Introduction
Bladder cancer is the most common malignant tumor in
the urinary system, which often occurs in the bladder mucosa.
According to the statistics of the World Health Organization (WHO),
bladder cancer is one of the world's top ten common tumors
(1,2).
Bladder urothelial carcinoma is divided into non-muscular and
myometrial infiltrating urothelial carcinoma. Patients with
non-muscular infiltrating urothelial carcinoma are often treated
with transurethral resection of bladder tumor, and intravesical
instillation therapy after operation is used to prevent recurrence
(3). Metastatic bladder cancer is
mainly treated with chemotherapy, and the maximal effective rate of
chemotherapy can reach 65% (4).
Cinobufacini is made from the refined fat-soluble components in the
dried toad skin extracted using a scientific method, which contains
toad aglucones and bufalin that have an extraordinary anticancer
effect, and the anticancer activity of the ingredients is ten times
or even a hundred times stronger than those of the currently
clinically used anticancer drugs such as paclitaxel, doxorubicin
and camptothecin with no side effects, bringing benefits to many
cancer patients (5,6). Oral administration of cinobufacini has
good curative effects in the treatment of liver, stomach, lung,
colon, esophageal, pancreatic cancer and acute leukemia (6–9). However,
there is no study on the effect of cinobufacini on human bladder
cancer. Autophagy is a lysosomal-dependent degradation way in
eukaryotic cells, which produces amino acids, fatty acids and
adenosine triphosadenine for cell re-use by degrading the injured
organelle proteins under hypoxia and other stress conditions, it is
the survival mechanism of cells (including tumor cells) under the
harsh environment (10), and it is
also one of the mechanisms of drug resistance of many tumor
cells.
The purpose of the present study was to elucidate
the value of cinobufacini for bladder cancer by investigating the
effect of cinobufacini on human bladder cancer cell line T24, its
effect on T24 cell apoptosis and its specific mechanism, thus
providing a theoretical basis for the clinical treatment of bladder
cancer.
Materials and methods
Instruments and materials
Human bladder cancer cell line T24 was purchased
from the Cell Bank, Shanghai Institute of Life Sciences, Chinese
Academy of Sciences (Shanghai, China);
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide
(MTT) kits (R&D Systems, Inc., Minneapolis, MN, USA), dimethyl
sulfoxide (DMSO), cinobufacini standards and rapamycin
(Sigma-Aldrich; Merck & Co., Inc., Whitehouse Station, NJ,
USA); rabbit anti human polyclonal Bcl-2 (dilution, 1:500; cat. no.
2872), Bax (dilution, 1:500; cat. no. 2774), cleaved caspase-3
(dilution, 1:500; cat. no. 9661), p62 (dilution, 1:500; cat. no.
5114), LC3B (dilution, 1:500; cat. no. 2775) and Atg7 (dilution,
1:500; cat. no. 2631) antibodies were purchased from Cell Signaling
Technology, Inc., (Danvers, MA, USA). Rabbit anti-human polyclonal
GAPDH antibody (dilution, 1:500; cat. no. SAB2100894) was purchased
from Sigma Aldrich; Merck & Co. Inc.. Horseradish
peroxidase-labeled goat anti-rabbit secondary polyclonal antibodies
(dilution, 1:2,000; cat. no. 7074) was from Cell Signaling
Technology, Inc.. Enhanced chemiluminescence (ECL) liquid
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
pipettes (Eppendorf, Hamburg, Germany), polymerase chain reaction
(PCR) instruments (Applied Biosystems; Thermo Fisher Scientific,
Inc.), ultraviolet imaging system (Biometra GmbH, Göttingen,
Germany), electronic balance (BP121S; Sartorius AG, Göttingen,
Germany), −80°C refrigerator and low temperature centrifuge (Thermo
Fisher Scientific, Inc.); microplate reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The study was approved by the Ethics
Committee of The Second Affiliated Hospital of Fujian Medical
University (Quanzhou, China).
Cell culture methods
The medium was replaced on an ultra-clean workbench
for the human bladder cancer cell line T24 after the purchase.
Cells were further cultured using Dulbecco's modified Eagle's
medium (DMEM) (ultra-high glucose; containing 10% inactivated fetal
bovine serum and 1% streptomycin) in an incubator with 5%
CO2 and saturated humidity at 37°C until the cells grew
to about 80% of the whole culture flask, and then they were
subcultured. After that, the subcultured cells were further
cultured under the above conditions until the cells grew again to
the logarithmic growth phase, and the cells were treated. Plates
were paved for follow-up experiments.
Detection of cell viability
Human bladder cancer T24 cells in the logarithmic
growth phase were prepared into 4×106 single cell
suspension and inoculated in 96-well plates at 100 µl/well. After
the cells were cultured in an incubator with 5% CO2 and
saturated humidity at 37°C for 24 h, 0.05, 0.1 and 0.5 mg/ml
cinobufacini were used for treatment (cinobufacini was dissolved
with 0.5% DMSO and diluted with medium), respectively. A total of
six repeated wells were set in each group, and the DMSO treatment
group was regarded as the negative control group. After the drug
acted on the cells for a certain period of time, the cell viability
detection test was performed according to the procedures of the MTT
reagent instructions. After the cell culture was terminated,
MTT-labeled reagent was added into the culture plates at 10 µl/well
(the final concentration was 0.5 mg/ml). The plates were placed in
an incubator with 5% CO2 for incubation at 37°C for 4 h.
