Introduction
Breast cancer is the most common type of cancer in
females in the United States (1) and
China (2) in 2015. A total of 882,900
cases were diagnosed and 324,300 mortalities occurred in developing
countries in 2012, accounting for 25% of all cancer cases and 15%
of all cancer-associated mortalities amongst females (3). Therapeutic treatments for breast cancer
typically include surgery, chemotherapy, radiotherapy, endocrine
therapy and molecular targeted therapy (4–6). Although
the prognosis for patients with early-stage breast cancer has
improved over previous years, the 5-year survival rate of patients
with metastatic breast cancer remains <20% in the United States
in 2016 (7). Notably, no targeted
therapies are available for the treatment of triple-negative breast
cancer (TNBC), which is defined by the lack of the expression of
estrogen receptor, progesterone receptor and human epidermal growth
factor receptor; therefore, chemotherapy remains the standard
treatment for this cancer (8). With
developments in the fields of molecular biology, immunology and
pharmacogenomics, immunotherapy is developing promising breast
cancer therapies, including cancer vaccines, bispecific antibodies
and immune checkpoint inhibitors (5,9).
The multiple advances in immunotherapeutic
strategies over previous years include the characterization,
cloning and expression of tumor-specific T cell receptors (TCRs)
derived from T cells ex vivo (10,11). At
present, the stimulation of expression of TCRs in T cells for
adoptive transfer is an established procedure (12,13).
Targeting cancer-testis antigens (CTAs), including NY-ESO-1,
melanoma-associated antigen 3 and glycoprotein 100, using
engineered T cells has demonstrated clinical efficacy in the
treatment of a number of tumor types (including synovial cell
sarcoma, multiple myeloma and melanoma) (10–13), but
has not yet been attempted in breast cancer.
The trophoblast-specific gene placenta-specific 1
(PLAC1) is ectopically expressed in multiple human malignancies
including ovarian cancer, hepatocellular carcinoma, colorectal
carcinoma and breast cancer (14–16);
however, the most frequent occurrence is in breast cancer, where it
is particularly associated with cancer cell proliferation,
migration and invasion, resulting in the classification of PLAC1 as
an oncoplacental protein (14–16). In
one previous study, a total of 51/62 (82%) primary breast cancer
samples scored positive for PLAC1 expression, with 15/62 (24%) with
low expression, 25/62 (40%) with intermediate expression and 11/62
(17%) with high expression (16). In
the same previous study, RNA interference-mediated silencing of
PLAC1 in two breast cancer cell lines, MCF-7 and BT-549, profoundly
impaired their motility, migration and invasion, in addition to
inducting G1-S cell cycle arrest with almost complete abrogation of
proliferation (16). Therefore, PLAC1
qualified as a promising candidate for targeted therapeutic methods
for breast cancer. Furthermore, the identification of
PLAC1-specific TCR-engineered T cells would be crucial to the
immunotherapy of breast cancer.
In the present study, a human leukocyte antigen
(HLA)-A*0201-restricted and PLAC1-specific TCR was isolated and
used for constructing lentiviral vectors. Normal CD8+ T cells
engineered using this TCR demonstrated potent antitumor effects
in vitro and in vivo in response to
HLA-A*0201+/PLAC1+ breast cancer cells. These results suggested
that the use of this TCR for adoptive immunotherapy may be valuable
for the treatment of patients with PLAC1-expressing breast
cancer.
Materials and methods
Cells and lentiviral vectors
Breast cancer cell lines (MCF-7, MDAMB-231 and T47D)
and the virus packaging cell line (293T) were purchased from the
American Type Culture Collection (Manassas, VA, USA). The cells
were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 U/ml streptomycin (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) in an incubator at 37°C and 5% CO2.
Peripheral blood mononuclear cells (PBMCs) were isolated from the
blood of healthy donors by Ficoll gradient centrifugation with 700
× g for 20 min at 4°C, subsequent to obtaining written informed
consent. The present study conformed to The Declaration of Helsinki
and was approved by the Ethics Committee of Shenzhen Beike Cell
Engineering Research Institute (Guangdong, China). Two male and two
female patients participated in present study, with an age range
from 25 to 40 years old and the mean age was 31.5 years old. The
blood of healthy donors was collected at the Hengze Clinic
(Shenzhen, Guangdong, China) on January 7, 2016. Subsequently, the
cytotoxic (CD8+) T cells from PBMCs were selected using a negative
selection procedure using a CD8+ T cell Isolation kit (Miltenyi
Biotec GmbH, Bergisch Gladbach, Germany) according to the
manufacturer's protocol. These isolated cells were cultured using
the T cell Activation/Expansion kit (Miltenyi Biotec GmbH) and RPMI
1640 medium supplemented with 10% human serum from donors of AB
blood group (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 400
U/ml interleukin (IL)-2 (Peprotech, Inc., Rocky Hill, NJ, USA) and
400 U/ml IL-15 (Peprotech, Inc., Rocky Hill, NJ, USA) at 37°C and
5% CO2.
The lentiviral vector pCDH-EF1-MCS-Thosea asigna
virus (T2A)-Puro and three packaging vectors, pLP1, pLP2 and
pLP-VSVG, were obtained from Addgene, Inc. (Cambridge, MA,
USA).
Generation of PLAC1-specific T
lymphocytes
In order to generate cytotoxic T-lymphocytes (CTLs),
the monocyte-derived dendritic cells (DC) of an HLA-A2+ donor were
loaded with 5 µg/ml PLAC128–36-peptide (VLCSIDWFM)
(GenScript, Jiangsu, China) for the stimulation of autologous CD8+
T cells. Following two stimulations, the
HLA-A2+/PLAC128–36-multimer+ CD8+ T cells were sorted by
flow cytometry (FACSAria; BD Biosciences, San Jose, CA, USA), and
the data was analyzed using BD FACSDiva software (version 6.0; BD
Biosciences). Subsequently, the sorted T cells were cloned using a
limiting dilution as previously described (17,18) and
after 2 weeks, screened for specific recognition of MCF-7 cells
(PLAC1+, HLA-A2+) based on specific interferon γ (IFN-γ) secretion
(data not shown), which was typically stimulated by antigen
encounters, and was ascribed to the major histocompatibility
complex (MHC)-dependent TCR activation (19). Then, the PLAC1-reactive clones were
expanded using the T cell Activation/Expansion kit and RPMI 1640
supplemented with 10% human serum from donors of AB blood group,
400 U/ml IL-2, and 400 U/ml IL-15 at 37°C and 5%
CO2.
To identify the sequences of the TCR genes, a
5′-RACE-PCR GeneRacer kit (Invitrogen; Thermo Fisher Scientific,
Inc.) amplifying the TCRα- and TCRβ-chains was performed using RNA
isolated from the T cell clones using an RNA Isolation Kit (cat.
no. R6731; Omega Bio-tek, Inc., Norcross, GA, USA) according to the
manufacturer's protocol. The RACE-PCR products were sequenced, the
TCRα- and TCRβ-chains were linked by a 2A peptide linker
(TCRα-T2A-TCRβ) as previously described (17,18), and
the whole constructs were codon-optimized and synthesized by
GenScript. These synthesized fragments were cloned into the
lentiviral vector pCDH-EF1-MCS-T2A-Puro via EcoRI and
BamHI restriction sites.
A virus packaging cell line, 293T, was seeded at a
density of 1×107 cells/150-mm dish and then incubated at
37°C with 5% CO2 for 24 h. After 24 h, the cells were
subjected to transfection at 37°C for 4 h with Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and 20.0 µg pCDH-TCR
construct (or pCDH-EF1-MCS-T2A-Puro empty vector) with the pLP1,
pLP2, and pLP-VSVG packaging vectors. After culturing the cells for
48 h, the filtered culture supernatants were used for infection.
The CD8+ T cells from the PBMCs of healthy donors were transduced
using the lentiviral vectors carrying TCR or control empty vector
[negative control (NC)] in the presence of 5 µg/ml polybrene
(Sigma-Aldrich; Merck KGaA) for 12 h, followed by incubation at
37°C for 48 h in RPMI 1640 supplemented with 10% human serum from
donors of AB blood group, 400 U/ml IL-2 and 400 U/ml IL-15. The
TCR-transduced CD8+ T cells were evaluated for the
expression of appropriate TCR by multimer staining at 4°C for 20
min and flow cytometry.
