Introduction
The process of liver regeneration is, on the
molecular level, an extremely complicated process that requires a
perfect interplay of cell-cell signaling and gene expression
continuity (1). Liver regeneration
has traditionally been divided into three phases: Initiation,
proliferation and termination (2).
The duration of these phases depends on the organism under
examination, including human, pig or rat, and the type of surgical
intervention, including partial hepatectomy, intoxication by drugs,
or hereditary predispositions (3).
The organisms most frequently used to investigate liver
regeneration are rats and mice, which are relatively
well-investigated model organisms. However, pigs (Sus
scrofa) are anatomically and physiologically closer to humans
than rodents, and therefore are attractive subjects for biomedical
research, despite the higher cost of maintenance (4). Budai et al (5) outlines a detailed comparison of existing
associating liver partition and portal vein ligation for staged
hepatectomy (ALPPS) animal models and their advantages.
Currently, it is known that application of
adipose-derived stem cells may positively modulate tissue
regeneration processes (6–8). There are a number of clinical studies
that are aimed at verifying the safety and effectiveness of this
form of treatment; however, the molecular mechanisms of action
remain largely unclear (9,10). It is likely to be the primarily
paracrine mechanism of action that produces growth factors and
cytokines, which positively modulate regenerative processes, such
as improved angiogenesis, and limit inflammatory processes
(9,10).
The present study analyzed the effect of the
application of stromal vascular fat tissue stem cells on liver
regeneration during the first stage of ALPPS procedure. ALPPS is a
relatively recent modification of the two-staged hepatectomy, first
described in 2010 (11). ALPPS
approach allows for surgery on severe liver tumor burden in two
associated steps. In the first step, tumor loci are removed from
less affected liver lobe, the two liver lobes are split by
parenchyma transection and the more metastatic region of the liver
is deportalized. Deportalization of one liver lobe stimulates the
second liver lobe to undergo hypertrophic regeneration (the future
liver remnant). The patient is then permitted 1 or 2 weeks to
recover. The second step removes the deportalized region of the
liver, while the hypertrophic future liver remnant is fully
functional (12). This approach
significantly increases possibility of curative treatment of severe
liver tumor diseases (13).
It is assumed that the application of stem cells
obtained from stromal vascular fat tissue accelerates the
regenerative process by allowing for improved angiogenesis and
modulation of inflammation, as has been previously observed in
animal-model studies (14,15); however, to the best of our knowledge,
this has not been demonstrated in direct connection with ALPPS
approach and Sus scrofa model organism. The aim of the
present study was to identify candidate genes that may be used as
screening markers for monitoring the process of liver regeneration
following the first stage of ALPPS.
Materials and methods
Animals
A total of six juvenile domestic swine (Polish white
pigs; 6 months; seven females and one castrated male; weight 30–50
kg; Instytut Zootechniki, Grodziec Ślaski, Poland) were included in
the present study. The pigs were housed in separated boxes at room
temperature (15–20°C), air humidity of 50–60%, normal atmosphere,
12 h light/dark cycles and access to food and water ad
libidum. Procedures were performed in the Center for
Cardiovascular Research and Development, American Heart of Poland
S.A. (Ustroń, Poland) between September and October 2014. Approval
from the Bioethical Committee from the Center for Cardiovascular
Research and Development, American Heart of Poland S.A. (Ustroń,
Poland) was obtained. Animals were assigned to two groups: n=3
without stem cell application (pig nos. 1–3) and n=3 with stem cell
application (pig nos. 4–6), based on their identification numbers.
All animals received an acclimation period of 3 days prior to any
procedures, during which and no premedication was administered.
Animals were anesthetized following an overnight fast (water was
not withheld) based on their body weight using ketamine (20 mg/kg),
xylazine (2 mg/kg) and atropine (1 mg/pig). Propofol was also
administered as a bolus (1 mg/kg) prior to intubation to induce
muscle relaxation. General anesthesia was maintained during
procedures with a constant infusion drip of propofol. Fentanyl (100
µg/pig) was administered at the beginning of each procedure to
potentiate anesthesia and as an analgesic, and all animals received
mechanical ventilation support throughout the procedures. At
pre-determined time-points the animals were euthanized with
pentobarbital solution (140 mg/kg), and livers were harvested for
histological and whole transcriptome analysis. Pigs were necropsied
and examined for abnormal findings, and were labeled with the
animal identification number, protocol number and date of
collection.
ALPPS first phase
Pigs were anaesthetized as aforementioned.
Laparotomy and investigation of the abdominal organs was performed,
and revision of the liver was conducted, with the preparation of
the liver hilus, identification of the portal vein and its
branching, identification of the bile duct and hepatic arteries.
Confirmation of the injection site was performed by venography
using contrast medium (iopromidum) and C-arm fluoroscopy. The entry
of hepatic veins into vena cava inferior was identified. The flow
of portal blood into four lobes of the liver was interrupted; only
the inflow of portal blood into the one selected hepatic lobe
(future liver remnant) was preserved. This procedure was followed
by splitting of liver between the lobe with preserved perfusion
through the portal vein and other lobes, to which the inflow of
portal blood was closed. Samples of liver tissue were harvested
from the future liver remnant lobes and were stored snap-frozen
using liquid nitrogen (−196°C) in a tissue bank. Furthermore, 15 ml
of the human adipose stem cells-stromal vascular fraction
concentrate (Cytori Therapeutics, Inc., San Diego, CA, USA) was
administered intra-arterially to the group of animals with planned
administration of stem cells via the hepatic artery during the
surgery procedure. For more information about characteristics of
this concentrate see a previous study by Lin et al (16). The animals in the group that did not
undergo stem cell application were administered 15 ml of saline via
an identical route of administration. Hydrocortisone was applied
intravenously prior to the administration of stem cells to prevent
an autoimmune reaction (rejection). The animals were monitored
postoperatively by measuring body temperature (per rectum)
and weight daily.
ALPPS second phase
Surgery was performed 9 days after the first stage.
Re-laparotomy and investigation of abdominal organs were performed,
together with liver revision and identification of pre-marked
structures in the hilus and entry of hepatic veins into the vena
cava inferior. In total, four liver lobes were removed with the
perfused lobe remaining in place.
Tissue sampling, RNA isolation and
whole transcriptome sequencing
All samples of liver tissue were collected into
separate 5 ml polypropylene tubes prefilled with equivalent volume
of RNA later solution and stored at −20°C. Isolation of total RNA
was performed using the QuickGene Mini 80 semiautomatic device and
appropriate RNA tissue kit SII (both from Kurabo Industries Ltd.,
Osaka, Japan). RNA concentration and integrity were determined
using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.,
Santa Clara, CA, USA). RNA-sequencing libraries preparation and
cDNA sequencing was performed by Macrogen, Inc. (Seoul, Republic of
Korea), resulting in a set of 101 nucleotide paired-end-read data
files.
Transcriptome data analysis
The quality of the raw sequencing data was assessed
using FastQC (v0.11.5) (17) and
aligned to a reference genome of Sus scrofa (Ensembl v82;
Sus scrofa 10.2) using the STAR aligner (v2.4.1b) (18). Up to five mapping reads were used for
subsequent analyses. Raw gene counts were obtained by calculating
reads mapping to exons and summarized by genes using reference gene
annotation (Ensembl v.82, Sus scrofa assembly, GTF) by
featureCounts (v1.4.6-p5) (19).
Differential gene expression was calculated using edgeR (v3.10.5)
(20). Two states (day 0 and 9)
within each experimental group of animals were compared. False
discovery rate (FDR) correction was used to correct the P-values
for multiple assessments. Genes were determined as differentially
expressed when the FDR adjusted P-value ≤0.1 and log2
fold-change (log2FC) ≥0.5. Pathway analyses were
performed in STRING (v10.5) (21,22),
Panther (23) with the aid of Kyoto
Encyclopedia of Genes and Genomes (KEGG) (24,25).