Then the dissolving solution was added into plates at 100 µl/well,
which were placed in an incubator with 5% CO2 at 37°C
for incubation overnight. Afterwards, the optical density of each
well was detected with 550 nm as the detection wavelength and 750
nm as the reference wavelength. According to the formula, cell
proliferation inhibition rate (%) = (1-average A value in the
medication well / average A value in the control well) ×100%. The
half maximal inhibitory concentration of cells (the concentration
required for inhibiting the growth of 50% of cells;
IC50) were calculated using the GraphPad Prism 6.0
(GraphPad Software, Inc. La Jolla, CA, USA) based on the effects of
different drug concentrations on the inhibition rate of cell
growth.
Detection of cell apoptosis rate by
Annex V/propidium iodide (PI) double staining
T24 cells were collected after they grew to the
logarithmic growth phase and then prepared into 8×106
single cell suspension. The cells were inoculated in 6-well plates
with 300 µl each and cultured in an incubator with 5%
CO2 and saturated humidity at 37°C for 24 h, after which
cinobufacini at different concentrations was added for treatment
for 24 h. The cells in each well were digested and washed with
pre-cooled phosphate-buffered saline (PBS) three times, and then
the above cell suspension was transferred to a flow tube. Annexin
V-fluorescein isothiocyanate and PI were added to each tube, tubes
were gently whirled, and the cells were incubated at room
temperature for 15 min. A cell flow cytometer was used to detect
the cells, and the detection in all the groups needed to be
finished within 1 h, otherwise it would affect the detection
effect.
Hoechst 33258 staining
T24 cells in the logarithmic growth phase were
digested and prepared into single cell suspension. The cells at the
density of 6×106 were inoculated in 24-well plates and
further incubated for 12 h in an incubator until they were in good
growth condition, and then cinobufacini at different concentrations
was added. At 24 h after the further culture, the upper culture
solution was carefully discarded, and pre-cooled PBS was used to
wash cells twice. After that, cells were fixed with 4%
paraformaldehyde for 10 min and washed with distilled water 3 times
with 5 min each time. After the washing, Hoechst 33258 staining
solution was added for staining away from light for 10 min. At the
end of the staining, cells were washed with distilled water twice,
after which the morphology of cells was observed under a
microscope, and the level of cell apoptosis in each group was
calculated.
Quantitative real-time PCR
Cinobufacini (0.5 mg/ml) was used to study the
effects of drugs on apoptosis and autophagy-related genes. The
control, cinobufacini (0.5 mg/ml) and rapamycin group (0.5 mg/ml
cinobufacini +50 nM rapamycin) were set, and cells treated in the
above groups were digested and washed with PBS 3 times. TRIzol
reagents (TRIzol kit; Thermo Fisher Scientific, Inc.) were added,
followed by the addition of chloroform for centrifugation. The
supernatant was obtained, and then isopropanol was added to extract
cell RNAs in each group, after which RNAs were treated with 75%
ethanol. After that, ethanol was discarded by centrifugation, and
RNAs were dried. Then 50 µl diethyl pyrocarbonate was added to
obtain RNA samples of each group. The complementary DNA chain was
obtained by reverse transcription using the reverse transcription
PCR kit and was taken as the template, after which primers, Taq
polymerase and buffer, deoxy-ribonucleoside triphosphate mixture
and double distilled water were added to perform PCR amplification
on the PCR instrument. Finally, the products were placed on a
quantitative PCR instrument to determine the messenger RNA (mRNA)
expression of target genes. The primers were synthesized by Tiangen
Biotech Co., Ltd. (Beijing, China). The primer sequences of each
gene are shown in Table I and
Bax/GAPDH, Bcl-2/GAPDH, cleaved caspase-3/GAPDH, p62/GAPDH,
LC3/GAPDH and LC3/GAPDH were used to measure the expression level
of related genes in each group.
 | Table I.Primer sequences of each gene. |
Table I.
Primer sequences of each gene.
| Genes | Sequence |
|---|
| Bax | F:
5′-CTGACGGCAACTTCAACTGG-3′ |
|
| R:
5′-GTGAGGAGGCTTGAGGAGTC-3′ |
| Bcl-2 | F:
5′-TGTGTGTGGAGAGCGTCAAC −3′ |
|
| R:
5′-GCCAGAGAAATCAAACAGAGG-3′ |
| p62 | F:
5′-AGCTGCCCTCAGCCCTCTA −3′ |
|
| R:
5′-GGCTTCTCTTCCCTCCATGTT-3′ |
| LC3 | F:
5′-AACATGAGCGAGTTGGTCAAG −3′ |
|
| R:
5′-GCTCGTAGATGTCCGCGAT-3′ |
| Cleaved | F:
5′-TCACCATTCGGTCAATCAGAGC −3′ |
| caspase-3 | R:
5′-ACCAAGGGAGAACCAGGAAACG-3′ |
| GAPDH | F:
5′-ACCCACTCCTCCACCTTTGAC −3′ |
|
| R:
5′-TCCACCACCCTGTTGCTGTAG-3′ |
Western blot analysis detection
The control, cinobufacini (0.5 mg/ml) and rapamycin
group (0.5 mg/ml cinobufacini +50 nM rapamycin) were set. Cells
treated with the above-mentioned drugs were digested, and the
lysate was added, followed by the centrifugation for 10 min at
1,200 × g at 4°C. The supernatant was the corresponding total
protein in each group. The total protein concentration was measured
using the bicinchoninic acid protein assay kit. Sodium dodecyl
sulfate polyacrylamide gel electrophoresis was conducted for
samples, which were then transferred to a polyvinylidene fluoride
membrane (IPVH00010; Millipore Corp., Billerica, MA, USA). After
that, Bax, Bcl-2, cleaved caspase-3, p62, LC3, Atg7 and GAPDH
primary antibodies (dilution, 1:1,000) were used for incubation at
4°C overnight. After the washing, secondary antibodies conjugated
with horseradish peroxidase (dilution, 1:5,000) were used for
incubation for 1 h. Then ECL mixture was added, and the tabletting
time was determined according to the fluorescence intensity of the
protein band. The fixation was conducted after color development,
bands were scanned, and ImageJ software (National Institutes of
Health, Bethesda, MD, USA) was used for gray value analysis.