Flow cytometry
A total of 1×106 T cells were collected
by centrifugation with 650 × g for 10 min at room temperature,
fixed with paraformaldehyde for 30 min at room temperature, washed
twice with PBS, and then blocked with fluorescence-activated cell
sorter (FACS) solution (PBS containing 2% FBS and 0.1%
NaN3) for 30 min at 4°C. Then cells were stained using
fluorescein isothiocyanate (FITC)-labeled monoclonal antibody (mAb)
against human CD8 (cat. no. 555366; dilution, 1:10; BD Biosciences,
San Jose, CA, USA), PerCP-Cy5.5-labeled mAb against human CD3 (cat
no. 560835; dilution, 1:10; BD Biosciences), and phycoerythrin
(PE)-labeled PLAC1-multimer (code, F2A-G; dilution, 1:10;
Proimmune, Ltd., Oxford, UK) diluted with FACS solution for 20 min
at 4°C. Cancer cells were digested with 0.25% trypsin for 3 min at
37°C, collected by centrifugation at 300 × g for 10 min at room
temperature, fixed with paraformaldehyde for 30 min at room
temperature and washed twice with PBS. Then, cells were blocked
with FACS solution for 30 min at 4°C. HLA-A2 expression in the
cultured cancer cells was evaluated by flow cytometry using the
PE-labeled HLA-A2 antibody (cat no. 558570; dilution, 1:10; BD
Biosciences) and PE-labeled isotype control (cat no. 555743;
dilution, 1:10; BD Biosciences) diluted with FACS solution for 20
min at 4°C. Isotype control antibody was used in order to eliminate
nonspecific combinations in a previous test (data not shown). The
cell was analyzed using flow cytometry (FACS Calibur; BD
Biosciences) and the data was analyzed using CellQuest Pro.app
software (version 6.0; BD Biosciences).
Cytokine release assay
TCR-, NC-transduced or untransduced CD8+ T cells
were serially diluted with RPMI 1640 medium supplemented with 10%
FBS and co-cultured with 1×104 target cells including
MCF-7, MDA-MB-231 and T47D in 100 µl RPMI 1640 medium supplemented
with 10% FBS, resulting in a gradient effector cell to target cell
(E:T) ratio (20:1, 10:1 and 5:1, respectively). After 4 h, the
culture supernatants were collected, and the level of IFN-γ and
tumor necrosis factor α (TNF-α) determined using a commercial
enzyme-linked immunosorbent assay kit (cat. no. DIF50 and DTA00C;
R&D Systems, Inc., Minneapolis, MN, USA) according to the
manufacturer's protocol.
Cytotoxicity assay
For evaluating the efficacy of TCR- or NC-transduced
CD8+ T cell-mediated lysis, the cells were serially diluted with
RPMI 1640 medium supplemented with 10% FBS and co-cultured with
1×104 target cells including MCF-7, MDA-MB-231 and T47D,
resulting in a gradient E:T ratio as described above. After 4 h of
co-culture at 37°C, the cytolysis was determined using a lactate
dehydrogenase (LDH) activity assay (Roche Diagnostics GmbH,
Mannheim, Germany) according to the manufacturer's protocol, as
previously described (20). The
absorbance was measured on a microplate reader at 490 nm (BioTek
Instruments, Inc., Winooski, VT, USA). Following background
subtraction, the percentage lysis was calculated using the
following equation: 100% × [(experimental release-effector
spontaneous release-target spontaneous release)/(target maximum
release-target spontaneous release)].
Immunofluorescence
Immunofluorescence was performed as described
previously (21,22). Briefly, MCF-7, MDA-MB-231 and T47D
cells were fixed with 4% paraformaldehyde for 30 min at room
temperature and permeabilized using 0.2% Triton X-100. Following
blocking with 2% bovine serum albumin for 1 h at 4°C, the cells
were incubated with a rabbit anti-PLAC1 antibody (cat. no.
ab105395; dilution, 1:100; Abcam, Cambridge, UK) or a rabbit IgG
(cat. no. A7016; dilution, 1:100; Beyotime Institute of
Biotechnology, Haimen, China) overnight at 4°C. Subsequently, the
cells were washed, followed by the addition of PE-conjugated goat
anti-rabbit IgG (cat. no. HS121-01; dilution, 1:100; TransGen,
Beijing, China) for 1 h at 37°C in the dark, and visualized using
an inverted fluorescence microscope (magnification, ×100) (Olympus
IX71; Olympus Corporation, Tokyo, Japan).
PLAC1 gene silencing with small
interfering RNA (siRNA)
One pair of siRNA oligonucleotides corresponding to
the target sequence for human PLAC1 (CACCTACCGTGTTACTGAA) were
designed and synthesized by RiboBio (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China). A negative-control siRNA (NC siRNA) (Guangzhou
RiboBio Co., Ltd.) was used as the control. Cells were plated to
achieve 40–60% confluency in RPMI 1640 containing 10% FBS without
antibiotics. The siRNA was transfected into MCF-7 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, the cells were analyzed 48 h after transfection, and
PLAC1 protein expression was determined using western blot
analysis.
Western blot analysis
Western blot analysis was performed as described
previously (23). Briefly, the
TCR-transduced or NC-transduced CD8+ T cells, MCF-7 cells and PLAC1
gene silencing- and NC-silencing-MCF-7 cells were lysed using
radioimmunoprecipitation assay buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) and the protein concentrations measured
using a BCA Assay kit (Beyotime Institute of Biotechnology,
Jiangsu, China). Equivalent amounts of protein (20 µg) were
separated using a 10% gel and SDS-PAGE and transferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were probed with primary antibodies against
phosphoinositide 3-kinase γ (PI3Kγ; cat. no. 5405S; 1:1,000),
protein kinase B (AKT; cat. no. 9272S; 1:1,000), phosphorylated
(p)-AKT (cat. no. 4060S; 1:1,000), extracellular signal-regulated
kinase (ERK; cat. no. 4695S; 1:1,000), p-ERK (cat. no. 4377S;
1:1,000), p38 (cat. no. 8690S; 1:1,000), p-p38 (cat. no. 4631S;
1:1,000), c-Jun N-terminal kinases (JNK; cat. no, 9252S; 1:1,000),
p-JNK (cat. no. 4668S; 1:1,000), nuclear factor-κB (NF-κB; cat. no.
4764S; 1:1,000), β-actin (cat. no. 4970L; 1:1,000) and GAPDH (cat.
no. 5174S; 1:1,000) (all from Cell Signaling Technology, Inc.,
Danvers, MA, USA), and PLAC1 (cat. no. ab105395; 1:1,000; Abcam,
Cambridge, UK) in TBS-Tween-20 (TBST) containing 5% non-fat milk
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 4°C overnight,
followed by three washes in TBST for 10 min per wash. Subsequently,
the membranes were incubated with the IRDye 800CW goat anti-rabbit
secondary antibody (1:5,000 dilution in TBST containing 5% non-fat
milk; cat. no. 926-32211; LI-COR Bioscience; Lincoln, Nebraska,
USA) for 2 h at room temperature. The protein bands were visualized
using an Infrared Imaging System Odyssey (LI-COR Bioscience;
Lincoln, Nebraska, USA). Quantity One software (version 4.62;
Bio-Rad Laboratories, Inc., Hercules, California, USA) was used to
quantify of the western blots.
In vivo tumor growth assays
A total of 15 subcutaneous transplant breast cancer
model female nude mice (BALB/c-nu) (4–5-week-old) weighing between
18 and 20 g, were purchased directly from the Medical Laboratory
Animal Center of Guangdong Province (Guangdong, China). The mice
were housed under controlled 12 h light-dark cycles, with constant
temperature (22–24°C) and humidity (55–60%), and were given
sterilized food and tap water ad libitum. Briefly, the mice were
pre-treated with 200 µg/kg estradiol valerate (Sigma-Aldrich; Merck
KGaA) by gastric perfusion, starting 1 week prior to cell
implantation as previously described (24). Subsequently, each of the mice were
administered ~5×106 MCF-7 cells (in 100 µl serum-free
media) mixed with 100 µl Matrigel (BD Biosciences) into the left
flank subcutaneously. Protocols for the treatment of nude mice were
approved by the Laboratory Animal Ethics Committee of the Medical
Laboratory Animal Center of Guangdong Province. When large tumors
reached a mean volume of 150 mm3, the mice were randomly
divided into 3 groups (5 animals/group) and subjected to
intravenous tail vein transplantation with normal saline,
1×107 TCR-, or NC-transduced CD8+ T cells twice with a 1
week interval. The tumors were measured once a week using
electronic calipers. The longest length and width were recorded and
the tumor volume was calculated according to the formula π/6
[(length × (width)2]. The mice were anesthetized prior
to cervical dislocation using 45 mg/kg pentobarbital sodium at the
culmination of the experiment.
