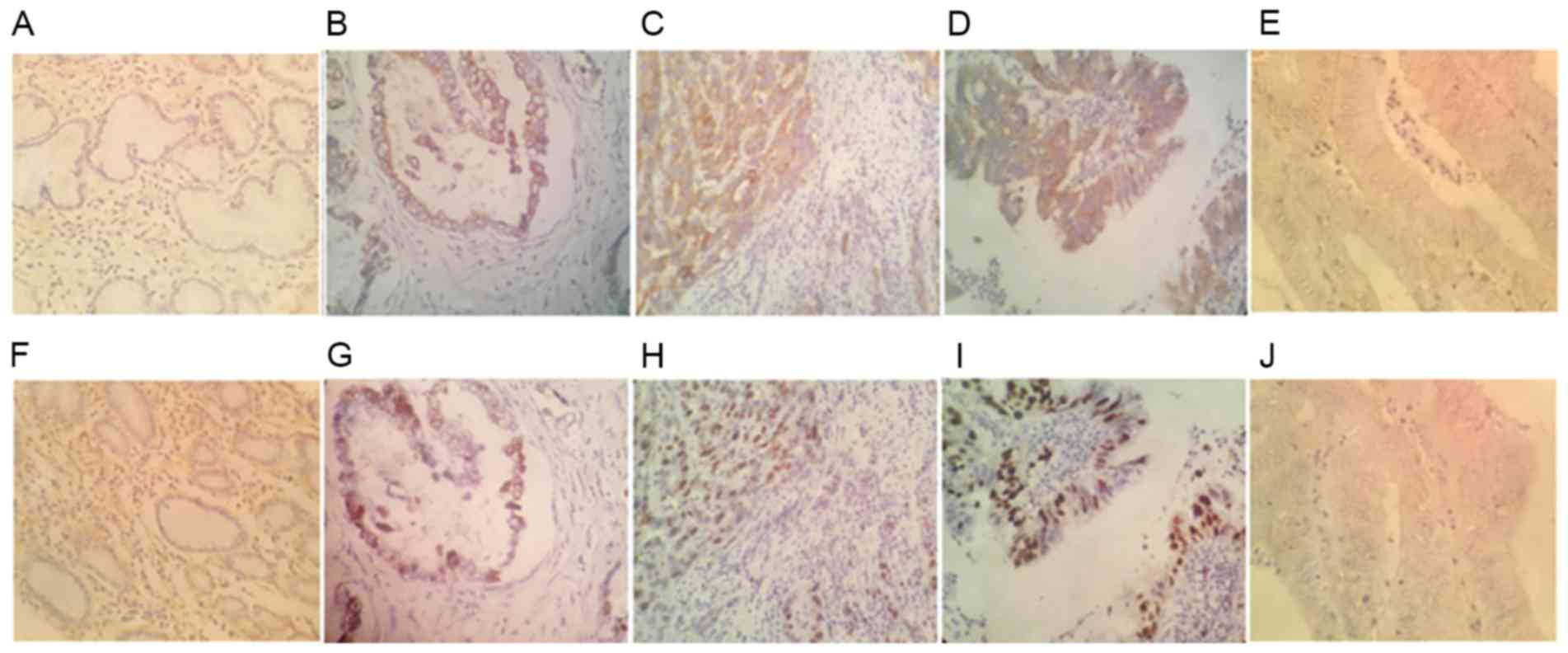
was distributed in the cytoplasm of gastric cancer cells (Fig. 1). Compared with the expression in
normal gastric mucosal tissues (Fig.
1A), 38 (63.3%) gastric cancer samples displayed
ZNF280B-positive staining (Fig.
1B-D).
To further elucidate the clinical significance of
ZNF280B in gastric cancer, the association between ZNF280B and
clinicopathological characteristics was assessed. ZNF280B
expression was identified in 21/26 (80.8%) cases of larger tumor
size (≤5 cm) whereas only 17/34 (50.0%) smaller tumors exhibited
positive ZNF280B immunoreactivity. Hence, ZNF280B expression was
associated with tumor size (P=0.017). In addition, only 20/41
(48.8%) cases in stage I and II exhibited positive ZNF280B
staining, whereas ZNF280B was positively stained in 18/19 (94.7%)
samples in stage III and IV (P<0.001). On the other hand, as
demonstrated in Table I, no
association existed between ZNF280B expression and other
clinicopathological variables, including sex, age and
differentiation degree (P>0.05).