Statistical analysis
The data of this study are expressed as mean ±
standard deviation. The data were analyzed by SPSS 19.0 software
(SPSS Inc., Chicago, IL, USA), and the t-test was used for
intergroup comparisons. Enumeration data were compared by
Chi-square test, and intergroup comparisons were detected using
analysis of variance (ANOVA) followed by post hoc test (least
significant difference). The analysis of homogeneity variance
showed that if the variance was homogeneous, pairwise comparisons
were conducted using Bonferroni method; if the variance was not
homogeneous, pairwise comparisons were conducted using Welch
method. Multiple comparisons were conducted using Dunnett's T3
method. P<0.05 was considered to indicate a statistically
significant difference.
Results
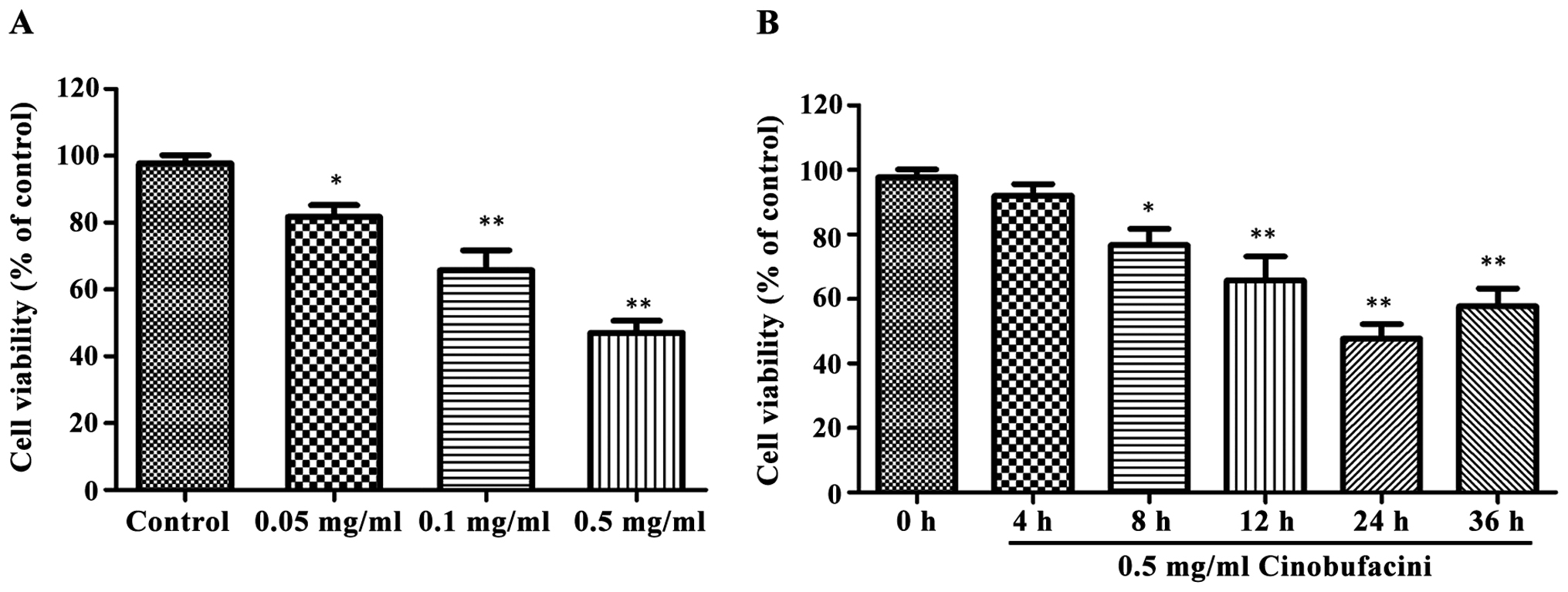
Detection of cell viability by MTT
assay
MTT assay was used to detect the effects of
cinobufacini at different concentrations on the viability of T24
cells. As shown in Fig. 1, after
cells were treated with 0.05, 0.1 and 0.5 mg/ml cinobufacini for 24
h, respectively, the cell viability was significantly decreased
(P<0.05, P<0.01). The effect of 0.5 mg/ml cinobufacini on
cell viability at different time was different and time-dependent,
and 24 h after the treatment, the cell viability was the lowest,
which was significantly lower than that of the control group
(P<0.01).