Volumetric measurements
All magnetic resonance imaging (MRI) experiments
were performed using a 1.5 T scanner (GE Healthcare, Chicago, IL,
USA), and an eight-channel phased array head coil was used. MRI
measurements were performed at baseline (day 0) and on day 9, prior
to second ALPPS stage.
Statistical analysis
The non-parametric paired Wilcoxon test was used for
statistical comparison of changes in liver volume between day 0 and
9. According to the experimental design, this comparison was
performed separately for the group that did not receive the
application of stem cells and for the group that did. Software R
was used for statistical analysis (version 3.4.1; The R Foundation
for Statistical Computing, Wien, Austria). P<0.05 was considered
to indicate a statistically significant difference. Data in the
barplots are presented as the mean ± standard deviation.
Results
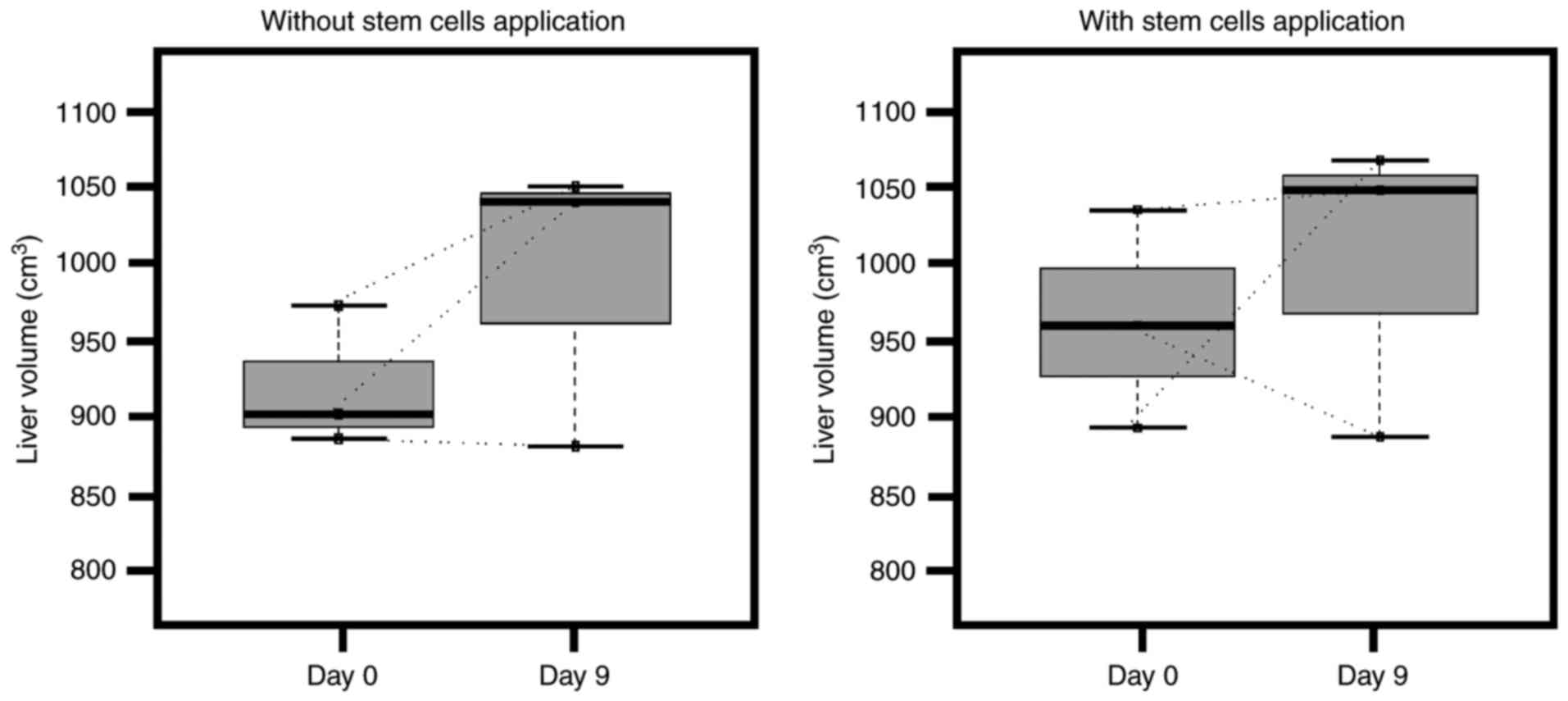
Although each step of the ALPPS procedure was
performed successfully, no significant changes in total liver
volume were observed following the first ALPPS stage (Fig. 1) (P=0.5 without application of stem
cells; P=0.75 with application of stem cells). This may be due to
the fact that only future liver remnants are expected to increase
in size over a longer period of time.
Comprehensive transcriptome analysis of samples from
future liver remnant was performed to examine for changes in the
gene expression between groups with and without application of stem
cells. We hypothesized that the application of stem cells would
accelerate liver regeneration by inhibition of undesirable
processes, such as fibrosis and inflammation.
A total of 39 significantly differentially expressed
genes were identified in the group without application of stem
cells between day 0 and 9 (Table I),
there of 37 genes were upregulated and two downregulated. In the
group with stem cell treatment there were no differentially
expressed genes between day 0 and 9. The highest significantly
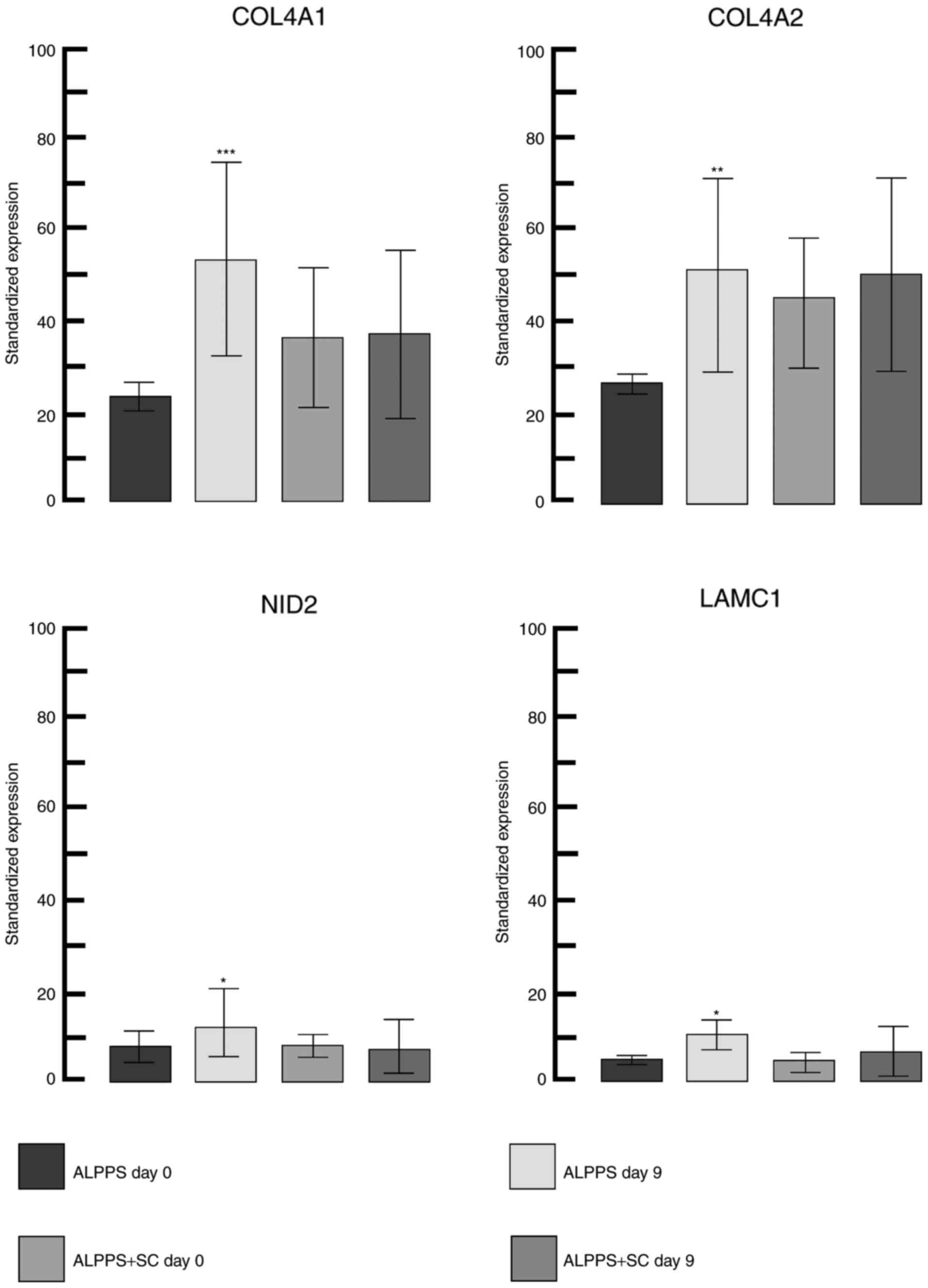
different gene expression was observed for collagen type IV α1
chain (COL4A1). COL4A1, COL4A2, laminin subunit γ1 and nidogen 2
(all of which were upregulated; Fig.