ZNF280B expression is associated with
the cell proliferation index (PI)
To study the effect of ZNF280B on the proliferation
of gastric cancer cells, the association between ZNF280B and PI was
investigated. PI was determined with Ki-67 immunohistochemical
staining. As demonstrated in Fig. 1,
the positive ZNF280B expression group PI was significantly higher
(38.8±6.2) compared with that in the negative ZNF280B expression
group (16.9±8.9; P<0.01). These results suggest that ZNF280B
expression may be associated with the rate of proliferation of
gastric cancer cells.
ZNF280B transfection enhances ZNF280B
protein expression in MGC-803 cells
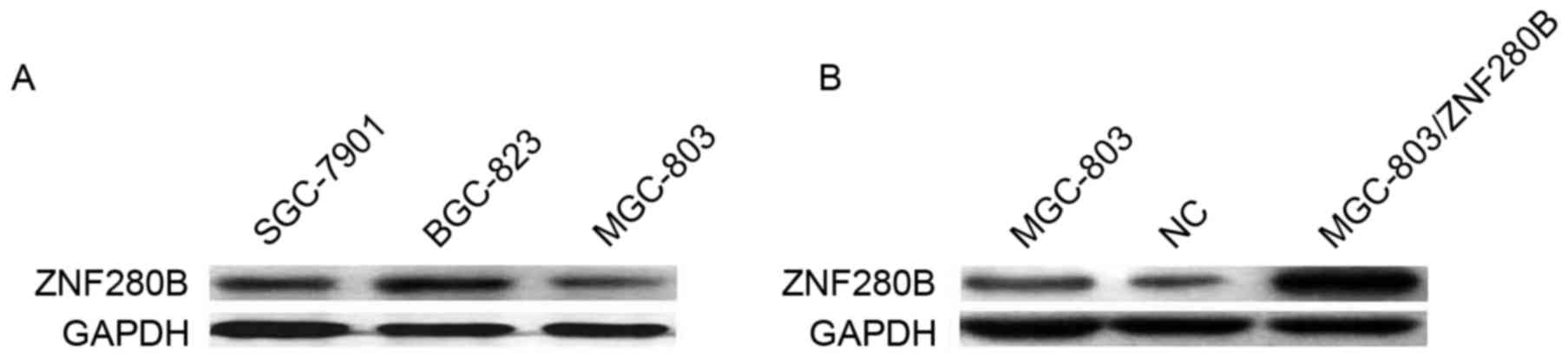
The endogenous expression of ZNF280B was assessed in
3 gastric cancer cell lines, including SGC-7901, BGC-823 and
MGC-803, and was revealed to be the lowest in MGC-803 cells, which
were used for the subsequent experiments (Fig. 2A). As demonstrated in Fig. 2B, 48 h after MGC-803/ZNF280B transient
transfection, ZNF280B protein expression was evidently increased in
MGC-803 cells. These results indicate that ZNF280B expression was
effectively upregulated subsequent to transfection.