Statistical analysis
Data were expressed as the mean ± standard
deviation. SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) was
used for analysis. All experiments (excluding the nude mice
subcutaneous transplant tumor model experiment) were repeated at
least 3 times. A one-way analysis of variance with Bonferroni
correction was used for analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
CD8+ T cells may be efficiently
engineered to express TCRs recognizing the HLA-A2-restricted PLAC1
peptides
cDNAs encoding TCRα- and β-chains specific for
HLA-A2-restricted PLAC1 were cloned from a CTL generated by the
in vitro stimulation of CD8+ T cells using a HLA-A2+ DC
loaded with PLAC128–36-peptide (VLCSIDWFM). The TCRα-
and TCRβ-chains were linked by a 2A peptide linker (TCRα-T2A-TCRβ),
and the whole constructs were codon-optimized and synthesized.
These synthesized fragments were then cloned into the lentiviral
vector pCDH-EF1-MCS-T2A-Puro via EcoRI and BamHI
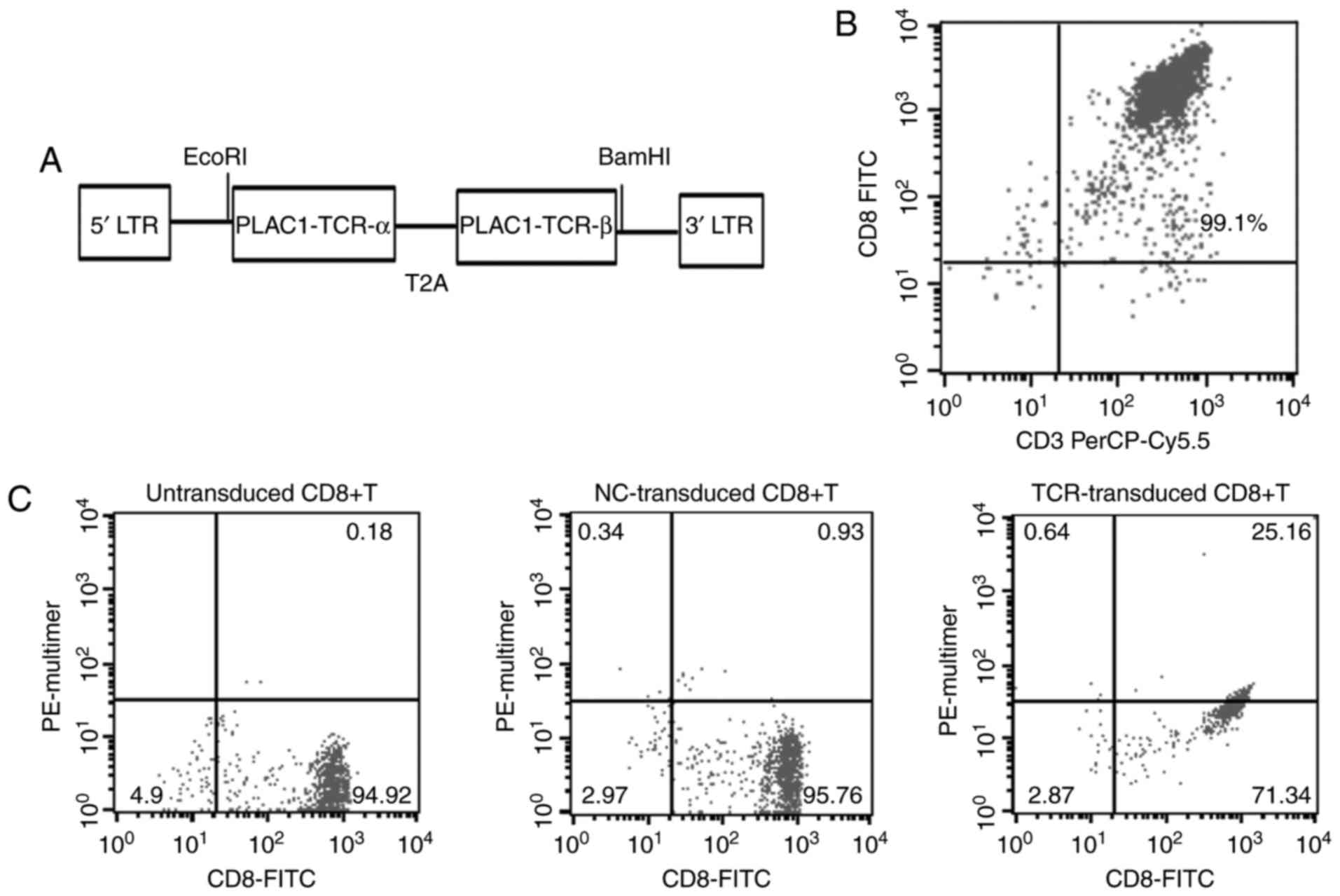
restriction sites (Fig. 1A). To
further characterize the TCR, the lentiviral vectors carrying TCR
or NC were used for the transduction of the CD8+ T cells, isolated
from PBMCs (99.1% purity), with a negative selection procedure
using magnetic beads (Fig. 1B). The
results revealed that the accurately expressing and matching
efficiency of TCRα- and TCRβ-chains in CD8+ T cells was up to
25.16%, as detected by FITC-labeled CD8 mAb and PE-labeled
PLAC1-multimers (Fig. 1C). These
HLA-A*0201-restricted and PLAC1-specific TCR-engineered CD8+ T
cells were then used for subsequent experiments.
Identification of PLAC1 and HLA-A2
serotype-positive breast cancer cell lines
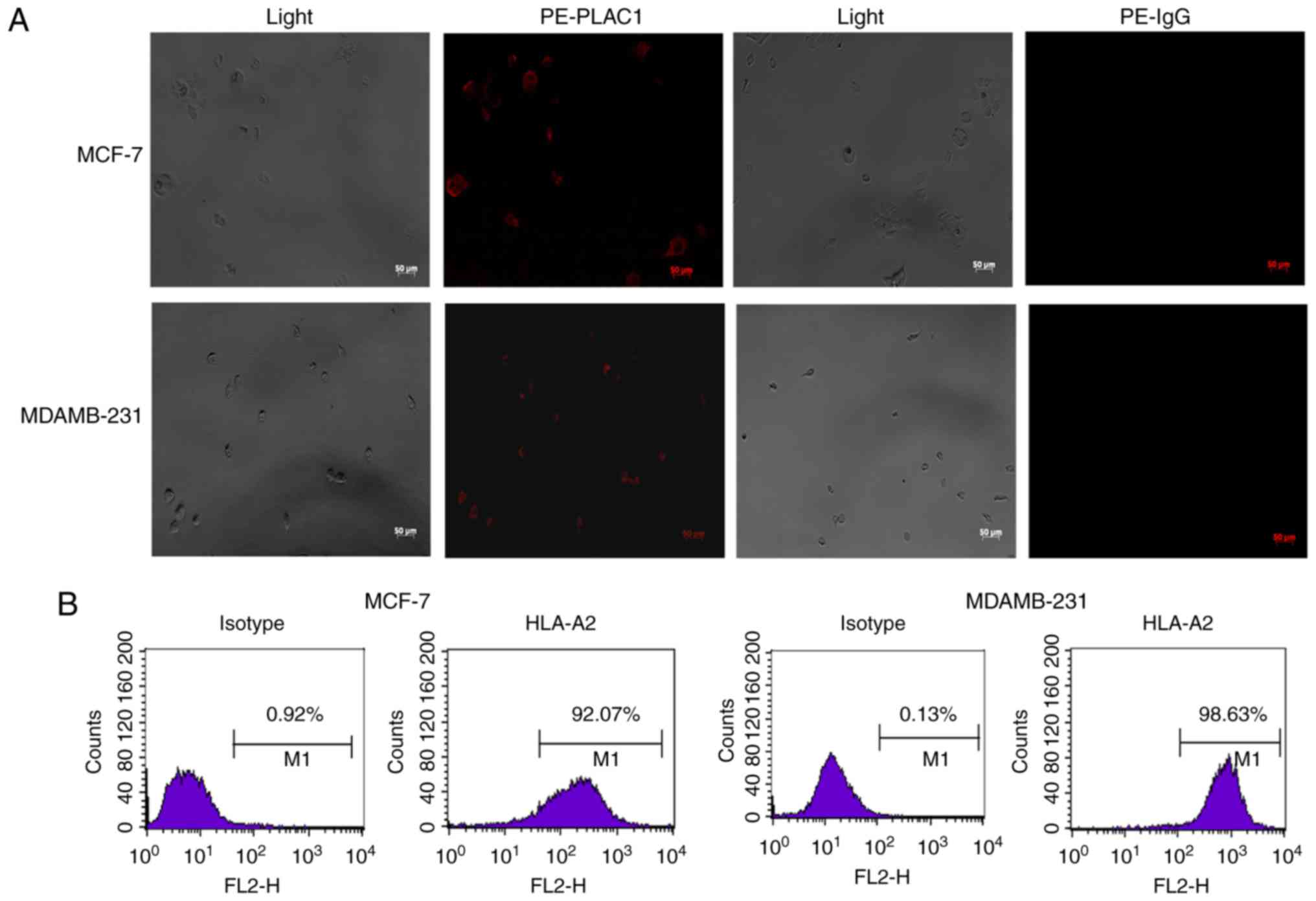
Human non-metastatic breast cancer cells (MCF-7) and
TNBC cells (MDAMB-231) were examined for the baseline expression of
PLAC1 by immunofluorescence. As presented in Fig. 2A, the two cell lines expressed PLAC1.