2) form major components of the basement membrane (with COL4A1
and COL4A2 constituting a functional heterotrimer with 2:1
stoichiometry) (26,27). The greatest positive change
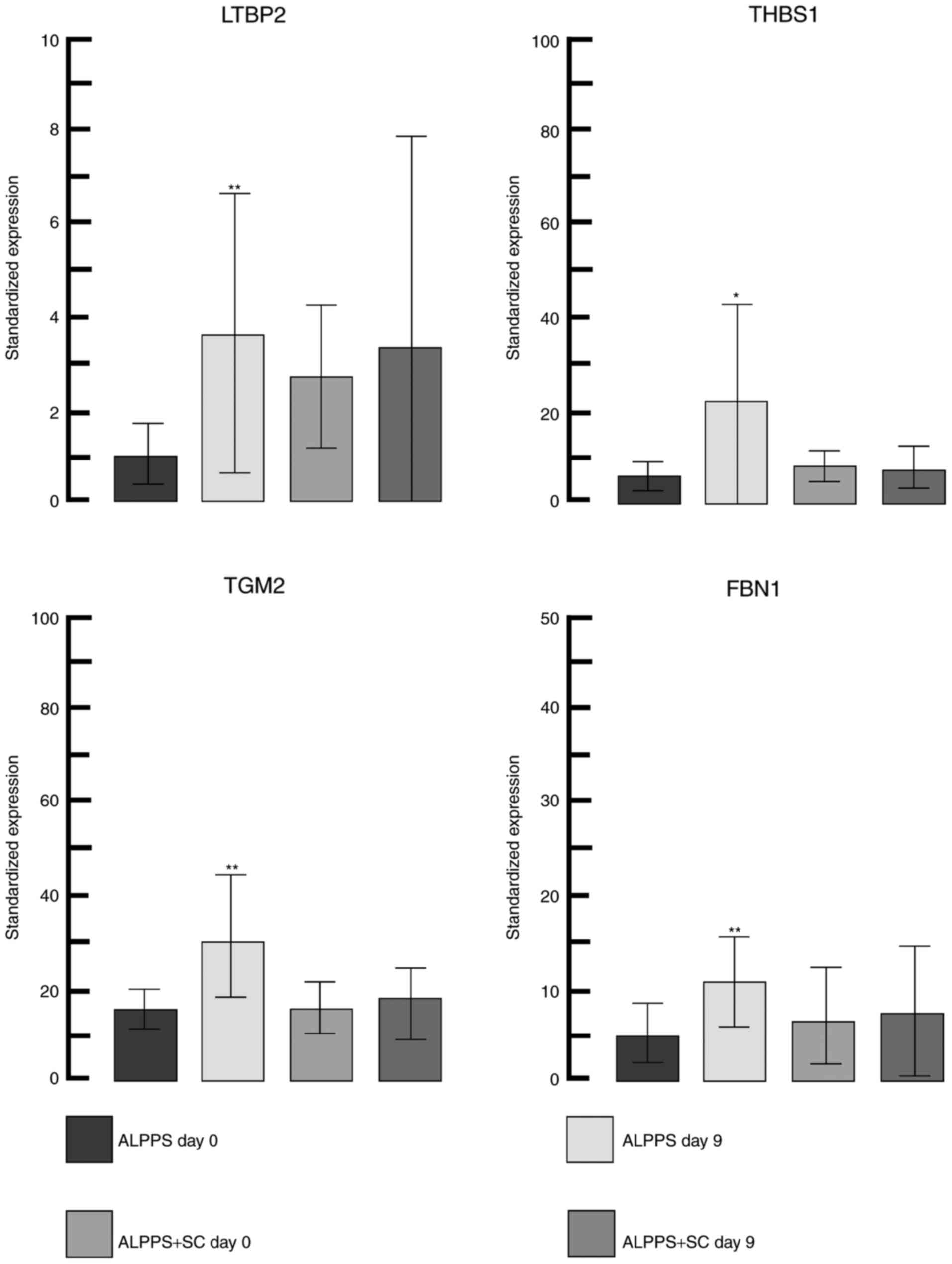
(upregulation) in the gene expression was for latent transforming
growth factor-β binding protein 2 (LTBP2) and the greatest negative
change (downregulation) was for heme binding protein 2. LTBP2
together with thrombospondin 1, transglutaminase 2 and fibrillin 1,
all of which were upregulated (detailed changes in gene expression
are depicted in Fig. 3) and serve an
important role in the transforming growth factor-β pathway in the
extracellular matrix remodeling process (28).
 | Table I.Significantly differentially
expressed genes in the group without application of stem cells
between day 0 and 9, sorted by lowest FDR value. |
Table I.
Significantly differentially
expressed genes in the group without application of stem cells
between day 0 and 9, sorted by lowest FDR value.