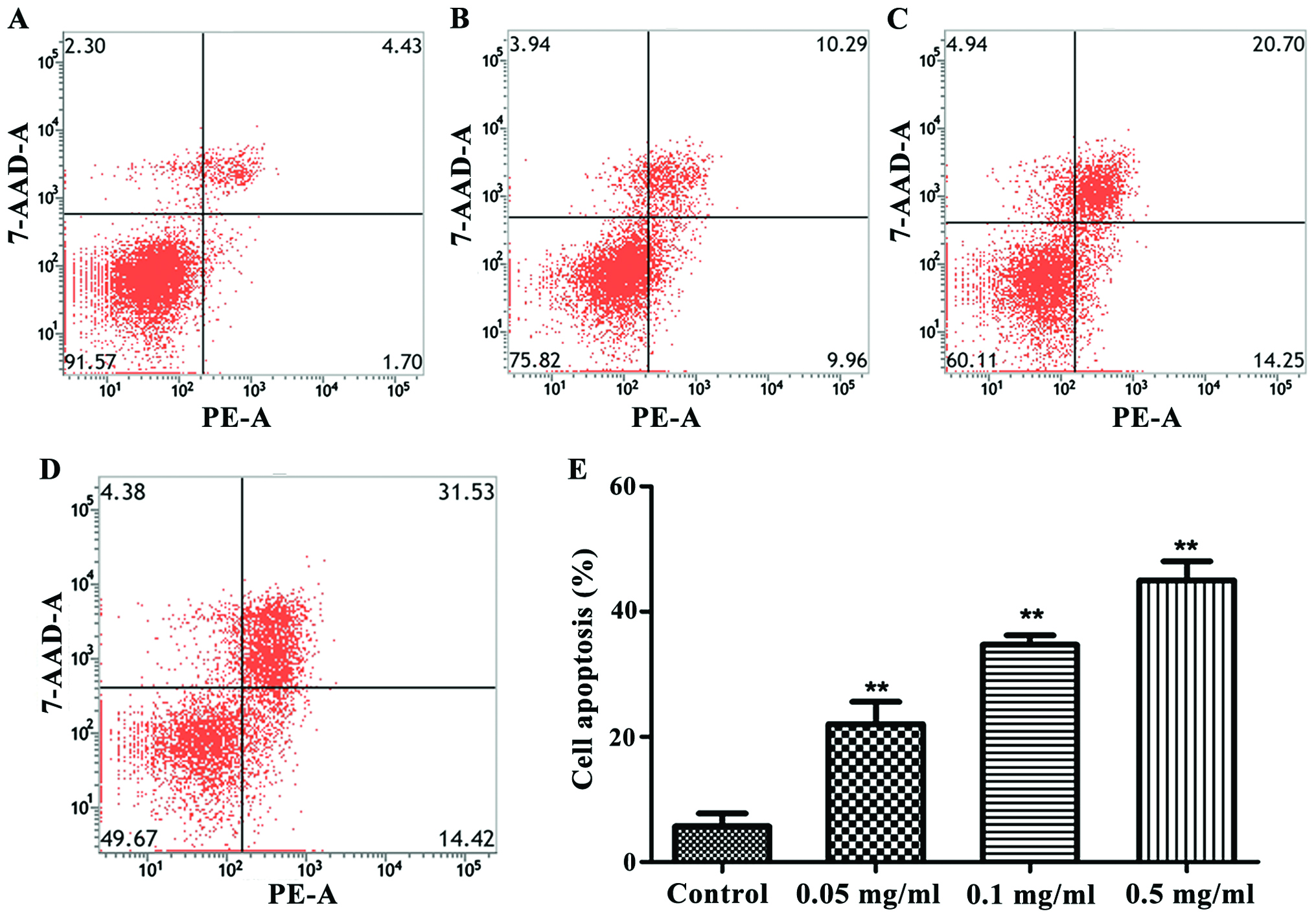
Detection of cell apoptosis by flow
cytometer
Apoptosis of cells was measured by a flow cytometer.
As shown in Fig. 2, the treatment
with 0.05, 0.1 and 0.5 mg/ml cinobufacini could increase the
apoptosis level of cells. The apoptosis level was the highest when
the concentration of cinobufacini was 0.5 mg/ml (P<0.01).