Next, these two different cancer cell lines were serotyped with an
HLA-A2 antibody by flow cytometry and were revealed to be HLA-A2
positive. The HLA-A2 expression efficiency in MCF-7 and MDAMB-231
cell lines reached 92.07 and 98.63%, respectively (Fig. 2B), thereby resulting in the selection
of these 2 cell lines for further studies.
Evaluation of the function of PLAC1
TCR-engineered CD8+ T cells
In order to evaluate the recognition of PLAC1 TCRs,
transduced CD8+ T cells were subjected to a co-culture assay with
MCF-7 (PLAC1+, HLA-A2+) and MDAMB-231 (PLAC1+, HLA-A2+) cells. A
significantly higher release of IFN-γ and TNF-α was observed in
TCR-engineered CD8+ T cells co-cultured with HLA-A2+/PLAC1+ MCF-7
and MDAMB-231 cell lines compared with NC-transduced and
untransduced CD8+ T cells co-cultured with these two cell lines
(P<0.01; Fig. 3A and B). As
NC-transduced and untransduced CD8+ T cells could not mediate the
release of effector cytokines when co-cultured with HLA-A2+/PLAC1+
MCF-7 and MDAMB-231 cell lines, only the NC-transduced CD8+T cells
were selected as the control in subsequent studies.
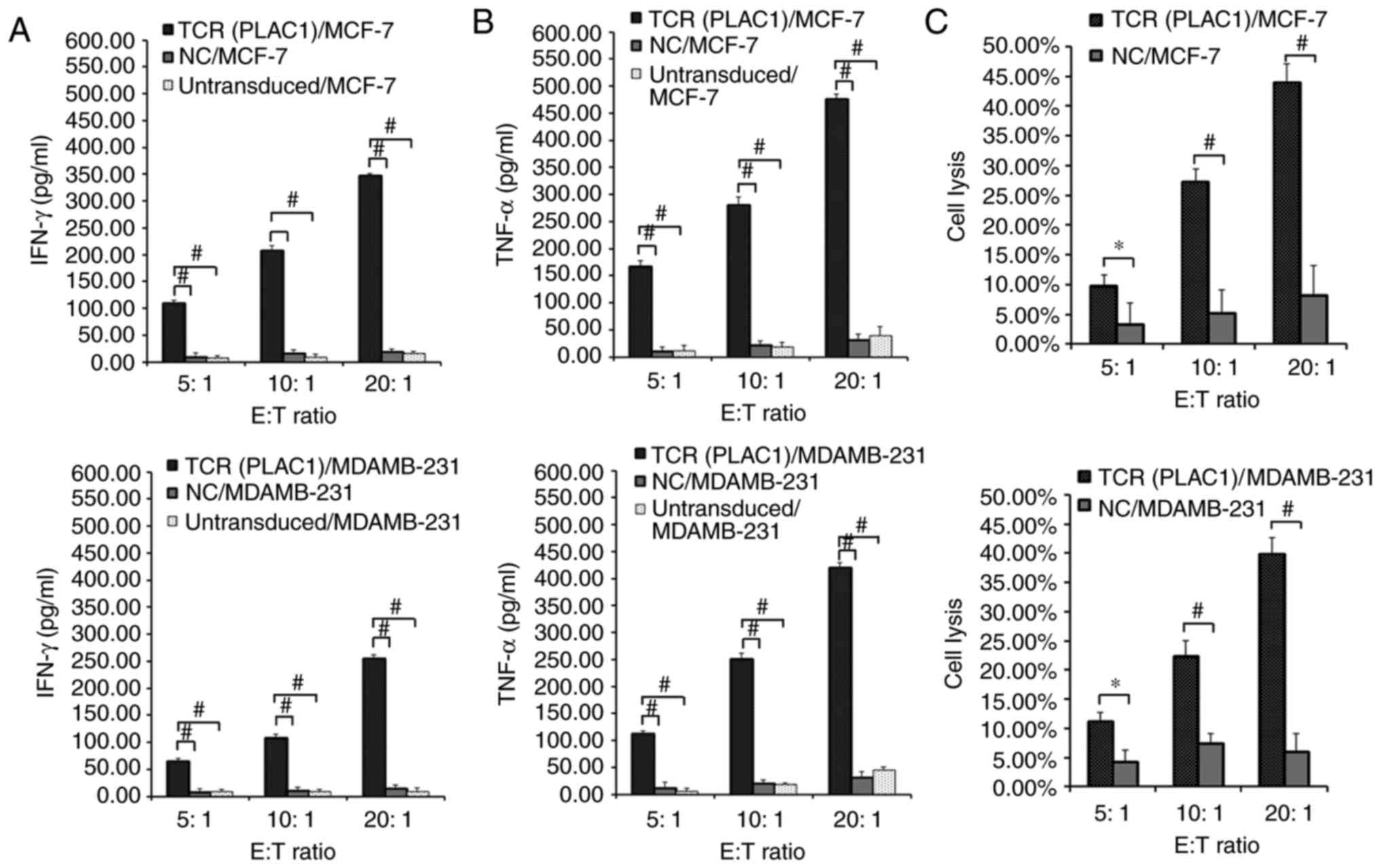 | Figure 3.Evaluation of the function of PLAC1
TCR-engineered CD8+ T cells. Human leukocyte antigen-A2-restricted
and PLAC1-specific TCR-, NC-transduced or untransduced CD8+ T cells
were co-cultured for 4 h with 1×104 MCF-7 and MDAMB-231
cells respectively at a range of E:T ratios (5:1, 10:1 and 20:1).
Concentration of (A) IFN-γ and (B) TNF-α secreted into the culture
medium were measured using an enzyme-linked immunosorbent assay.