| Identifier | Symbol | Gene name | log2
FC | FDR |
|---|
|
ENSSSCG00000009544 | COL4A1 | Collagen, type IV,
α1 | 0.93 | 0.00 |
|
ENSSSCG00000007000 | FAT1 | FAT tumor
suppressor homolog 1 (Drosophila) | 0.78 | 0.01 |
|
ENSSSCG00000000712 | VWF | Von Willebrand
factor | 1.15 | 0.01 |
|
ENSSSCG00000023522 | TGM2 | Transglutaminase
2 | 0.75 | 0.01 |
|
ENSSSCG00000004658 | FBN1 | Fibrillin 1 | 0.94 | 0.01 |
|
ENSSSCG00000011859 | HEG1 | HEG homolog 1
(zebrafish) | 0.95 | 0.01 |
|
ENSSSCG00000001725 | GPR116 | G protein-coupled
receptor 116 | 0.69 | 0.01 |
|
ENSSSCG00000009545 | COL4A2 | Collagen, type IV,
α2 | 0.84 | 0.01 |
|
ENSSSCG00000014442 | PDGFRB | Platelet-derived
growth factor receptor, β-polypeptide | 0.82 | 0.01 |
|
ENSSSCG00000028022 | COL6A2 | Collagen, type VI,
α2 | 0.74 | 0.02 |
|
ENSSSCG00000002368 | LTBP2 | Latent transforming
growth factor-β binding protein 2 | 1.64 | 0.02 |
|
ENSSSCG00000004150 | HEBP2 | Heme binding
protein 2 | −1.09 | 0.02 |
|
ENSSSCG00000008749 | SLIT2 | Slit homolog 2
(Drosophila) | 1.28 | 0.02 |
|
ENSSSCG00000011443 | STAB1 | Stabilin 1 | 0.84 | 0.02 |
|
ENSSSCG00000005751 | COL5A1 | Collagen, type V,
α1 | 0.76 | 0.03 |
|
ENSSSCG00000009045 | HHIP | Hedgehog
interacting protein | 0.71 | 0.03 |
|
ENSSSCG00000004387 | FOXO3A | Forkhead box
O3 | 0.65 | 0.04 |
|
ENSSSCG00000001834 | MFGE8 | Milk fat
globule-EGF factor 8 protein | 0.74 | 0.04 |
|
ENSSSCG00000027969 | AHNAK | AHNAK
nucleoprotein | 0.91 | 0.04 |
|
ENSSSCG00000009320 | FLT1 | Fms-related
tyrosine kinase 1 | 0.85 | 0.04 |
|
ENSSSCG00000004091 | AKAP12 | A kinase (PRKA)
anchor protein 12 | 0.81 | 0.04 |
|
ENSSSCG00000028239 | FBXL7 | F-box and
leucine-rich repeat protein 7 | 1.10 | 0.04 |
|
ENSSSCG00000011075 | KIAA1217 | Kiaa1217 | 0.61 | 0.04 |
|
ENSSSCG00000022000 | COL1A2 | Collagen, type I,
α2 | 0.80 | 0.05 |
|
ENSSSCG00000029189 | DCHS1 | Dachsous 1
(Drosophila) | 0.91 | 0.05 |
|
ENSSSCG00000017548 | NGFR | Nerve growth factor
receptor | 0.79 | 0.05 |
|
ENSSSCG00000009111 | SYNPO2 | Synaptopodin 2 | 0.91 | 0.06 |
|
ENSSSCG00000015068 | APOA4 | Apolipoprotein
A-IV | −0.62 | 0.06 |
|
ENSSSCG00000015555 | LAMC1 | Laminin, γ1 | 0.74 | 0.07 |
|
ENSSSCG00000005030 | NID2 | Nidogen 2
(osteonidogen) | 0.68 | 0.07 |
|
ENSSSCG00000011102 | NRP1 | Neuropilin 1 | 0.55 | 0.08 |
|
ENSSSCG00000026383 | NRP2 | Neuropilin 2 | 0.78 | 0.08 |
|
ENSSSCG00000015326 | COL1A2 | Collagen, type I,
α2 | 0.78 | 0.09 |
|
ENSSSCG00000027331 | COL6A3 | Collagen, type VI,
α3 | 0.71 | 0.09 |
|
ENSSSCG00000011743 | MECOM | MDS1 and EVI1
complex locus | 1.28 | 0.09 |
|
ENSSSCG00000005494 | TNC | Tenascin C | 1.40 | 0.10 |
|
ENSSSCG00000015426 | RELN | Reelin | 0.61 | 0.10 |
|
ENSSSCG00000016035 | COL5A2 | Collagen, type V,
α2 | 0.67 | 0.10 |
|
ENSSSCG00000004789 | THBS1 | Thrombospondin
1 | 1.10 | 0.10 |
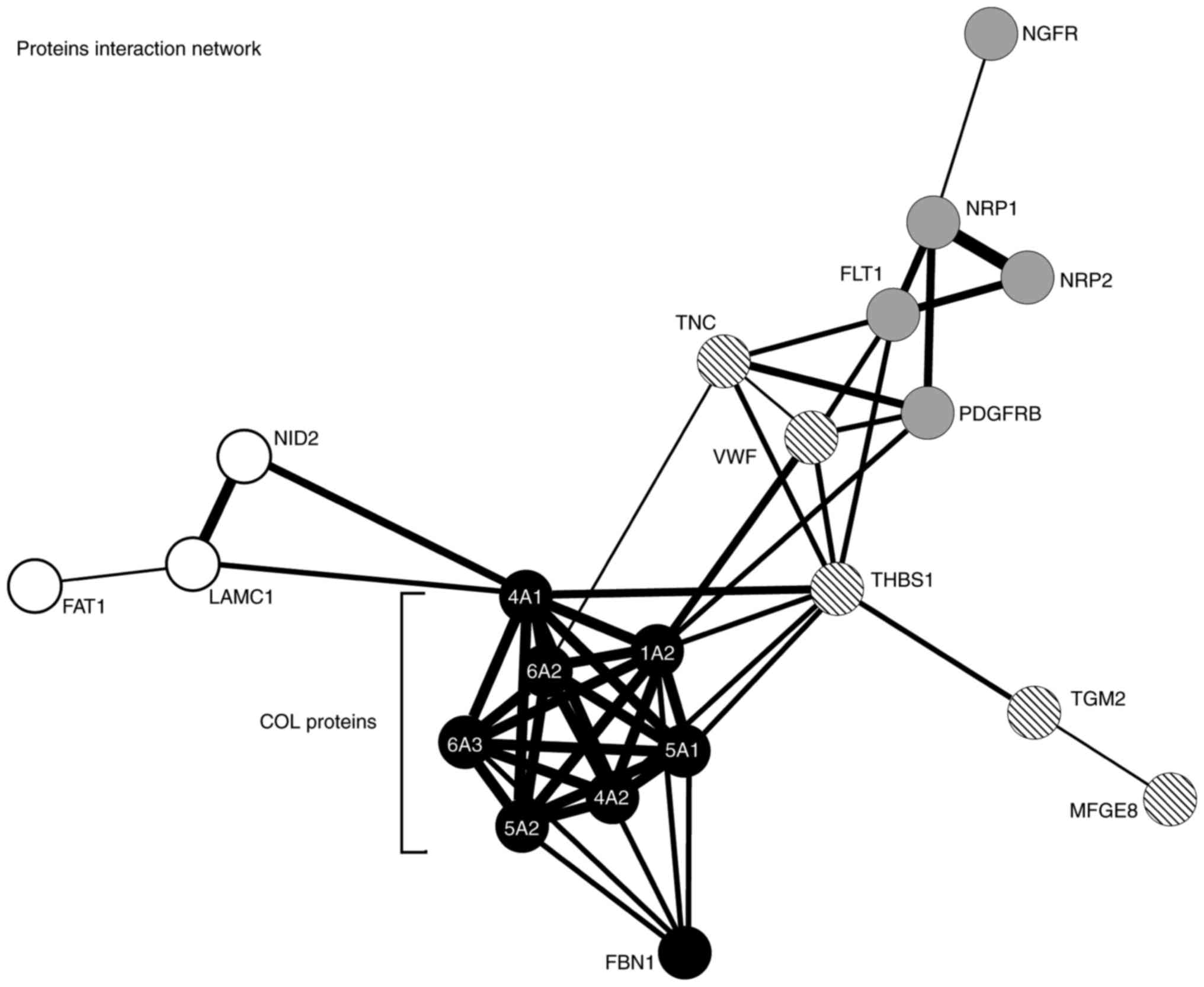
Functional classification revealed that the majority
of differentially expressed genes from the group of pigs that
received the application of stem cells are associated with their
functional interactions and localization (primarily in the
extracellular matrix and cytoplasmic membrane); Fig. 4 contains a detailed interactome, with
mainly collagens making up a strong interaction network. Analysis
of molecular functions revealed 19 significantly enrichment
categories, as ‘growth factor binding’, ‘extracellular matrix
structural constituent’ or ‘semaphorin receptor activity’ (Table II). This is in congruence with a
previous study by Rychtrmoc et al (29), where they observed changes in
expression in a number of genes involved in extracellular matrix
remodeling pathways in liver regeneration termination using
microarray and reverse transcription-quantitative polymerase chain
reaction analysis in a rat model (29). At the level of biological processes
the most relevant significantly enriched categories were
‘anatomical structure morphogenesis’, ‘circulatory system
development’ and ‘axon development’ (Table III). The most enriched KEGG pathways
were ‘PI3K-Akt signaling pathway’, ‘Focal adhesion’ and
‘ECM-receptor interaction’ (Table
IV). The phosphoinositide 3-kinase (PI3K)-RAC
serine/threonine-protein kinase (Akt) signaling pathway is likely
to drive forward liver regeneration via hepatocyte growth factor
stimulation, as observed on rat oval cells in vitro
(30). Inhibition of the PI3K-Akt
pathway disturbed liver regeneration in mice (31).
 | Table II.Molecular function enrichment in the
group without application of stem cells between day 0 and 9, sorted
by FDR value. |
Table II.