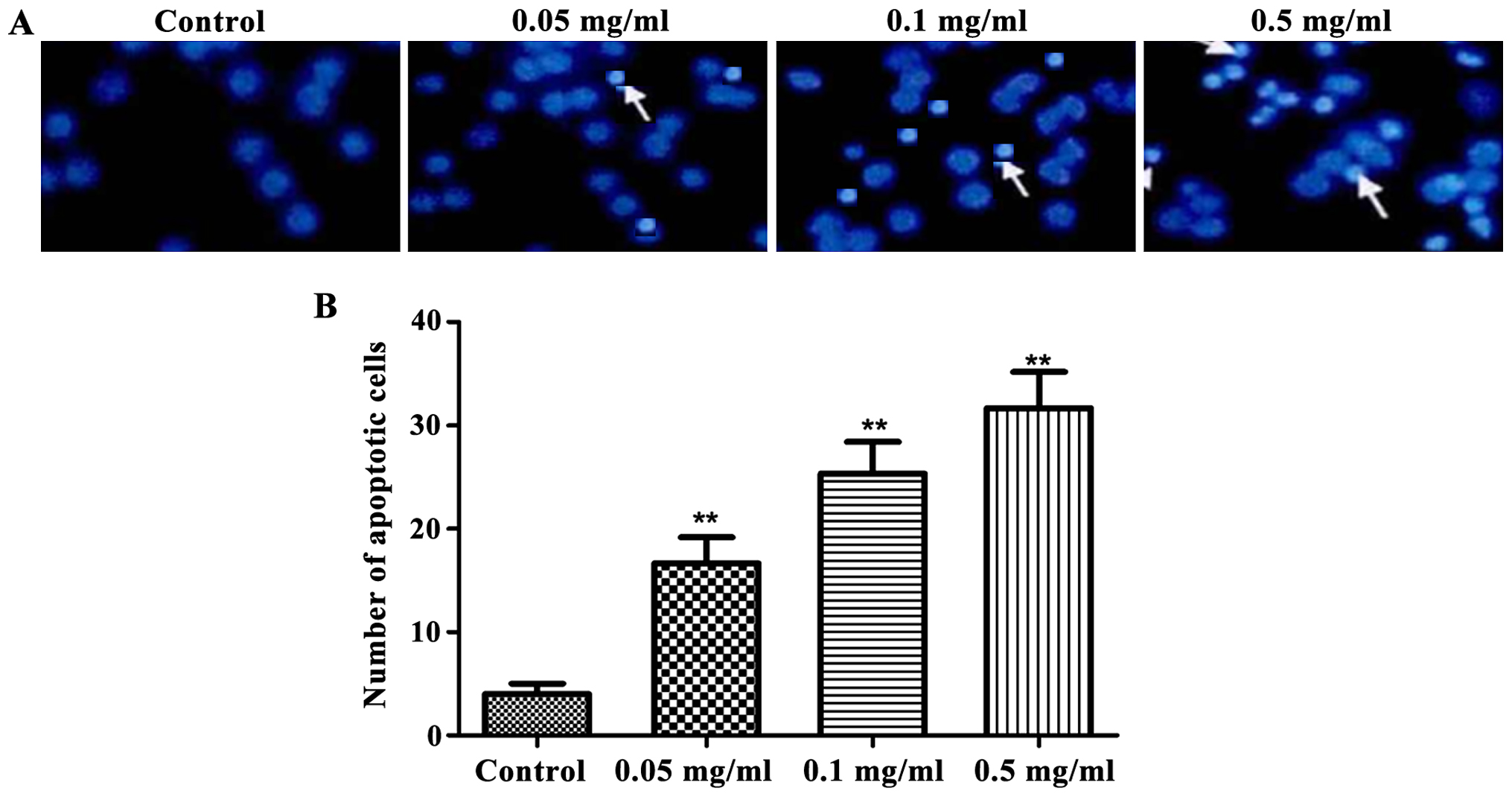
Detection of cell apoptosis by Hoechst
staining
The effects of cinobufacini on apoptosis of human
bladder cancer cell line T24 was detected by Hoechst 33258. As
shown in Fig. 3, cinobufacini
significantly increased the amount of apoptotic cells
(P<0.01).
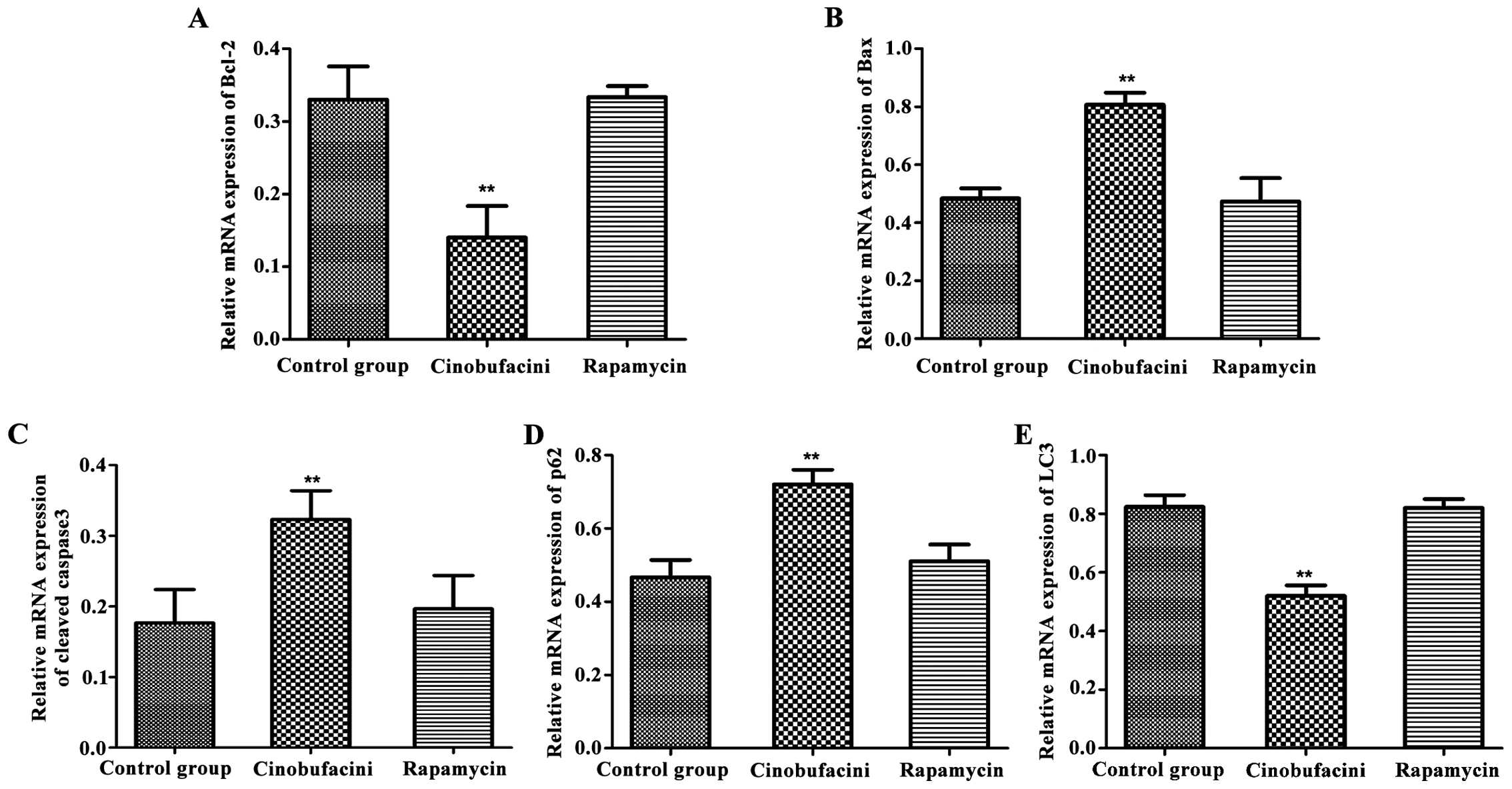
mRNA expression level
The relative expression levels of apoptosis- and
autophagy-related genes were detected by real-time quantitative PCR
instrument. As shown in Fig. 4, after
administration of cinobufacini, the expression levels of
apoptosis-related genes, Bax and cleaved caspase-3 in T24 cells
were increased (P<0.01), that of Bcl-2 was decreased
(P<0.01), that of autophagy-related protein p62 was upregulated
(P<0.01), and that of LC3 was decreased (P<0.01) compared
with those in the control group. Protein changes caused by
cinobufacini in the above conditions could be reversed after
administration of rapamycin.
Protein expression level
Western blot analysis was used to detect the
expression levels of apoptosis- and autophagy-related proteins in
each group. As shown in Fig. 5, after
administration of cinobufacini, the expression levels of
apoptosis-related proteins, Bax and cleaved caspase-3 in T24 cells
were significantly increased (P<0.01), and that of Bcl-2 was
decreased (P<0.01) compared with those in the control group.
After administration of rapamycin, the expression levels of Bcl-2,
Bax and cleaved caspase-3 were comparable to those in the control
group. After administration of cinobufacini, the expression levels
of the autophagy-related protein p62 was upregulated (P<0.01),
the ratio of LC3-II/I was decreased (P<0.01), and the expression
level of Atg7 was decreased (P<0.01), indicating that rapamycin
can reverse the above-mentioned changes induced by
cinobufacini.
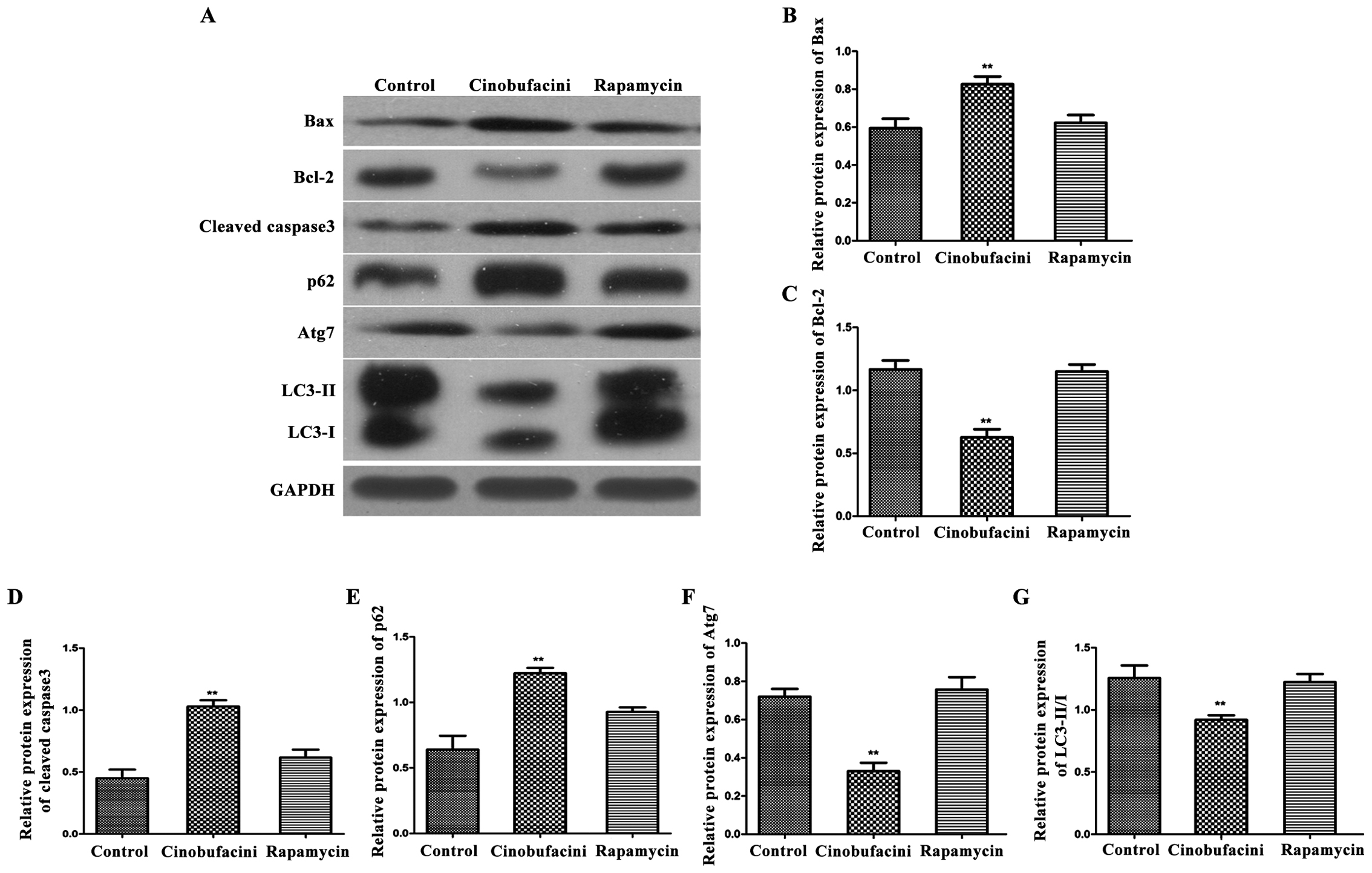 | Figure 5.Detection of the expression levels of
apoptosis- and autophagy-related proteins in each group by western
blot analysis. (A) Protein band graphs of each group; and (B-G)
statistical graph of each protein band. Compared with those in the
control group, the expression level of Bcl-2 is decreased, those of
Bax and cleaved caspase-3 are increased, that of p62 is