(C) TCR- or NC-transduced CD8+ T cells were co-cultured for 4 h
with 1×104 MCF-7 and MDAMB-231 cells. Cytolysis was
determined using a lactate dehydrogenase activity assay. Subsequent
to background subtraction, the percentage lysis was calculated by
100% × [(experimental release-effector spontaneous release-target
spontaneous release)/(target maximum release-target spontaneous
release)]. *P<0.05 and #P<0.01 with comparisons
shown by lines. PLAC1, placenta-specific 1; TCR, T cell receptor;
CD8+ T cell, cytotoxic T cell; NC, negative control; E:T, effector
cell to target cell; IFN-γ, interferon γ; TNF-α, tumor necrosis
factor α. |
Subsequently, the specific lysis of breast cancer
cell lines by the engineered CD8+ T cells was measured using an LDH
assay. PLAC1 TCR-transduced CD8+ T cells demonstrated an
significantly increased lytic function against HLA-A2+/PLAC1+ MCF-7
and MDAMB-231 cells compared with NC-transduced CD8+ T cells, which
demonstrated little reactivity against any of the target cells
(P<0.05; Fig. 3C). PLAC1
TCR-transduced CD8+ T cells were not capable of lysing T47D cells,
which were PLAC1-positive, however, this cell line was identified
to be HLA-A2 negative in the immunofluorescence and flow cytometry
assays (Fig. 4A-C). As PLAC1 is
generally expressed in the breast cancer cells, it is difficult to
identify a PLAC1-negative cell line. Therefore, the PLAC1 gene was
silenced using siRNA in MCF-7 cells, to further detect the function
of PLAC1 TCR-engineered CD8+ T cells. The results revealed that the
significant PLAC1 gene silencing in MCF-7 cells compared with NC
MCF-7 cells (P<0.01; Fig. 4D)
resulted in the significant decrease in the specific lysis of
breast cancer cells by the engineered CD8+ T cells compared with NC
cells (P<0.05; Fig. 4E). These
results indicated that the PLAC1 TCR-engineered CD8+ T cells may
efficiently and specifically recognize and kill breast cancer
cells.
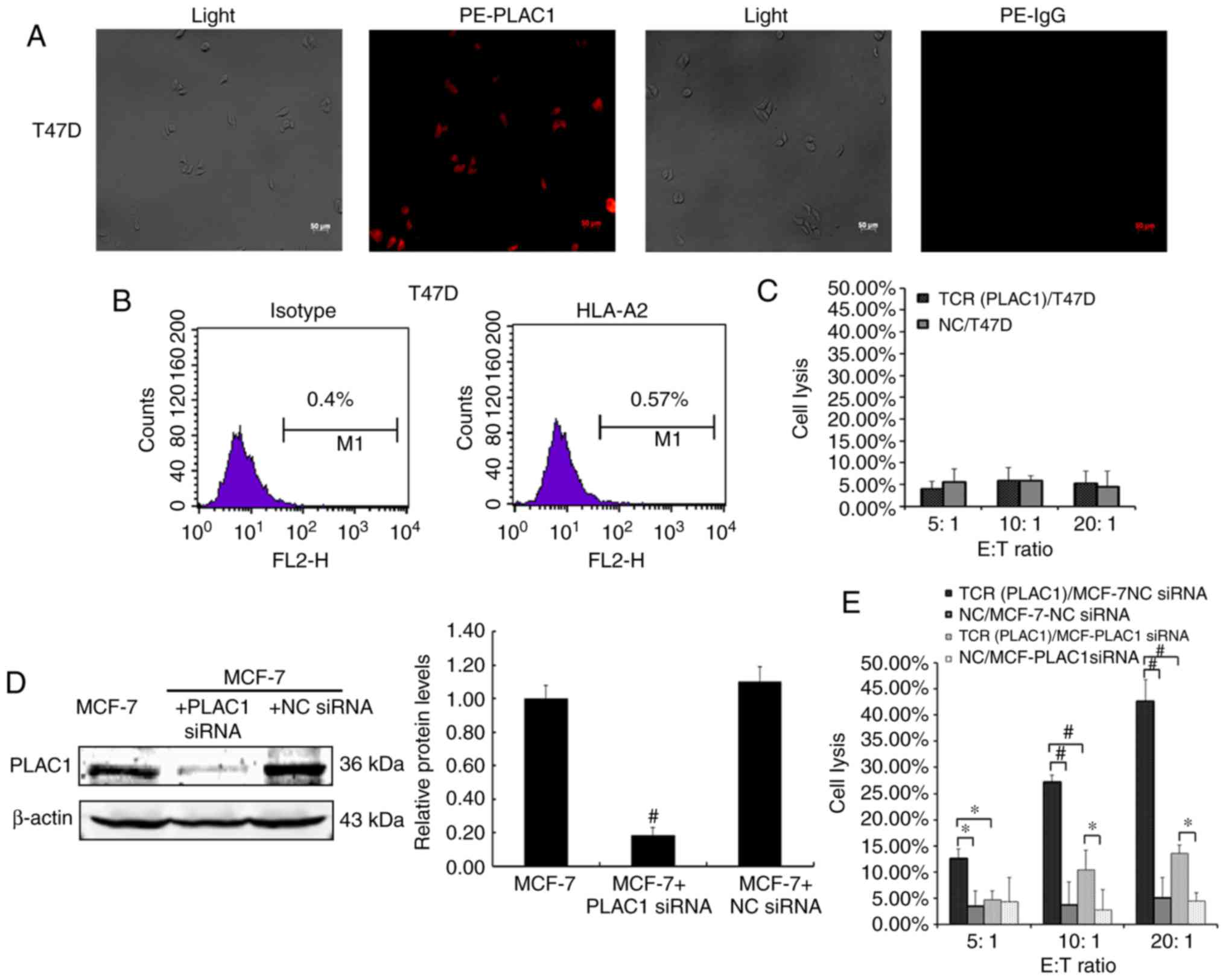 | Figure 4.PLAC1 TCR-engineered CD8+ T cells
specifically recognize and kill breast cancer cells. (A)
Immunofluorescence staining with rabbit anti-PLAC1 primary antibody
or a rabbit IgG and PE-conjugated secondary antibody in the human
breast cancer cell lines T47D. Scale bar indicates 50 µm. The
results are representative of three independent experiments. (B)
T47D cells were analyzed using flow cytometry for HLA-A2 expression
using the PE-labeled HLA-A2 antibody and PE-labeled isotype
control. The results are representative of three independent
experiments. (C) Lysis of T47D breast cancer cells by the
engineered CD8+ T cells was determined using a LDH activity assay.
Data are expressed as the means ± SD of three independent
experiments. (D) MCF-7 cells were transfected with PLAC1 siRNA or
NC siRNA and then analyzed using western blot analysis for PLAC1
expression following normalization to β-actin. The results are
representative of three independent experiments. Bars represent the
relative protein level as compared with the MCF-7 cells alone
group. #P<0.01 vs. MCF-7 + NC siRNA group. (E) Lysis
of MCF-7-NC siRNA and MCF-7-PLAC1 siRNA cells by the engineered
CD8+ T cells was determined using a LDH activity assay. Data are
expressed as the means ± SD of three independent experiments.
*P<0.05 and #P<0.01 with comparisons shown by
lines. PLAC1, placenta-specific 1; PE, phycoerythrin; IgG,
immunoglobulin G; E:T, effector cell to target cell; TCR, T cell
receptor; NC, negative control; siRNA, small interfering RNA; CD8+
T cell, cytotoxic T cell; HLA, human leukocyte antigen; SD,
standard deviation; LDH, lactate dehydrogenase. |
Mechanism of enhanced activation of
TCR-transduced CD8+ T cells
A previous study demonstrated that PI3Kγ kinase
activity is essential for optimal T cell activation and
differentiation, in addition to an efficient T cell-mediated immune
response (25). Upon TCR engagement,
multiple signaling mechanisms, including mitogen-activated protein
kinase (MAPK) and NF-κB, modulated by the PI3K/AKT signaling
pathway, are activated during the process of T cell activation,
which results in gene induction and cell cycle progression
(26–29). In order to investigate the mechanism
of the enhanced activation of TCR-transduced CD8+ T cells, western
blot analysis was performed and demonstrated that the expression of
PI3Kγ, and the phosphorylation of AKT (p-AKT) and ERK1/2 (p-ERK) in
addition to the expression of NF-κB, but not the expression of JNK,
p-JNK, P-38 and p-p38 in TCR-transduced CD8+ T cells were enhanced
as compared with the NC-transduced CD8+ T cells (Fig. 5). These results suggested that the
PI3K pathway possessed the capacity to develop the T cells in order
to respond to the engineered TCR stimulation.