Molecular function enrichment in the
group without application of stem cells between day 0 and 9, sorted
by FDR value.
| Pathway ID | Pathway
description | Observed gene
count | FDR | Matching
proteins |
|---|
| GO.0019838 | Growth factor
binding | 8 |
5.25×10−9 | COL1A2, COL4A1,
COL5A1, FLT1, NRP1, NRP2, DGFRB, THBS1 |
| GO.0048407 | Platelet-derived
growth factor binding | 4 |
4.28×10−6 | COL1A2, COL4A1,
COL5A1, PDGFRB |
| GO.0005539 | Glycosaminoglycan
binding | 7 |
3.18×10−5 | COL5A1, LTBP2,
NRP1, NRP2, SLIT2, STAB1, THBS1 |
| GO.0005201 | Extracellular
matrix structural constituent | 5 |
6.82×10−5 | COL1A2, COL4A1,
COL4A2, COL5A1, FBN1 |
| GO.0097493 | Structural molecule
activity conferring elasticity | 3 |
6.88×10−5 | AHNAK, COL4A1,
FBN1 |
| GO.0005021 | Vascular
endothelial growth factor-activated receptor activity | 3 |
8.59×10−5 | FLT1, NRP1,
NRP2 |
| GO.0008201 | Heparin
binding | 6 |
8.59×10−5 | COL5A1, LTBP2,
NRP1, NRP2, SLIT2, THBS1 |
| GO.0005515 | Protein
binding | 21 | 0.000139 | AHNAK, AKAP12,
APOA4, COL1A2, COL4A1, COL5A1, FBN1, FLT1, FOXO3, HHIP, MECOM,
NGFR, NID2, NRP1, NRP2, PDGFRB, RELN, SLIT2, SYNPO2, THBS1,
TNC |
| GO.0005509 | Calcium ion
binding | 9 | 0.00039 | DCHS1, FAT1, FBN1,
HEG1, LTBP2, MECOM, NID2, SLIT2, THBS1 |
| GO.0043394 | Proteoglycan
binding | 3 | 0.000866 | COL5A1, SLIT2,
THBS1 |
| GO.0004714 | Transmembrane
receptor protein tyrosine kinase activity | 4 | 0.00106 | FLT1, NRP1, NRP2,
PDGFRB |
| GO.0030023 | Extracellular
matrix constituent conferring elasticity | 2 | 0.0044 | COL4A1, FBN1 |
| GO.0038085 | Vascular
endothelial growth factor binding | 2 | 0.0044 | NRP1, PDGFRB |
| GO.0046872 | Metal ion
binding | 17 | 0.0117 | APOA4, COL1A2,
COL5A1, DCHS1, FAT1, FBN1, HEG1, HHIP, LTBP2, MECOM, NID2, NRP1,
NRP2, RELN, SLIT2, TGM2, THBS1 |
| GO.0017154 | Semaphorin receptor
activity | 2 | 0.0259 | NRP1, NRP2 |
| GO.0019955 | Cytokine
binding | 3 | 0.0335 | NRP1, NRP2,
THBS1 |
| GO.0005178 | Integrin
binding | 3 | 0.0461 | COL5A1, FBN1,
THBS1 |
| GO.0005198 | Structural molecule
activity | 6 | 0.0461 | AHNAK, COL1A2,
COL4A1, COL4A2, COL5A1, FBN1 |
| GO.0030169 | Low-density
lipoprotein particle binding | 2 | 0.0476 | STAB1, THBS1 |
 | Table III.Biological process enrichment in the
group without application of stem cells between day 0 and 9, sorted
by lowest FDR value. |
Table III.
Biological process enrichment in the
group without application of stem cells between day 0 and 9, sorted
by lowest FDR value.
| Pathway ID | Pathway
description | Observed gene
count | FDR | Matching
proteins |
|---|
| GO.0009653 | Anatomical
structure morphogenesis | 21 |
7.36×10−10 | COL1A2, COL4A1,
COL4A2, COL6A2, COL6A3, DCHS1, FAT1, FBN1, FLT1, FOXO3, HEG1, HHIP,
MECOM, NGFR, NRP1, NRP2, PDGFRB, SLIT2, TGM2, THBS1, TNC |
| GO.0072358 | Cardiovascular
system development | 14 |
1.8×10−8 | COL1A2, COL4A1,
COL4A2, COL5A1, DCHS1, FBN1, FLT1, HEG1, MECOM, NRP1, NRP2, PDGFRB,
SLIT2, THBS1 |
| GO.0072359 | Circulatory system
development | 14 |
1.8×10−8 | COL1A2, COL4A1,
COL4A2, COL5A1, DCHS1, FBN1, FLT1, HEG1, MECOM, NRP1, NRP2, PDGFRB,
SLIT2, THBS1 |
| GO.0044243 | Multicellular
organismal catabolic process | 7 |
1.38×10−7 | APOA4, COL1A2,
COL4A1, COL4A2, COL5A1, COL6A2, COL6A3 |
| GO.0001568 | Blood vessel
development | 11 |
1.51×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, FLT1, HEG1, NRP1, NRP2, PDGFRB, SLIT2, THBS1 |
| GO.0001944 | Vasculature
development | 11 |
1.58×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, FLT1, HEG1, NRP1, NRP2, PDGFRB, SLIT2, THBS1 |
| GO.0006935 | Chemotaxis | 12 |
1.58×10−7 | COL4A1, COL4A2,
COL5A1, COL6A2, COL6A3, FLT1, NGFR, NRP1, NRP2, PDGFRB, RELN,
SLIT2 |
| GO.0030198 | Extracellular
matrix organization | 10 |
1.72×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FBN1, NID2, THBS1, TNC |
| GO.0061564 | Axon
development | 11 |
2.00×10−7 | COL4A1, COL4A2,
COL5A1, COL6A2, COL6A3, NGFR, NRP1, NRP2, RELN, SLIT2, TNC |
| GO.0007411 | Axon guidance | 10 |
3.15×10−7 | COL4A1, COL4A2,
COL5A1, COL6A2, COL6A3, NGFR, NRP1, NRP2, RELN, SLIT2 |
| GO.0022617 | Extracellular
matrix disassembly | 7 |
5.34×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FBN1 |
| GO.0040011 | Locomotion | 14 |
6.1×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FAT1, FLT1, NGFR, NRP1, PDGFRB,
SLIT2, THBS1 |
| GO.0048666 | Neuron
development | 12 |
1.04×10−6 | APOA4, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, MECOM, NGFR, NRP1, NRP2, SLIT2,
TNC |
| GO.0030574 | Collagen catabolic
process | 6 |
1.23×10−6 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3 |
| GO.0071363 | Cellular response
to growth factor stimulus | 11 |
1.3×10−6 | COL1A2, COL4A2,
FBN1, FLT1, FOXO3, LTBP2, MECOM, NGFR, NRP1, NRP2, PDGFRB |
| GO.0000904 | Cell morphogenesis
involved in differentiation | 11 |
1.57×10−6 | COL4A1, COL4A2,
COL5A1, COL6A2, COL6A3, HEG1, NGFR, NRP1, NRP2, RELN, SLIT2 |
| GO.0007409 | Axonogenesis | 10 |
1.57×10−6 | COL4A1, COL4A2,
COL5A1, COL6A2, COL6A3, NGFR, NRP1, NRP2, RELN, SLIT2 |
| GO.0031175 | Neuron projection
development | 11 |
1.57×10−6 | APOA4, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, NGFR, NRP1, NRP2, SLIT2, TNC |
| GO.0006928 | Movement of cell or
subcellular component | 14 |
1.58×10−6 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FAT1, FLT1, NGFR, NRP1, NRP2,
PDGFRB, SLIT2, THBS1 |
| GO.0048468 | Cell
development | 15 |
1.79×10−6 | APOA4, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FOXO3, HEG1, MECOM, NGFR, NRP1,
NRP2, PDGFRB, SLIT2, TNC |
 | Table IV.Kyoto Encyclopedia of Genes and
Genomes pathway enrichment in the group without application of stem
cells between day 0 and 9, sorted by lowest FDR value. |
Table IV.