upregulated, that of Atg7 is decreased, and the ratio of LC3-II/I
is reduced, indicating that after administration of rapamycin,
above changes in proteins can be reversed and the protein
expression level is similar to that in the control group,
**P<0.01. Bcl-2, B-cell lymphoma-2; Atg7, autophagy-related
protein 7; LC3, light chain 3. |
Discussion
Surgery combined with chemotherapy can significantly
reduce the recurrence and deterioration of bladder cancer, but the
current clinically commonly used chemotherapy drugs such as
paclitaxel have a strong toxicity and irritation, limiting the
treatment of patients and affecting the prognosis, thus reducing
the effective rate of the treatment for bladder cancer (11,12).
Cinobufacini, as a traditional Chinese medicine preparation made
from the toad skin, has been found to have significant heat
clearing, detoxification, stasis and disintegrating mass effects,
and it has less toxicity and irritation to patients (13). At present, a large number of research
data show that cinobufacini significantly inhibits the
proliferation of a variety of tumor cells and induces apoptosis
(14). Ye et al (15) found that cinobufacini can block the
cycle of leukemia cell lines, thus inducing the expression of
apoptotic proteins and leading to cell apoptosis so as to inhibit
cell proliferation. Wu et al (16) found that cinobufacini promotes
apoptosis of liver cancer cells to a certain degree, but the
mechanism still needs to be further studied. In the present study,
it was found that cinobufacini could effectively inhibit the
proliferation of bladder cancer cells and was dose- and
time-dependent to some extent. Results of the flow cytometer and
Hoechst staining revealed that cinobufacini could significantly
increase the apoptosis level of bladder cancer cells. The above
results suggested that the effect of cinobufacini on the
proliferation of bladder cancer cells may be achieved by increasing
apoptosis. Quantitative real-time PCR and western blot analysis
were used to detect the expression levels of apoptosis- and
autophagy-related proteins, respectively, which revealed that after
the treatment with cinobufacini, the expression level of Bcl-2 was
significantly decreased, while those of Bax and cleaved caspase-3