ZNF280B overexpression enhances the
proliferation and colony formation ability of MGC-803 cells
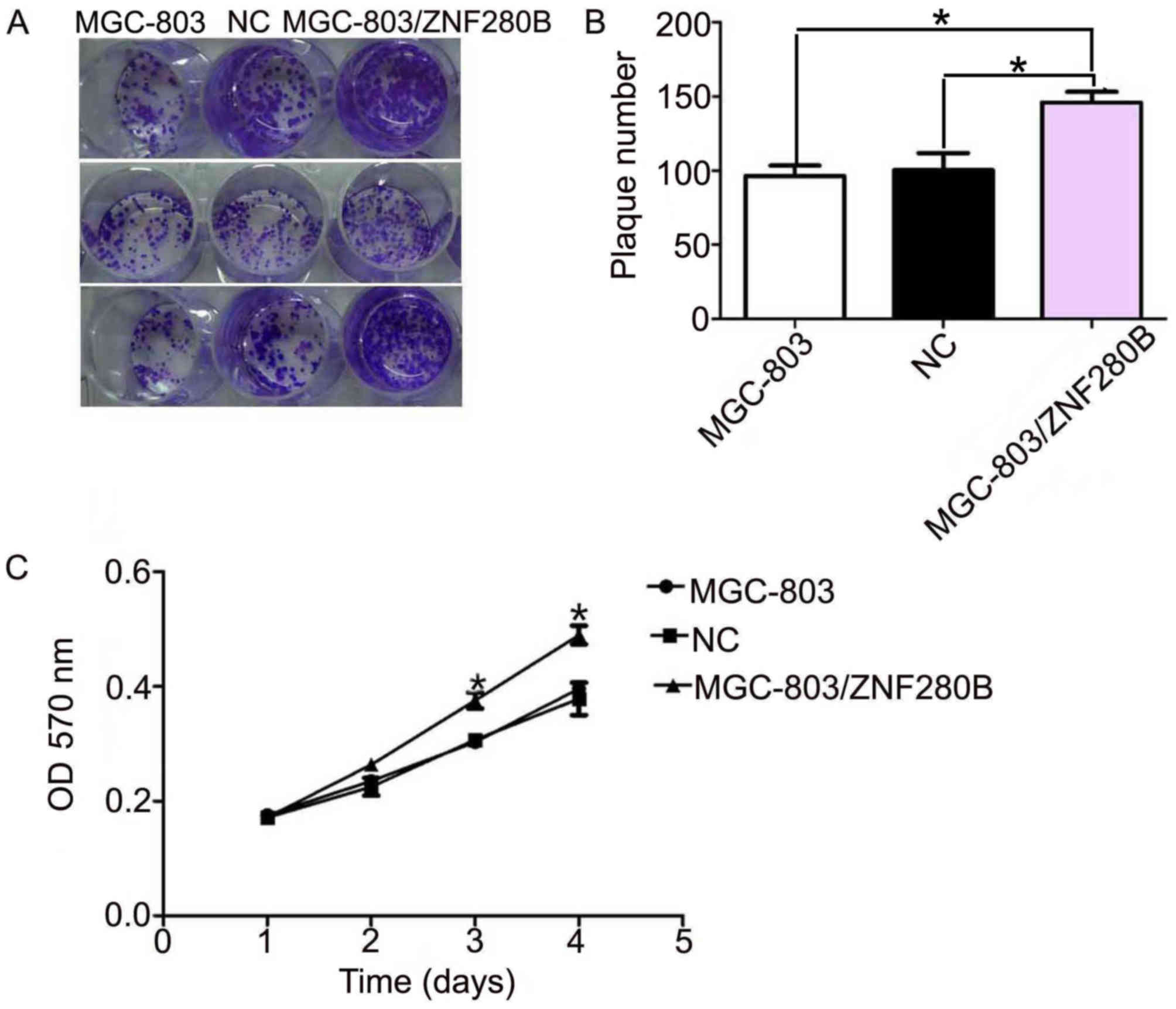
A colony formation assay indicated that the number
of MGC-803/ZNF280B colonies (146±5.8) was significantly higher than
that of the MGC-803 (97±5.1) and NC groups (101±6.5; P=0.039 and
P=0.042, respectively). However, there was no significant
difference between the MGC-803 and NC groups (P=0.369) (Fig. 3A and B).
An MTT assay was performed to study the role of
ZNF280B in the proliferation of MGC-803 cells. As demonstrated in
Fig. 3C, ZNF280B significantly
enhanced the proliferation of MGC-803 cells at days 3 and 4
(P<0.05). These results support the previous finding that
ZNF280B expression is associated with tumor size.