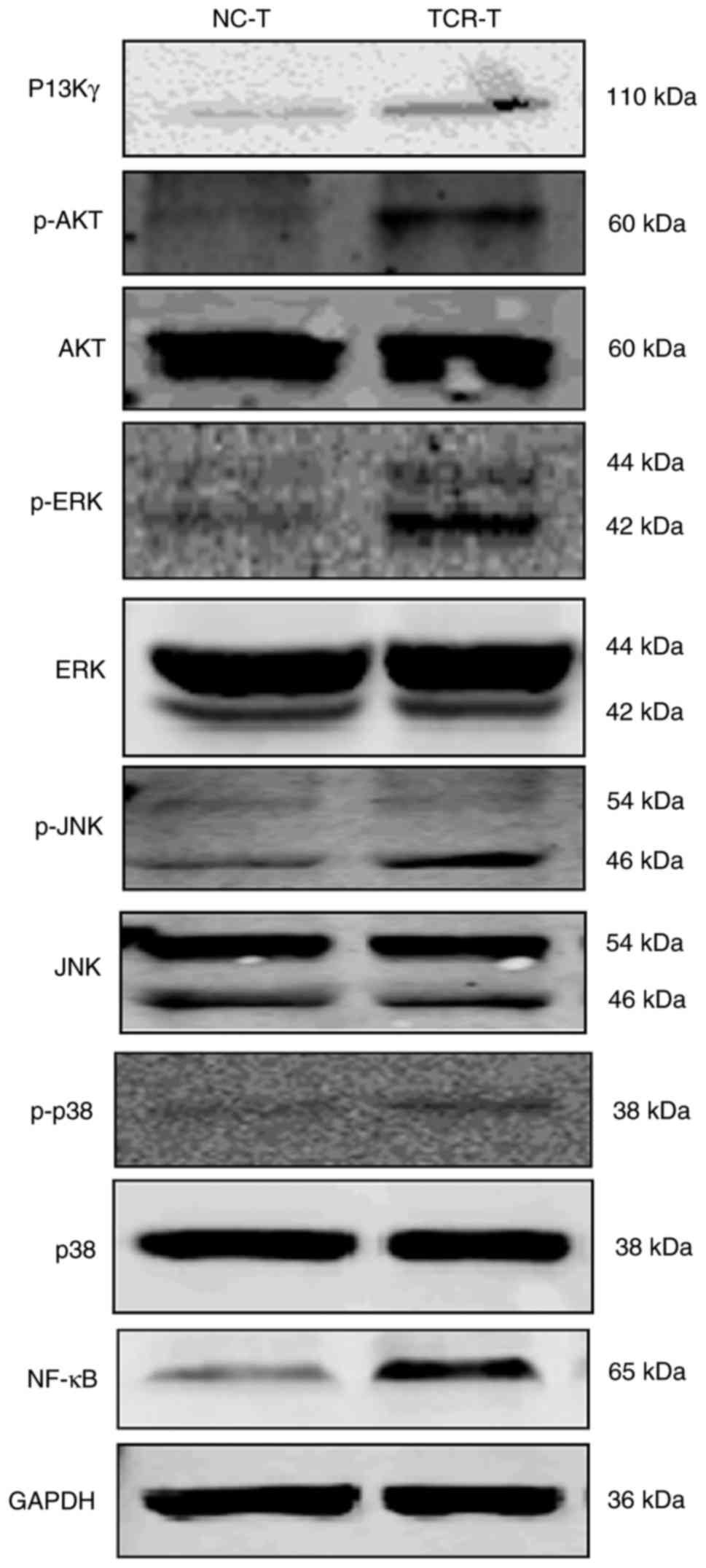 | Figure 5.Mechanism of enhanced activation of
TCR-transduced CD8+ T cells. Cell lysates of TCR- or NC-transduced
CD8+ T cells were analyzed by western blot analysis for the
expression of PI3Kγ, AKT, p-AKT, ERK1/2, p-ERK1/2, JNK, p-JNK, p38,
p-p38, NF-κB and GAPDH. The results are representative of three
independent experiments. NC, negative control; TCR, T cell
receptor; CD8+ T cells, cytotoxic T cells; PI3Kγ, phosphoinositide
3-kinase γ; AKT, protein kinase B; ERK1/2, extracellular
signal-regulated kinase; JNK, c-Jun N-terminal kinases; NF-κB,
nuclear factor κB; p-, phosphorylated. |
TCR-transduced CD8+ T cells inhibit
tumor growth in vivo
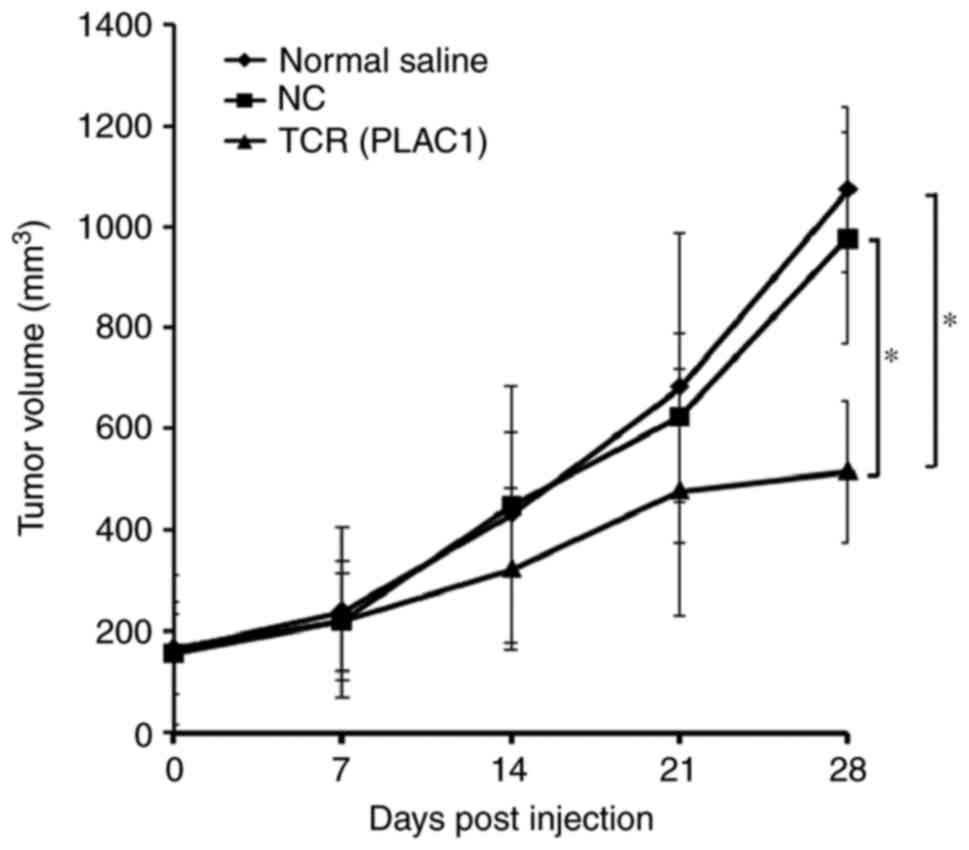
To examine the in vivo efficacy of PLAC1
TCR-transgenic CD8+ T cells, 5×106 MCF-7 cells were
inoculated into BALB/c-nu mice to establish a subcutaneous
transplant tumor model. Upon achieving a mean tumor volume of 150
mm3, the mice were randomly divided into 3 groups (5
animals/group) and subjected to intravenous tail vein
transplantation of either normal saline, 1×107 PLAC1
TCR-transduced CD8+ T cells or NC-transduced CD8+ T cells, twice
with a 1 week interval. Slowing of tumor growth was observed 14
days post-injection. At day 28, the tumors in the PLAC1
TCR-transduced cells group were significantly smaller, ~50% smaller
compared with the normal saline or NC-transduced cells group
(P<0.05; Fig. 6). These results
indicated that TCR-transduced CD8+ T cells inhibited the tumor
growth in vivo.
Discussion
Toxicity to normal tissues is a potential negative
side-effect in TCR-based adoptive immunotherapy targeting tumor
antigens that are additionally expressed in certain normal tissues
(12,30). Adoptive immunotherapy should strive to
produce T cells that target tumor-specific antigens that are not
expressed in normal tissues. Therefore, CTA is the ideal candidate
due to its overexpression in multiple tumor types and limited
expression in normal tissue (31). As
a novel member of the CTA group, PLAC1 has been revealed to be
expressed in a wide range of human malignancies including ovarian
cancer, hepatocellular carcinoma, colorectal carcinoma and breast
cancer (14–16). PLAC1 was identified to be frequently
expressed in breast cancer, but not in normal tissues except the
testes and placenta, and was involved in the proliferation,
migration and invasion of breast cancer (32), thereby indicating its key function in
cancer tumorigenesis. Germ cells do not present antigens due to
their lack of expression of MHC molecules. Therefore, germ cells
cannot be targeted by TCR-engineered T cells (33). Thus, the immune responses directed
against CTAs by PLAC1 are unlikely to result in the recognition of
these tissues. In addition, HLA-A*0201 is the most widely expressed
HLA-I molecule in the Chinese population (34). Therefore, the identification of
HLA-A*0201-restricted and PLAC1-specific TCR-engineered T cells
would be critical to the immunotherapy of breast cancer; however,
it has not yet been well-studied. In the present study, an
HLA-A*0201-restricted and PLAC1-specific TCR was identified, which
was transduced into CD8+T cells in order to investigate the
antitumor effects of PLAC1-specific TCR-engineered CD8+T cells in
breast cancer cells in vitro and in vivo.