were significantly increased, indicating cinobufacini can induce
the occurrence of autophagy, that is, this can further confirm that
the antitumor effect of cinobufacini is achieved by promoting
apoptosis. The above results are consistent with the mechanism of
antitumor effect of cinobufacini in pancreatic cancer found by Yin
et al (17). The increased
Bcl-2 expression level can inhibit apoptosis of cells, while the
increased Bax expression level can promote apoptosis, which is
achieved by inhibiting the activity of Bcl-2 (18).
Autophagy plays an important role in maintaining
cell homeostasis, removing excess or necrotic organelles and so on.
In previous years, it was found that the activation or inhibition
of autophagy has important significance for the survival of tumor
cells (19). In the present study, it
was found that after the treatment with 0.5 mg/ml cinobufacini, the
expression level of autophagy-related gene p62 was increased and
that of Atg7 was decreased in the bladder cancer cell line T24.
Western blot analysis showed that the expression level of
autophagy-related protein p62 was increased, the ratio of LC3-II/I
was decreased, and the expression level of Atg7 was decreased. The
above results suggested that cinobufacini can effectively reduce
the level of autophagy in bladder cancer cells and reduce the
occurrence of autophagy. After administration of rapamycin, the
levels of the above autophagy- and apoptosis-related proteins were
reversed, which were comparable to those of the control group.
Apoptosis of bladder cancer cells induced by cinobufacini might be
caused by the autophagy pathway. The inhibition of autophagy by
cinobufacini could significantly reduce the expression of Bax,
which led to the increased expression of Bcl-2, thus increasing the
death of tumor cells.
In conclusion, cinobufacini can effectively reduce
the survival rate and promote apoptosis of bladder cancer cells.
Its mechanism may be to regulate the occurrence of apoptosis
through the inhibition of autophagy; cinobufacini has the potential
to be developed into a drug for the treatment of bladder cancer,
but its other molecular mechanisms in the treatment of bladder
cancer still need to be further studied.
Acknowledgements
Not applicable.