Kyoto Encyclopedia of Genes and
Genomes pathway enrichment in the group without application of stem
cells between day 0 and 9, sorted by lowest FDR value.
| Pathway ID | Pathway
description | Observed gene
count | FDR | Matching
proteins |
|---|
| 4151 | PI3K-Akt signaling
pathway | 13 |
9.81×10−13 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FLT1, FOXO3, NGFR, PDGFRB, RELN,
HBS1, TNC |
| 4510 | Focal adhesion | 11 |
1.49×10−12 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, FLT1, PDGFRB, RELN, THBS1, TNC |
| 4512 | ECM-receptor
interaction | 9 |
1.49×10−12 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3, RELN, THBS1, TNC |
| 4974 | Protein digestion
and absorption | 6 |
3.52×10−7 | COL1A2, COL4A1,
COL4A2, COL5A1, COL6A2, COL6A3 |
| 5146 | Amoebiasis | 4 | 0.00149 | COL1A2, COL4A1,
COL4A2, COL5A1 |
| 5200 | Pathways in
cancer | 5 | 0.00832 | COL4A1, COL4A2,
HHIP, MECOM, PDGFRB |
| 4015 | Rap1 signaling
pathway | 4 | 0.0144 | FLT1, NGFR, PDGFRB,
THBS1 |
A more detailed examination of gene expression in
specific pigs between day 0 and 9 revealed certain notable facts
(only values with a log2 FC ± 3 with >4 normalized
edgeR counts were taken into account). Only certain genes in pig
nos. 4 and 6 (that received stem cell treatment) met these more
stringent criteria (Table V). In pig
no. 6, there was an extremely large increase in the expression of
the RNA component of RNase P and 7S kinase (7SK) RNA. According to
Reiner et al (32), RNase P
may serve an important role in transcription of a number of
non-coding RNAs that are transcribed by RNA polymerase III. 7SK RNA
is one of the genes transcribed by RNA polymerase III. It is
therefore likely that in pig no. 6 there was co-expression of these
two genes, which are localized on the same chromosome (RNase P RNA
component, chromosome 7:83, 579, 873–83, 580, 200 forward strand;
7SK RNA, chromosome 7:134, 400, 749–134, 401, 079 forward strand).
There were also three overexpressed genes for Metazoan signal
recognition particle RNA (also transcribed by RNA polymerase III).
Interleukin-13 receptor subunit α2 was also downregulated in pig
no. 6. However, these results for individual pigs cannot
conclusively inform on the mode of action of the applied stem
cells, but serve as a source of hypotheses for subsequent
studies.
 | Table V.Differentially expressed genes in pig
nos. 4 and 6 (that received stem cell treatment) between the day 0
and 9, sorted by highest Log2FC value. |
Table V.
Differentially expressed genes in pig
nos. 4 and 6 (that received stem cell treatment) between the day 0
and 9, sorted by highest Log2FC value.
| Identifier | Symbol | Gene name |
log2FC |
|---|
|
ENSSSCG00000019556 | 7SK | 7SK RNA | 4.11 |
|
ENSSSCG00000020439 | RNaseP_nuc | Nuclear RNase
P | 4.02 |
|
ENSSSCG00000024699 | Metazoa_SRP | Metazoan signal
recognition particle RNA | 3.48 |
|
ENSSSCG00000029839 | Metazoa_SRP | Metazoan signal
recognition particle RNA | 3.47 |
|
ENSSSCG00000029605 | Metazoa_SRP | Metazoan signal
recognition particle RNA | 3.06 |
|
ENSSSCG00000012594 | IL13RA2 | Interleukin 13
receptor subunit α2 | −3.09 |
|
ENSSSCG00000029023 | ARL5B | ADP-ribosylation
factor-like 5B | −5.43 |
|
ENSSSCG00000008595 | APOB | Apolipoprotein
B | −3.73 |
|
ENSSSCG00000002387 | GPATCH2L | G patch domain
containing 2-like | −3.59 |
|
ENSSSCG00000030247 | EPM2AIP1 | EPM2A (laforin)
interacting protein 1 | −3.57 |
|
ENSSSCG00000024674 | ABL2 | v-abl Abelson
murine leukemia viral oncogene homolog2 | −3.48 |
|
ENSSSCG00000030726 | CH242-150C11.4 | CH242-150C11.4 | −3.46 |
|
ENSSSCG00000005466 | ROD1 |
PTBP3-polypyrimidine tract binding protein
3 | −3.23 |
|
ENSSSCG00000016510 | UBN2 | Ubinuclein 2 | −3.19 |
|
ENSSSCG00000008909 | CLOCK | Clock homolog
(mouse) | −3.17 |
|
ENSSSCG00000004616 | ONECUT1 | One cut homeobox
1 | −3.12 |
|
ENSSSCG00000015284 | MDM4 | Mdm4 p53 binding
protein homolog (mouse) | −3.10 |
|
ENSSSCG00000008292 | TET3 | Tet methylcytosine
dioxygenase 3 | −3.09 |
|
ENSSSCG00000016119 | RAPH1 | Ras association
(RalGDS/AF-6) and pleckstrin homology domains 1 | −3.09 |
|
ENSSSCG00000004106 | LATS1 | LATS, large tumor
suppressor, homolog 1 (Drosophila) | −3.08 |
|
ENSSSCG00000010604 | SH3PXD2A | SH3 and PX domains
2A | −3.07 |
|
ENSSSCG00000025182 | ELK4 | ELK4, ETS-domain
protein (SRF accessory protein 1) | −3.04 |
|
ENSSSCG00000002755 | NFAT5 | Nuclear factor of
activated T-cells 5, tonicity-responsive | −3.03 |
|
ENSSSCG00000016031 | CRLR | Calcitonin
receptor-like | −3.02 |
|
ENSSSCG00000005285 | GNAQ | Guanine nucleotide
binding protein (G protein), q polypeptide | −3.02 |
Discussion
Although the liver has the ability to regenerate
itself, the application of stem cells speeds up the process; this
has been demonstrated in the present study via the presence of
fewer differentially expressed genes in the presence of stem cells,
indicating that the regeneration process is finished or is in the
late phase. Timing is crucial in the ALPPS procedure, so faster
liver regeneration between stages is highly beneficial. According
to the experimental design, no significant changes to liver
morphology were expected; as 9 days is too short a period to
observe liver fibrosis (33–37), gene expression analyses were
performed, which reliably identify expression changes in collagen
and other fibrogenic factors before they become visible via
microscopy. Previous animal studies demonstrated that microscopic
changes to liver structure following intervention were not observed
for several weeks (33–37).
Differentially expressed genes in the group of pigs
that did not receive stem cell application (between day 0 and 9)
encode proteins primarily involved in extracellular matrix
remodeling, angiogenic and neurogenic processes. Owing to the fact
that in the group that underwent the application of stem cells,
there were no differentially expressed genes between day 0 and 9,
the application of stem cells seemingly positively modulated the
regenerative processes by accelerating regeneration, and preventing
an unwanted fibrosis and inflammation processes. To provide more
precise interpretation a larger number of biological replicates and
more time-points are required (ideally on day 0, 3, 5, 7, 9, 11 and
20 to observe upward/downward trends in gene expression in broader
time scale), although in a large animal model, such an approach is
limited by financial costs.
Angiogenesis is a process that accompanies liver
regeneration process and serves an important role in restoration of
vascular networks in the place of liver damage. This process is
driven by several pro-angiogenic growth factors. A number of the
primary pro-angiogenic factors are vascular endothelial growth
factors that bind to their membrane receptors, including
Fms-related tyrosine kinase-1 (Flt-1), fetal liver kinase-1 or
Flt-4. The present study observed the increased expression of Flt-1
receptor in the group without application of stem cells between day
0 and 9, which is in congruence of former study in a rat model, in
which expression of Flt-1 was significantly increased between day 4
and 10 following 70% hepatectomy (38).