Upregulated expression of ZNF280B
promotes growth of gastric cancer cells in vivo
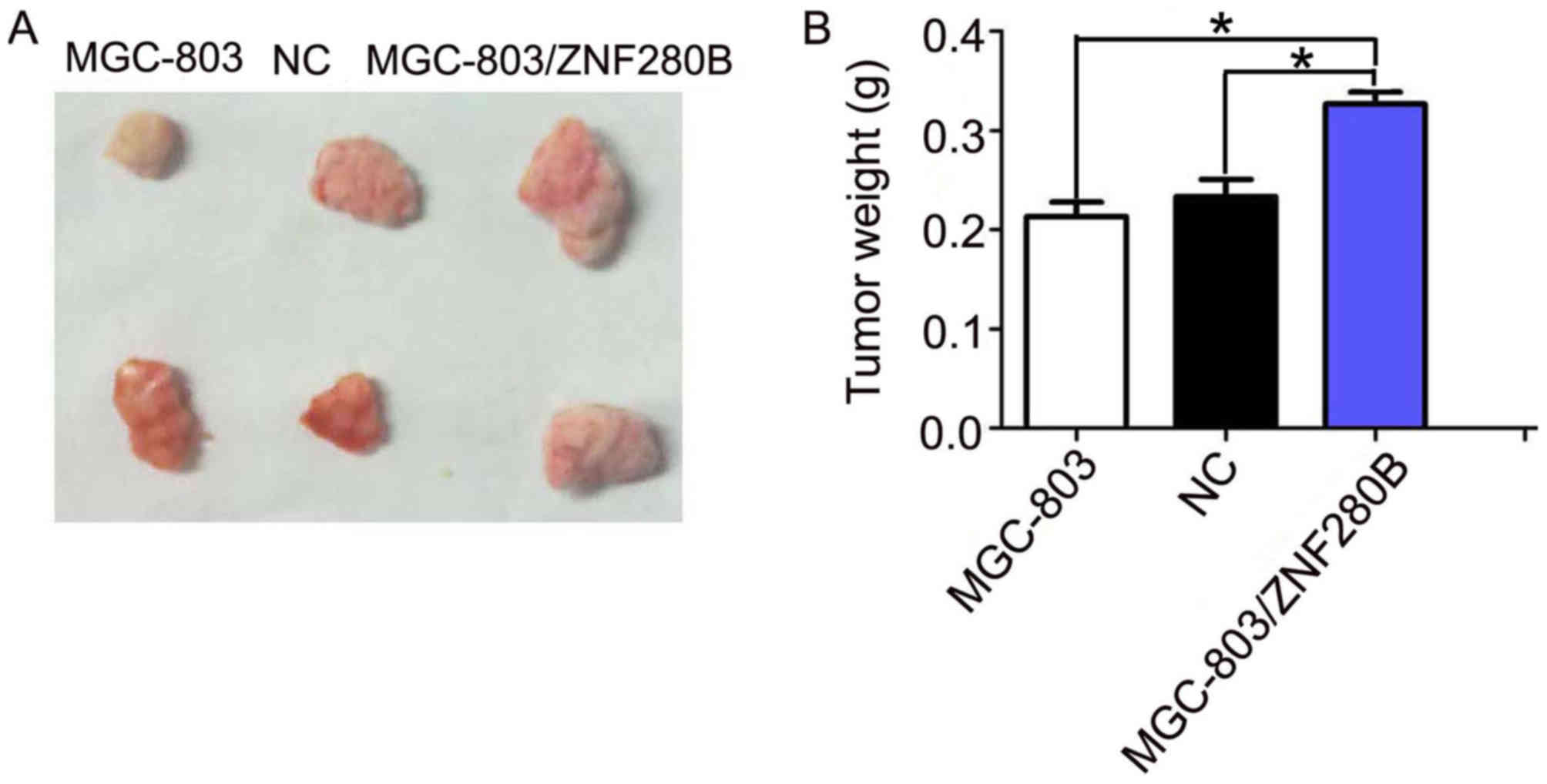
It was identified that ZNF280B acts as an oncogene
in gastric cancer by promoting cell proliferation. It was further
investigated whether ZNF280B would have a similar effect in
vivo. MGC-803, MGC-803/ZNF280B and NC cells were subcutaneously
injected into nude mice. It was found that upregulated ZNF280B
expression significantly promoted tumor growth, compared with the
MGC-803 and NC groups (Fig. 4). In
addition, compared with the MGC-803 and NC groups, tumor weight was
significantly heavier in the MGC-803/ZNF280B group (P<0.05).