Funding
The present study was supported by Youth Research
Project, Health and Family Planning Commission (Fujian, China),
2015 (Grant no. 2015-1-59) and Science and Technology Planning
Project, Quanzhou (Fujian, China) 2016 (Grant no. 2016Z044). YL
received funding support.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Author's contributions
DC designed the study and wrote the manuscript. JC
contributed to cell culture. YG made substantial contributions to
analysis of data. YL gave the final approval of the version to be
published. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Second Affiliated Hospital of Fujian Medical University
(Quanzhou, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Figueroa JD, Ye Y, Siddiq A, Garcia-Closas
M, Chatterjee N, Prokunina-Olsson L, Cortessis VK, Kooperberg C,
Cussenot O, Benhamou S, et al: Genome-wide association study
identifies multiple loci associated with bladder cancer risk. Hum
Mol Genet. 23:1387–1398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Egbers L, Grotenhuis AJ, Aben KK, Witjes
Alfred J, Kiemeney LA and Vermeulen SH: The prognostic value of
family history among patients with urinary bladder cancer. Int J
Cancer. 136:1117–1124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chung W, Bondaruk J, Jelinek J, Lotan Y,
Liang S, Czerniak B and Issa JP: Detection of bladder cancer using
novel DNA methylation biomarkers in urine sediments. Cancer
Epidemiol Biomarkers Prev. 20:1483–1491. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tseng-Rogenski S, Gee J, Ignatoski KW,
Kunju LP, Bucheit A, Kintner HJ, Morris D, Tallman C, Evron J, Wood
CG, et al: Loss of 15-hydroxyprostaglandin dehydrogenase expression
contributes to bladder cancer progression. Am J Pathol.
176:1462–1468. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong J, Zhai X, Chen Z, Liu Q, Ye H, Chen
W and Ling C: Treatment of huge hepatocellular carcinoma using
cinobufacini injection in transarterial chemoembolization: A
retrospective study. Evid Based Complement Alternat Med. May
17–2016.(Epub ahead of print). doi:10.1155/2016/2754542. View Article : Google Scholar
|
|
6
|
Chen T, Yuan S, Wan XN, Zhan L, Yu XQ,
Zeng JH, Li H, Zhang W, Hu XY, Ye YF, et al: Chinese herb
cinobufagin-reduced cancer pain is associated with increased
peripheral opioids by invaded CD3/4/8 lymphocytes. Oncotarget.
8:11425–11441. 2017.PubMed/NCBI
|
|
7
|
Kau MM, Wang JR, Tsai SC, Yu CH and Wang
PS: Inhibitory effect of bufalin and cinobufagin on steroidogenesis
via the activation of ERK in human adrenocortical cells. Br J
Pharmacol. 165:1868–1876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang ZJ, Sun L and Heinbockel T:
Resibufogenin and cinobufagin activate central neurons through an
ouabain-like action. PLoS One. 9:e1132722014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma K, Zhang C, Huang MY, Li WY and Hu GQ:
Cinobufagin induces autophagy-mediated cell death in human
osteosarcoma U2OS cells through the ROS/JNK/p38 signaling pathway.
Oncol Rep. 36:90–98. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lapierre LR, Kumsta C, Sandri M, Ballabio
A and Hansen M: Transcriptional and epigenetic regulation of
autophagy in aging. Autophagy. 11:867–880. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao L, Tian X, Duan X, Ye Y, Sun M and
Huang J: Association of body mass index with bladder cancer risk: A
dose-response meta-analysis of prospective cohort studies.
Oncotarget. 8:33990–34000. 2017.PubMed/NCBI
|
|
12
|
Franzen CA, Simms PE, Van Huis AF, Foreman
KE, Kuo PC and Gupta GN: Characterization of uptake and
internalization of exosomes by bladder cancer cells. Biomed Res
Int. 2014:6198292014.doi: 10.1155/2014/619829. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu XS, Qiao YB, Li Y, Yang B, Chen MB and
Xing CG: Preclinical study of cinobufagin as a promising
anti-colorectal cancer agent. Oncotarget. 8:988–998.
2017.PubMed/NCBI
|
|
14
|
Yu Y, Wang H, Meng X, Hao L, Fu Y, Fang L,
Shen D, Yu X and Li J: Immunomodulatory effects of cinobufagin on
murine lymphocytes and macrophages. Evid Based Complement Alternat
Med. 2015:8352632015.doi: 10.1155/2015/835263. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye M, Qu G, Guo H and Guo D: Specific 12
β-hydroxylation of cinobufagin by filamentous fungi. Appl Environ
Microbiol. 70:3521–3527. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu Q, Lin WD, Liao GQ, Zhang LG, Wen SQ
and Lin JY: Antiproliferative effects of cinobufacini on human
hepatocellular carcinoma HepG2 cells detected by atomic force
microscopy. World J Gastroenterol. 21:854–861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yin JH, Zhu XY, Shi WD and Liu LM:
Huachansu injection inhibits metastasis of pancreatic cancer in
mice model of human tumor xenograft. BMC Complement Altern Med.
14:4832014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salvador-Gallego R, Mund M, Cosentino K,
Schneider J, Unsay J, Schraermeyer U, Engelhardt J, Ries J and
García-Sáez AJ: Bax assembly into rings and arcs in apoptotic
mitochondria is linked to membrane pores. EMBO J. 35:389–401. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tiwari M, Sharma LK, Vanegas D, Callaway
DA, Bai Y, Lechleiter JD and Herman B: A nonapoptotic role for
CASP2/caspase 2: Modulation of autophagy. Autophagy. 10:1054–1070.
2014. View Article : Google Scholar : PubMed/NCBI
|