In order to generate PLAC1-specific T cell clones,
protocols similar to the method described by Sommermeyer et
al (18) were employed:
Autologous CD8+ T cells were stimulated with HLA-A2+ DC and loaded
with PLAC128–36-peptide (VLCSIDWFM). Liu et al
(34) demonstrated that the
HLA-A*0201-restricted T cell epitope, PLAC128–36-peptide
(VLCSIDWFM), may induce the most potent peptide-specific CTLs and
serve as a novel candidate epitope for the development of peptide
vaccines against PLAC1-positive breast cancer. In the present
study, CTL clones against PLAC1 (VLCSIDWFM) were additionally
isolated successfully. Furthermore, the codon-optimized PLAC1-TCR
gene was isolated and synthesized. The TCRα- and TCRβ-chains were
linked by a 2A peptide linker (TCRα-T2A-TCRβ). T2A and porcine
teschovirus-1 (P2A) are small ‘self-cleaving’ peptides. The first
small ‘self-cleaving’ peptide F2A was identified in the
foot-and-mouth disease virus, a member of the picornavirus group
(35,36). Subsequently, ‘2A like’ peptides from
equine rhinitis A virus, P2A and T2A were identified, and their
activities in proteolytic cleavage were certified in various in
vitro and in vivo eukaryotic systems (35,36).
Subsequently, the PLAC1-TCR gene was expressed in primary human
CD8+T cells using a negative selection process using magnetic
beads. The 25.16% expressing and matching efficiency of TCRα- and
TCRβ-chains detected by PLAC1-multimers indicated the accurate
expression of TCR by CD8+ T cells. It was hypothesized that a
longer time would be required to enrich T cells when the puromycin
resistance gene carried by the lentiviral vector
pCDH-EF1-MCS-T2A-Puro were used compared to those without the
puromycin resistance gene, while the proliferation ability of T
cells decreased with the increase in days (data not shown).
Therefore, the puromycin resistance gene was not used in the
present study. The puromycin resistance gene and the concentrated
lentiviral particles will be used to improve the transduction
efficiency in future studies. Furthermore, a limitation of the
present study is that the mispairing between the TCRα- and β chain
was not detected, as the accuracy of the expression and matching of
TCRα- and TCRβ-chains important for the functions of TCR (37). Studies by Mizote et al
(37) and Rapoport et al
(10) additionally detected the
structure of TCR using Streptamer or Dextramer reagents. The
percentage of variable region of β-chains positive cells and
mispairing will be examined in future studies.
The activation of T cell antigens triggers the
intracellular pathways that result in cytotoxic activity and/or the
production of immunomodulatory cytokines (including IFN-γ and
TNF-α) (38–40). In the present study, as the CD8+ T
cells were transduced with lentiviral vectors expressing the
HLA-A2-restricted and PLAC1-specific TCR, the effector T cell
functions were measured via cytokine release and cell lysis in
vitro subsequent to co-culturing with PLAC1+/HLA-A2+,
PLAC1+/HLA-A2- and PLAC1 gene silenced/HLA-A2+ target cells. The
TCR-transduced CD8+ T cells, when co-cultured with a human
non-metastatic breast cancer cell line (MCF-7; PLAC1+ and HLA-A2+)
and a TNBC cell line (MDAMB-231; PLAC1+ and HLA-A2+) produced IFN-γ
and TNF-α, suggesting TCR stimulation. Although the E:T ratio (5:1)
was low, the enhanced ability to lyse tumor cells of TCR T cells
compared to NC-transduced T-cells were detected, demonstrating the
notable properties of these engineered T cells. Furthermore, a
limitation of the present study was that TCR affinity was not
examined. The present study only evaluated the functions of PLAC1
TCR-transduced CD8+ T cells and original T cells by detecting the
specific release of IFN-γ and TNF-α when the TCR-engineered CD8+ T
cells were co-cultured with HLA-A2+/PLAC1+ MCF-7 and MDAMB-231 cell
lines (Fig. 3A and B). Experiments
associated with TCR affinity will be performed in future studies.
Upon TCR engagement, multiple signaling mechanisms, including MAPK
and NF-κB modulated by the PI3K/AKT signaling pathway, are
activated during T cell stimulation that result in gene induction
and cell cycle progression (26–29). In
the present study, whether the PI3K pathway may result in a
responsiveness in T cells towards the engineered TCR stimulation
was preliminarily detected. It was observed that the expression of
PI3Kγ, the phosphorylation of AKT and ERK1/2, and the expression of
NF-κB in TCR-transduced CD8+ T cells were enhanced compared with
the NC-transduced CD8+ T cells. Therefore, these results confirmed
that the PI3K pathway may effectuate a response in T cells towards
the engineered TCR stimulation; the underlying mechanism of TCR
activating the PI3K pathway post-transfection into T cell will be
illustrated in future studies.
To examine the in vivo efficacy of
TCR-transgenic T cells, xenograft mouse assays were performed.
Following transplantation, the result demonstrated that
PLAC1-TCR-transduced CD8+ T cells significantly delayed the tumor
progression in mice bearing breast cancer compared with normal
saline or N-transduced mice (P<0.05). These experiments will be
repeated with a larger number of animals in future studies. As T
cells may infiltrate the melanoma xenograft tumor masses in mice
(41) and the IFN-γ and TNF-α
production by TCR-T cells were enhanced (Fig. 3A and B), it was hypothesized that
PLAC1-TCR T cells may hone in on the tumor site, kill the tumor
cells and modulate the microenvironment by producing IFN-γ and
TNF-α. Furthermore, the results of the present study revealed that
TCR T cells may substantially inhibit the growth of small and
medium tumor types, while shortening the anti-tumor effect on
larger tumor types, thereby indicating that these PLAC1-TCR T cells
may exert a superior anti-tumor effect on early breast cancer
compared with that on malignant breast cancer in clinical
therapy.
To conclude, a novel HLA-A*0201-restricted and
PLAC1-specific TCR was identified. The present study demonstrated
that PLAC1 was a putative target in breast cancer treatment, and
the PLAC1-specific TCR-engineered T cells is a robust and novel
strategy for the treatment of PLAC1-positive breast cancer.
Acknowledgements
The present study was supported by the Special Funds
of Shenzhen For The Development of Strategic New Industries (grant
no. CXZZ20150430152511042).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang X, Tang H and Chen T: Epidemiology
of gynecologic cancers in China. J Gynecol Oncol. 29:e72018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistic, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mondal D and Sharma DN: External beam
radiation techniques for breast cancer in the new millennium: New
challenging perspectives. J Egypt Natl Canc Inst. 28:211–218. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu LY, Tang J, Zhang CM, Zeng WJ, Yan H,
Li MP and Chen XP: New immunotherapy strategies in breast cancer.
Int J Environ Res Public Health. 14:E682017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bernal-Estévez D, Sanchez R, Tejada RE and
Parra-López C: Chemotherapy and radiation therapy elicits tumor
specific T cell responses in a breast cancer patient. BMC Cancer.
16:5912016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakai K, Hung MC and Yamaguchi H: A
perspective on anti-EGFR therapies targeting triple-negative breast
cancer. Am J Cancer Res. 6:1609–1623. 2016.PubMed/NCBI
|
|
8
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Migali C, Milano M, Trapani D,
Criscitiello C, Esposito A, Locatelli M, Minchella I and Curigliano
G: Strategies to modulate the immune system in breast cancer:
Checkpoint inhibitors and beyond. Ther Adv Med Oncol. 8:360–374.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rapoport AP, Stadtmauer EA, Binder-Scholl
GK, Goloubeva O, Vogl DT, Lacey SF, Badros AZ, Garfall A, Weiss B,
Finklestein J, et al: NY-ESO-1-specific TCR-engineered T cells
mediate sustained antigen-specific antitumor effects in myeloma.