The process of axon guidance in liver regeneration
may be mediated by secreted third class semaphorins (Sema3A-G),
which bind to a membrane receptor complex whose main component is a
transmembrane glycoprotein neuropillin 1 or neuropillin 2, or a
heterodimer of the two (39). The
interaction between the semaphorins 3A and neuropillin 1 is also
notable in the angiogenic processes (40). The present study revealed increased
expression of neuropillin 1 and neuropillin 2 in the group without
application of stem cells between day 0 and 9.
The remodeling of extracellular matrix serves an
important role in the process of liver regeneration. In the
initiation stage of liver regeneration, the extracellular matrix is
broken down to allow for the proliferation of hepatocytes.
Subsequently, the extracellular matrix requires rebuilding to
ensure physical support is provided to endothelial cells.
Production of extracellular matrix is primarily provided by the
population of stellar liver cells. Restoration of the extracellular
matrix is manifested by an increased synthesis of collagen,
structural glycoproteins and proteoglycans, which occurs mainly
between day 3 and 5 following partial hepatectomy in a rat model
(41). The present study observed an
elevated expression of a number of genes associated with
extracellular matrix remodeling between day 0 and day 9 day in the
group without application of stem cells.
The application of stem cells in pig no. 6 (that
received the application of stem cells) likely decreased the
expression of interleukin 13 receptor subunit α2 (IL13RA2).
Functional IL13RA2 was overexpressed in activated hepatic stellate
cells in rat livers (42). Activated
hepatic stellate cells are associated with unwanted liver fibrosis
(43). The anti-fibrotic effect of
xenogeneic adipose mesenchymal stem cells was recently observed by
Maria et al (44), whereby a
mouse model of systemic sclerosis was used. It would be necessary
to use more biological replicates than in the present study to
determine more accurately the number of pigs in which this effect
occurred. In pig no. 6, rapid co-expression of RNAseP and 7SK
functional RNAs (>16 times higher expression) was observed. It
would be interesting to examine this observation in similar
experiments in the future. However, owing to the limited number of
biological replicates, clear interpretation cannot be performed. It
is possible, that RNAseP may serve as a major inductor of 7SK RNA
expression, as, according to Reiner et al (32), RNAseP activates the transcription of
RNA polymerase III.
The change in gene expression in pig no. 4 that
underwent application of stem cells likely demonstrates the
termination of proliferative processes, characterized by the
downregulation of Mdm4 p53 binding protein homolog (mouse) and LATS
large tumor suppressor homolog 1 (Drosophila) and thereby
stabilization of the p53 suppressor protein. This also reflects the
decreased expression of other transcription factors, including one
cut homeobox 1 (ONECUT1) or heart development protein with EGF like
domains 1. The overexpression of ONECUT1 was observed in early
stages of liver regeneration in a rat model (45). SH3 and PX domains 2A is apparently
involved in the production of free radicals as a member of the
NADPH oxidase complex complex (46).
This finding indicates that the proliferative processes in pig no.
4 were accelerated owing to the application of stem cells and
similarly, the formation of undesirable free radicals was
limited.
RNA sequencing studies aided the evaluation of gene
expression in animal models of variety human clinical conditions,
including in the study by Arvaniti et al (47), which revealed numerous previously
unknown genes associated with renal fibrosis using a mouse model
(47). Although the present study
encountered limitations including the mortality of one pig due to
source contamination and also the corruption of one sequenation
data file. These limitations resulted in decreased animal numbers;
however, the results obtained may provide insight and could be
validated by future studies that build on these findings. Certain
differentially expressed genes identified in the present study may
serve as molecular markers for monitoring the progress of liver
regeneration generally, not only during ALPPS, in human patients.
Analysis of differentially expressed genes indicates that the
application of stem cells elicited a positive effect in the
acceleration of regenerative processes; however, there is a
requirement for further experiments to be conducted with more
biological replicates and tissue sampling time-points.
Acknowledgements
The authors would like to acknowledge the CF New
Generation Sequencing Bioinformatics supported by the CIISB
research infrastructure (grant no. LM2015043, funded by MEYS CR)
for their support with obtaining scientific data presented in the
present study. The authors would also like to acknowledge access to
computing and storage facilities owned by parties and projects
contributing to the National Grid Infrastructure MetaCentrum,
provided under the program ‘Projects of Large Research,
Development, and Innovations Infrastructures’ (grant no. CESNET
LM2015042). The authors would like to thank Dr. Philip J. Coates
(Masaryk Memorial Cancer Institute, Brno, Czech Republic) for
proofreading and editing the study.
Funding
The present study was financially supported by the
Ministry of Education, Youth and Sports of the Czech Republic in
the ‘National Feasibility Program I’, (grant no. LO1208) ‘TEWEP’,
EU structural funding Operational Program Research and Development
for Innovation, (grant no. CZ.1.05/2.1.00/19.0388), OU and by the
Ministry of Health, Czech Republic, Conceptual Development of
Research Organization, University Hospital in Ostrava (grant nos.
SGS17/PrF/2016 and SGS17/PrF/2017) and by the Student Grant
Competition Faculty of Medicine, University of Ostrava (no.
SGS07/LF/2014).
Availability of data and materials
Preprocessed RNA sequencing datasets generated
during the present study are available from the corresponding
author on reasonable request.
Authors' contributions
MB, JC, MP and PP designed the study. MP, PV, PZ and
VP performed the experiments with animals. MB and JC performed
molecular biology experiments. MB, JO, VB and PP analysed the data.
MB, JO, VB and PP wrote the text.
Ethics approval and consent to
participate
Approval from the Bioethical Committee from the
Center for Cardiovascular Research and Development, American Heart
of Poland S.A. (Ustroń, Poland) was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ALPPS
|
associating liver partition and portal
vein ligation for staged hepatectomy
|
|
FDR
|
false discovery rate
|
|
log2 FC
|
log2 fold-change
|
References
|
1
|
Michalopoulos GK and DeFrances MC: Liver
regeneration. Science. 276:60–66. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zimmermann A: Regulation of liver
regeneration. Nephrol Dial Transplant. 19 Suppl 4:iv6–iv10. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palmes D and Spiegel HU: Animal models of
liver regeneration. Biomaterials. 25:1601–1611. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bendixen E, Danielsen M, Larsen K and
Bendixen C: Advances in porcine genomics and proteomics-a toolbox
for developing the pig as a model organism for molecular biomedical
research. Brief Funct Genomics. 9:208–219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Budai A, Fulop A, Hahn O, Onody P, Kovacs
T, Nemeth T, Dunay M and Szijarto A: Animal models for associating
liver partition and portal vein ligation for staged hepatectomy
(ALPPS): Achievements and future perspectives. Eur Surg Res.
58:140–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gimble JM, Katz AJ and Bunnell BA:
Adipose-derived stem cells for regenerative medicine. Circ Res.
100:1249–1260. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mizuno H, Tobita M and Uysal AC: Concise
review: Adipose-derived stem cells as a novel tool for future
regenerative medicine. Stem Cells. 30:804–810. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pak J, Lee JH, Kartolo WA and Lee SH:
Cartilage regeneration in human with adipose tissue-derived stem
cells: Current status in clinical implications. Biomed Res Int.
2016:47026742016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Premaratne GU, Ma LP, Fujita M, Lin X,
Bollano E and Fu M: Stromal vascular fraction transplantation as an
alternative therapy for ischemic heart failure: Anti-inflammatory
role. J Cardiothorac Surg. 6:432011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koh YJ, Koh BI, Kim H, Joo HJ, Jin HK,
Jeon J, Choi C, Lee DH, Chung JH, Cho CH, et al: Stromal vascular
fraction from adipose tissue forms profound vascular network
through the dynamic reassembly of blood endothelial cells.