These results imply that upregulated ZNF280B expression promotes
the growth of gastric cancer in vivo.
Discussion
There exist a number of publications describing the
correlation between the expression of ZNF280B and the growth of
various types of cancer (5–22). For example, Gunn et al
(22) reported that ZNF280B acts as a
tumor suppressor in chronic lymphocytic leukemia. However, the
biological roles of ZNF280B in carcinogenesis remain poorly
characterized. In the present study, the overexpression of ZNF280B
protein was confirmed by the immunohistochemical analysis of
gastric cancer samples. It was identified that ZNF280B expression
was associated with a larger tumor size. In addition, the present
study demonstrated a positive association between ZNF280B and
Ki-67, suggesting that ZNF280B is associated with the proliferation
of gastric cancer cells. Furthermore, ZNF280B expression was
significantly higher in stages III and IV compared with stages I
and II, suggesting that it may serve a vital role in the
progression of gastric cancer.
To further investigate the role of ZNF280B in
tumorigenesis of gastric cancer, the effects of ZNF280B on the
proliferation of gastric cancer cells were studied in vitro
and in vivo. The results of an MTT assay indicated that
upregulated ZNF280B expression effectively promoted the
proliferation of MGC-803 cells. Furthermore, colony formation was
greatly increased following the transfection with a ZNF280B
plasmid.
Finally, the present study confirmed that
upregulated ZNF280B expression promoted tumor growth in
vivo. Similarly, Gao et al (20) identified that ZNF280B served
pro-growth and pro-survival functions in prostate cancer with
knockdown and overexpression experiments. Additionally, ZNF280B
induces the expression of Mdm2, thereby controlling the subcellular
localization and stability of p53; the pro-cancer functions of
ZNF280B may be mediated by the downregulation of p53 in prostate
cancer cells (20).
To the best of our knowledge, this is the first
study that evaluates the association between ZNF280B and gastric
cancer, and although further confirmation using large cohort
samples is required, the data of the present study demonstrate that
ZNF280B may promote the progression of gastric cancer. Hence,
ZNF280B may be a promising target for treatment of gastric
cancer.
References
|
1
|
Wagner AD, Grothe W, Haerting J, Kleber G,
Grothey A and Fleig WE: Chemotherapy in advanced gastric cancer: A
systematic review and meta-analysis based on aggregate data. J Clin
Oncol. 24:2903–2909. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parrales A and Iwakuma T: Targeting
oncogenic mutant p53 for cancer therapy. Front Oncol.
21:2882015.
|
|
3
|
Kamada R, Toguchi Y, Nomura T, Imagawa T
and Sakaguchi K: Tetramer formation of tumor suppressor protein
p53: Structure, function, and applications. Biopolymers.
106:598–612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krig SR, Miller JK, Frietze S, Beckett LA,
Neve RM, Farnham PJ, Yaswen PI and Sweeney CA: ZNF217, a candidate
breast cancer oncogene amplified at 20q13, regulates expression of
the ErbB3 receptor tyrosine kinase in breast cancer cells.
Oncogene. 29:5500–5510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Z, Wang D, Zhu C, Shao H, Sun C, Qiu
H, Xue L, Xu J, Guo M and Li W: Aberrant alternative splicing of
human ZNF gene ZNF268 in human hematological malignancy. Oncol Rep.