Nat Med. 21:914–921. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kunert A, Straetemans T, Govers C, Lamers
C, Mathijssen R, Sleijfer S and Debets R: TCR-Engineered T cells
meet new challenges to treat solid tumors: Choice of antigen, t
cell fitness and sensitization of tumor milieu. Front Immunol.
4:3632013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chinnasamy N, Wargo JA, Yu Z, Rao M,
Frankel TL, Riley JP, Hong JJ, Parkhurst MR, Feldman SA, Schrump
DS, et al: A TCR targeting the HLA-A*0201-restricted epitope of
MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen
superfamily in several types of cancer. J Immunol. 186:685–696.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chodon T, Comin-Anduix B, Chmielowski B,
Koya RC, Wu Z, Auerbach M, Ng C, Avramis E, Seja E, Villanueva A,
et al: Adoptive transfer of MART-1 T cell receptor transgenic
lymphocytes and dendritic cell vaccination in patients with
metastatic melanoma. Clin Cancer Res. 20:2457–2465. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koslowski M, Tureci O, Biesterfeld S,
Seitz G, Huber C and Sahin U: Selective activation of
trophoblast-specific PLAC1 in breast cancer by
CCAAT/enhancer-binding protein beta (C/EBPbeta) isoform 2. J Biol
Chem. 284:28607–28615. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Old LJ: Cancer is a somatic cell
pregnancy. Cancer Immun. 7:192007.PubMed/NCBI
|
|
16
|
Koslowski M, Sahin U, Mitnacht-Kraus R,
Seitz G, Huber C and Tureci O: A placenta-specific gene ectopically
activated in many human cancers is essentially involved in
malignant cell processes. Cancer Res. 67:9528–9534. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leisegang M, Engels B, Meyerhuber P,
Kieback E, Sommermeyer D, Xue SA, Reuss S, Stauss H and Uckert W:
Enhanced functionality of T cell receptor-redirected T cells is
defined by the transgene cassette. J Mol Med (Berl). 86:573–583.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sommermeyer D, Conrad H, Krönig H, Gelfort
H, Bernhard H and Uckert W: NY-ESO-1 antigen-reactive T cell
receptors exhibit diverse therapeutic capability. Int J Cancer.
132:1360–1367. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rosati SF, Parkhurst MR, Hong Y, Zheng Z,
Feldman SA, Rao M, Abate-Daga D, Beard RE, Xu H, Black MA, et al: A
Novel murine T cell receptor targeting NY-ESO-1. J Immunother.
37:135–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SY and Sin JI: MC32 tumor cells
acquire Ag-specific CTL resistance through the loss of CEA in a
colon cancer model. Hum Vaccin Immunother. 11:2012–2020. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Q, Liu G, Shao D, Wang J, Yuan H, Chen
T, Zhai R, Ni W and Tai G: Mucin1 mediates autocrine transforming
growth factor beta signaling through activating the c-Jun
N-terminal kinase/activator protein 1 pathway in human
hepatocellular carcinoma cells. Int J Biochem Cell Biol.
59:116–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang F, Li Q, Ni W, Fang F, Sun X, Xie F,
Wang J, Wang F, Gao S and Tai G: Expression of human full-length
MUC1 inhibits the proliferation and migration of a B16 mouse
melanoma cell line. Oncol Rep. 30:260–268. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Q, Wang F, Liu G, Yuan H, Chen T, Wang
J, Xie F, Zhai R, Wang F, Guo Y, et al: Impact of Mucin1 knockdown
on the phenotypic characteristics of the human hepatocellular
carcinoma cell line SMMC-7721. Oncol Rep. 31:2811–2819. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yun BL, Cho N, Li M, Jang MH, Park SY,
Kang HC, Kim B, Song IC and Moon WK: Intratumoral heterogeneity of
breast cancer xenograft models: Texture analysis of
diffusion-weighted MR imaging. Korean J Radiol. 15:591–604. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ladygina N, Gottipati S, Ngo K, Castro G,
Ma JY, Banie H, Rao TS and Fung-Leung WP: PI3Kγ kinase activity is
required for optimal T cell activation and differentiation. Eur J
Immunol. 43:3183–3196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huse M: The T cell-receptor signaling
network. J Cell Sci. 122:1269–1273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Prinz PU, Mendler AN, Masouris I, Durner
L, Oberneder R and Noessner E: High DGK-α and disabled MAPK
pathways cause dysfunction of human tumor-infiltrating CD8+ T cells
that is reversible by pharmacologic intervention. J Immunol.
188:5990–6000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Robertson LK, Mireau LR and Ostergaard HL:
A role for phosphatidylinositol 3-kinase in TCR-stimulated ERK
activation leading to paxillin phosphorylation and CTL
degranulation. J Immunol. 175:8138–8145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Radoja S, Frey AB and Vukmanovic S: T cell
receptor signaling events triggering granule exocytosis. Crit Rev
Immunol. 26:265–290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Adamina M: When gene therapy meets
adoptive cell therapy: Better days ahead for cancer immunotherapy?
Expert Rev Vaccines. 9:359–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stevenson BJ, Iseli C, Panji S, Zahn-Zabal
M, Hide W, Old LJ, Simpson AJ and Jongeneel V: Rapid evolution of
cancer/testis genes on the X chromosome. BMC Genomics. 8:1292007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cocchia M, Huber R, Pantano S, Chen EY, Ma
P, Forabosco A, Ko MS and Schlessinger D: PLAC1, an Xq26 gene with
placenta-specific expression. Genomics. 68:305–312. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gjerstorff MF, Andersen MH and Ditzel HJ:
Oncogenic cancer/testis antigens: Prime candidates for
immunotherapy. Oncotarget. 6:15772–15787. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu W, Zhai M, Wu Z, Qi Y, Wu Y, Dai C,
Sun M, Li L and Gao Y: Identification of a novel HLA-A2-restricted
cytotoxic T lymphocyte epitope from cancer-testis antigen PLAC1 in
breast cancer. Amino Acids. 42:2257–2265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
De FP, Luke GA, Hughes LE, Gani D, Halpin
C and Ryan MD: E unum pluribus: Multiple proteins from a
self-processing polyprotein. Trends Biotechnol. 24:68–75. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang X, Liu X, Tao G, Qin M, Yin G, Suo J
and Suo X: ‘Self cleaving’ 2A peptide from porcine teschovirus 1
mediates cleavage of dual fluorescent proteins in transgenic
Eimeria tenella. Vet Res. 47:682016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mizote Y, Uenaka A, Isobe M, Wada H,
Kakimi K, Saika T, Kita S, Koide Y, Oka M and Nakayama E:
Production of NY-ESO-1 peptide/DRB1*08:03 tetramers and ex vivo
detection of CD4 T cell responses in vaccinated cancer patients.
Vaccine. 32:957–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Malyguine AM, Strobl S, Dunham K, Shurin
MR and Sayers TJ: ELISPOT assay for monitoring cytotoxic T
lymphocytes (CTL) activity in cancer vaccine clinical trials.
Cells. 1:111–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Colagiovanni A, Di Renzo L, Sarlo F,
Schiavino D and De Lorenzo A: Role of TNF-alpha polymorphism in
patients with nickel allergy: A marker of susceptibility to contact
polysensitization. Eur Rev Med Pharmacol Sci. 20:2663–2666.
2016.PubMed/NCBI
|
|
40
|
Wang S, Campos J, Gallotta M, Gong M,
Crain C, Naik E, Coffman RL and Guiducci C: Intratumoral injection
of a CpG oligonucleotide reverts resistance to PD-1 blockade by
expanding multifunctional CD8+ T cells. Proc Natl Acad Sci USA.
113:pp. E7240–e7249. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gammaitoni L, Giraudo L, Leuci V,
Todorovic M, Mesiano G, Picciotto F, Pisacane A, Zaccagna A, Volpe
MG, Gallo S, et al: Effective activity of cytokine-induced killer
cells against autologous metastatic melanoma including cells with
stemness features. Clin Cancer Res. 19:4347–4358. 2013. View Article : Google Scholar : PubMed/NCBI
|