Arterioscler Thromb Vasc Biol. 31:1141–1150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schnitzbauer A, Lang SA, Fichtner-Feigl S,
et al: In situ split with portal vein ligation induces rapid left
lateral lobe hypertrophy enabling two-staged extended right hepatic
resection. Berl Oral Presentation. 35:2010.
|
|
12
|
Schnitzbauer AA, Lang SA, Goessmann H,
Nadalin S, Baumgart J, Farkas SA, Fichtner-Feigl S, Lorf T,
Goralcyk A, Hörbelt R, et al: Right portal vein ligation combined
with in situ splitting induces rapid left lateral liver lobe
hypertrophy enabling 2-staged extended right hepatic resection in
small-for-size settings. Ann Surg. 255:405–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schadde E, Raptis DA, Schnitzbauer AA,
Ardiles V, Tschuor C, Lesurtel M, Abdalla EK, Hernandez-Alejandro
R, Jovine E, Machado M, et al: Prediction of mortality after ALPPS
stage-1: An analysis of 320 patients from the international ALPPS
registry. Ann Surg. 262:780–786. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saidi RF, Rajeshkumar B, Shariftabrizi A,
Bogdanov AA, Zheng S, Dresser K and Walter O: Human adipose-derived
mesenchymal stem cells attenuate liver ischemia-reperfusion injury
and promote liver regeneration. Surgery. 156:1225–1231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pascual-Miguelañez I, Salinas-Gomez J,
Fernandez-Luengas D, Villar-Zarra K, Clemente LV, Garcia-Arranz M
and Olmo DG: Systemic treatment of acute liver failure with adipose
derived stem cells. J Invest Surg. 28:120–126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin K, Matsubara Y, Masuda Y, Togashi K,
Ohno T, Tamura T, Toyoshima Y, Sugimachi K, Toyoda M, Marc H and
Douglas A: Characterization of adipose tissue-derived cells
isolated with the Celution system. Cytotherapy. 10:417–426. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. Anim Sci. 2010, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
|
|
18
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liao Y, Smyth GK and Shi W: featureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database Issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from gene ontology and Reactome pathways, and data
analysis tool enhancements. Nucleic Acids Res. 45:D183–D189. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40(Database
Issue): D109–D114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hahn E, Wick G, Pencev D and Timpl R:
Distribution of basement membrane proteins in normal and fibrotic
human liver: Collagen type IV, laminin, and fibronectin. Gut.
21:63–71. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pöschl E, Schlötzer-Schrehardt U,
Brachvogel B, Saito K, Ninomiya Y and Mayer U: Collagen IV is
essential for basement membrane stability but dispensable for
initiation of its assembly during early development. Development.
131:1619–1628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gressner OA, Rizk MS, Kovalenko E,
Weiskirchen R and Gressner AM: Changing the pathogenetic roadmap of
liver fibrosis? Where did it start; where will it go? J
Gastroenterol Hepatol. 23:1024–1035. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rychtrmoc D, Hubálková L, Víšková A, Libra
A, Bunček M and Červinková Z: Transcriptome temporal and functional
analysis of liver regeneration termination. Physiol Res. 61 Suppl
2:S77–S92. 2012.PubMed/NCBI
|
|
30
|
Okano J, Shiota G, Matsumoto K, Yasui S,
Kurimasa A, Hisatome I, Steinberg P and Murawaki Y: Hepatocyte
growth factor exerts a proliferative effect on oval cells through
the PI3K/AKT signaling pathway. Biochem Biophys Res Commun.
309:298–304. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jackson LN, Larson SD, Silva SR, Rychahou
PG, Chen LA, Qiu S, Rajaraman S and Evers BM: PI3K/Akt activation
is critical for early hepatic regeneration after partial
hepatectomy. Am J Physiol Gastrointest Liver Physiol.
294:G1401–G1410. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Reiner R, Ben-Asouli Y, Krilovetzky I and
Jarrous N: A role for the catalytic ribonucleoprotein RNase P in
RNA polymerase III transcription. Genes Dev. 20:1621–1635. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Veidal SS, Karsdal MA, Vassiliadis E,
Nawrocki A, Larsen MR, Nguyen QH, Hägglund P, Luo Y, Zheng Q,
Vainer B and Leeming DJ: MMP mediated degradation of type VI
collagen is highly associated with liver fibrosis-identification
and validation of a novel biochemical marker assay. PLoS One.
6:e247532011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng W, Xiao L, Ainiwaer A, Wang Y, Wu G,
Mao R, Yang Y and Bao Y: Molecular responses of radiation-induced
liver damage in rats. Mol Med Rep. 11:2592–2600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Zhang H, Zhao Z, Lv M, Jia J,
Zhang L, Tian X, Chen Y, Li B, Liu M, et al: Enhanced expression of
glucose-regulated protein 78 correlates with malondialdehyde levels
during the formation of liver cirrhosis in rats. Exp Ther Med.
10:2119–2125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chuang HM, Su HL, Li C, Lin SZ, Yen SY,
Huang MH, Ho LI, Chiou TW and Harn HJ: The role of
butylidenephthalide in targeting the microenvironment which
contributes to liver fibrosis amelioration. Front Pharmacol.
7:1122016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kongphat W, Pudgerd A and Sridurongrit S:
Hepatocyte-specific expression of constitutively active Alk5
exacerbates thioacetamide-induced liver injury in mice. Heliyon.
3:e003052017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ross MA, Sander CM, Kleeb TB, Watkins SC
and Stolz DB: Spatiotemporal expression of angiogenesis growth
factor receptors during the revascularization of regenerating rat
liver. Hepatology. 34:1135–1148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koncina E, Roth L, Gonthier B and Bagnard
D: Role of semaphorins during axon growth and guidance. Adv Exp Med
Biol. 621:50–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu L, Kitamura T, Iwabuchi K, Ichinose S,
Yanagida M, Ogawa H, Watanabe S, Maruyama T, Suyama M and Takamori
K: Interplay of neuropilin-1 and semaphorin 3A after partial
hepatectomy in rats. World J Gastroenterol. 18:5034–5041. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamamoto H, Murawaki Y and Kawasaki H:
Hepatic collagen synthesis and degradation during liver
regeneration after partial hepatectomy. Hepatology. 21:155–161.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shimamura T, Fujisawa T, Husain SR, Kioi
M, Nakajima A and Puri RK: Novel role of IL-13 in fibrosis induced
by nonalcoholic steatohepatitis and its amelioration by
IL-13R-directed cytotoxin in a rat model. J Immunol. 181:4656–4665.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mederacke I, Hsu CC, Troeger JS, Huebener
P, Mu X, Dapito DH, Pradere JP and Schwabe RF: Fate tracing reveals
hepatic stellate cells as dominant contributors to liver fibrosis
independent of its aetiology. Nat Commun. 4:28232013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maria AT, Toupet K, Maumus M, Fonteneau G,
Le Quellec A, Jorgensen C, Guilpain P and Noël D: Human adipose
mesenchymal stem cells as potent anti-fibrosis therapy for systemic
sclerosis. J Autoimmun. 70:31–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tan Y, Yoshida Y, Hughes DE and Costa RH:
Increased expression of hepatocyte nuclear factor 6 stimulates
hepatocyte proliferation during mouse liver regeneration.
Gastroenterology. 130:1283–1300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Diaz B, Shani G, Pass I, Anderson D,
Quintavalle M and Courtneidge SA: Tks5-dependent, nox-mediated
generation of reactive oxygen species is necessary for invadopodia
formation. Sci Signal. 2:ra532009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Arvaniti E, Moulos P, Vakrakou A,
Chatziantoniou C, Chadjichristos C, Kavvadas P, Charonis A and
Politis PK: Whole-transcriptome analysis of UUO mouse model of
renal fibrosis reveals new molecular players in kidney diseases.
Sci Rep. 6:262352016. View Article : Google Scholar : PubMed/NCBI
|