20:1243–1248. 2008.PubMed/NCBI
|
|
6
|
Deng MJ, Li XB, Peng H and Zhang JW:
Identification of the trans-activation domain and the nuclear
location signals of human zinc finger protein HZF1 (ZNF16). Mol
Biotechnol. 44:83–89. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng H, Begg GE, Harper SL, Friedman JR,
Speicher DW and Rauscher FJ II: Biochemical analysis of the
Kruppel-associated Box (KRAB) transcriptional repression domain. J
Biol Chem. 275:18000–18010. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Liang Q, Wen YQ, Chen LL, Wang LT,
Liu YL, Luo CQ, Liang HZ, Li MT and Li Z: Comparative proteomics
analysis of human osteosarcomas and benign tumor of bone. Cancer
Genet Cytogenet. 198:97–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bar-Shira A, Pinthus JH, Rozovsky U,
Goldstein M, Sellers WR, Yaron Y, Eshhar Z and Orr-Urtreger A:
Multiple genes in human 20q13 chromosomal region are involved in an
advanced prostate cancer xenograft. Cancer Res. 62:6803–6807.
2002.PubMed/NCBI
|
|
10
|
Huang W, Li N, Hu J and Wang L: Inhibitory
effect of RNA-mediated knockdown of zinc finger protein 91
pseudogene on pancreatic cancer cell growth and invasion. Oncol
Lett. 12:1343–1348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jen J and Wang YC: ZNF proteins in cancer
progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Liu G, Wu S, Jiang F, Xie J and
Wang Y: Zinc finger E-box-binding homeobox 1: Its clinical
significance and functional role in human thyroid cancer. Onco
Targets Ther. 9:1303–1310. 2016.PubMed/NCBI
|
|
13
|
Gaykalova DA, Vatapalli R, Wei Y, Tsai HL,
Wang H, Zhang C, Hennessey PT, Guo T, Tan M, Li R, et al: Outlier
analysis defines zinc finger gene family DNA methylation in tumors
and saliva of head and neck cancer patients. PLoS One.
10:e01421482015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hao YJ, Li Y, Fan LQ, Zhao Q, Tan BB, Jiao
ZK, Zhao XF, Zhang ZD and Wang D: Role of RNA-interference induced
zinc finger protein 139 suppression in gastric cancer cell
sensitivity to chemotherapeutic agents. Oncol Lett. 10:1333–1338.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang J and Liu LY: ZNF protein X-linked
is overexpressed in colorectal cancer and is associated with poor
prognosis. Oncol Lett. 10:810–814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang G, Ma F, Zhong M, Fang L, Peng Y, Xin
X, Zhong J, Yuan F, Gu H, Zhu W and Zhang Y: ZNF703 acts as an
oncogene that promotes progression in gastric cancer. Oncol Rep.
31:1877–1882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang L, Hamilton SR, Sood A, Kuwai T,
Ellis L, Sanguino A, Lopez-Berestein G and Boyd DD: The previously
undescribed ZKSCAN3 (ZNF306) is a novel ‘driver’ of colorectal
cancer progression. Cancer Res. 68:4321–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu R, Peng G, Dai H, Breuer EK,
Stemke-Hale K, Li K, Gonzalez-Angulo AM, Mills GB and Lin SY:
ZNF668 functions as a tumor suppressor by regulating p53 stability
and function in breast cancer. Cancer Res. 71:6524–6534. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Wang Y, Fan X, Mo X, Wang Z, Li Y,
Yin Z, Deng Y, Luo N, Zhu C, et al: ZNF307, a novel zinc finger
gene suppresses p53 and p21 pathway. Biochem Biophys Res Commun.
363:895–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao S, Hsieh CL, Zhou J and Shemshedini L:
Zinc finger 280B regulates sGCα1 and p53 in prostate cancer cells.
PLoS One. 8:e787662013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y and Mou L: A risk score system to
preoperatively predict TNM stages in gastric cancer. Am J Clin
Oncol. 34:130–134. 2011.PubMed/NCBI
|
|
22
|
Gunn SR, Bolla AR, Barron LL, Gorre ME,
Mohammed MS, Bahler DW, Mellink CH, van Oers MH, Keating MJ,
Ferrajoli A, et al: Array CGH analysis of chronic lymphocytic
leukemia reveals frequent cryptic monoallelic and biallelic
deletions of chromosome 22q11 that include the PRAME gene. Leuk
Res. 33:1276–1281. 2009. View Article : Google Scholar : PubMed/NCBI
|