|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Dicker D, Pain A, Hamavid H,
Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, et
al: The global burden of cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamane B and Weber S: Liver-directed
treatment modalities for primary and secondary hepatic tumors. Surg
Clin North Am. 89:97–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishikawa T: Strategy for improving
survival and reducing recurrence of HCV-related hepatocellular
carcinoma. World J Gastroenterol. 19:6127–6130. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu H, Lin CC, Li YY and Zhao Z: Dynamic
protein interaction modules in human hepatocellular carcinoma
progression. BMC Syst Biol. 5 7 Suppl:S22013. View Article : Google Scholar
|
|
6
|
Choi JK, Yu U, Yoo OJ and Kim S:
Differential coexpression analysis using microarray data and its
application to human cancer. Bioinformatics. 21:4348–4355. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee HK, Hsu AK, Sajdak J, Qin J and
Pavlidis P: Coexpression analysis of human genes across many
microarray data sets. Genome Res. 14:1085–1094. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He B, Zhang H and Shi T: A comprehensive
analysis of the dynamic biological networks in HCV induced
hepatocarcinogenesis. PLoS One. 6:e185162011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Varelas X, Bouchie MP and Kukuruzinska MA:
Protein N-glycosylation in oral cancer: Dysregulated cellular
networks among DPAGT1, E-cadherin adhesion and canonical Wnt
signaling. Glycobiology. 24:579–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mentzen WI, Floris M and de la Fuente A:
Dissecting the dynamics of dysregulation of cellular processes in
mouse mammary gland tumor. BMC Genomics. 10:6012009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Southworth LK, Owen AB and Kim SK: Aging
mice show a decreasing correlation of gene expression within
genetic modules. PLoS Genet. 5:e10007762009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Banerjee A, Ray RB and Ray R: Oncogenic
potential of hepatitis C virus proteins. Viruses. 2:2108–2133.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
McGivern DR and Lemon SM: Virus-specific
mechanisms of carcinogenesis in hepatitis C virus associated liver
cancer. Oncogene. 30:1969–1983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M,
Marshall KA, et al: NCBI GEO: Archive for high-throughput
functional genomic data. Nucleic Acids Res. 37:(Database Issue).
D885–D890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article172005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herrero J, Valencia A and Dopazo J: A
hierarchical unsupervised growing neural network for clustering
gene expression patterns. Bioinformatics. 17:126–136. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zuberi K, Franz M, Rodriguez H, Montojo J,
Lopes CT, Bader GD and Morris Q: GeneMANIA prediction server 2013
update. Nucleic Acids Res. 41:W115–W122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fukushima A: DiffCorr: An R package to
analyze and visualize differential correlations in biological
networks. Gene. 518:209–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database Issue). D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de Chassey B, Navratil V, Tafforeau L,
Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria
BF, Chantier T, et al: Hepatitis C virus infection protein network.
Mol Syst Biol. 4:2302008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Allocco DJ, Kohane IS and Butte AJ:
Quantifying the relationship between co-expression, co-regulation
and gene function. BMC Bioinformatics. 5:182004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
De Giorgi V, Buonaguro L, Worschech A,
Tornesello ML, Izzo F, Marincola FM, Wang E and Buonaguro FM:
Molecular signatures associated with HCV-induced hepatocellular
carcinoma and liver metastasis. PLoS One. 8:e561532013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
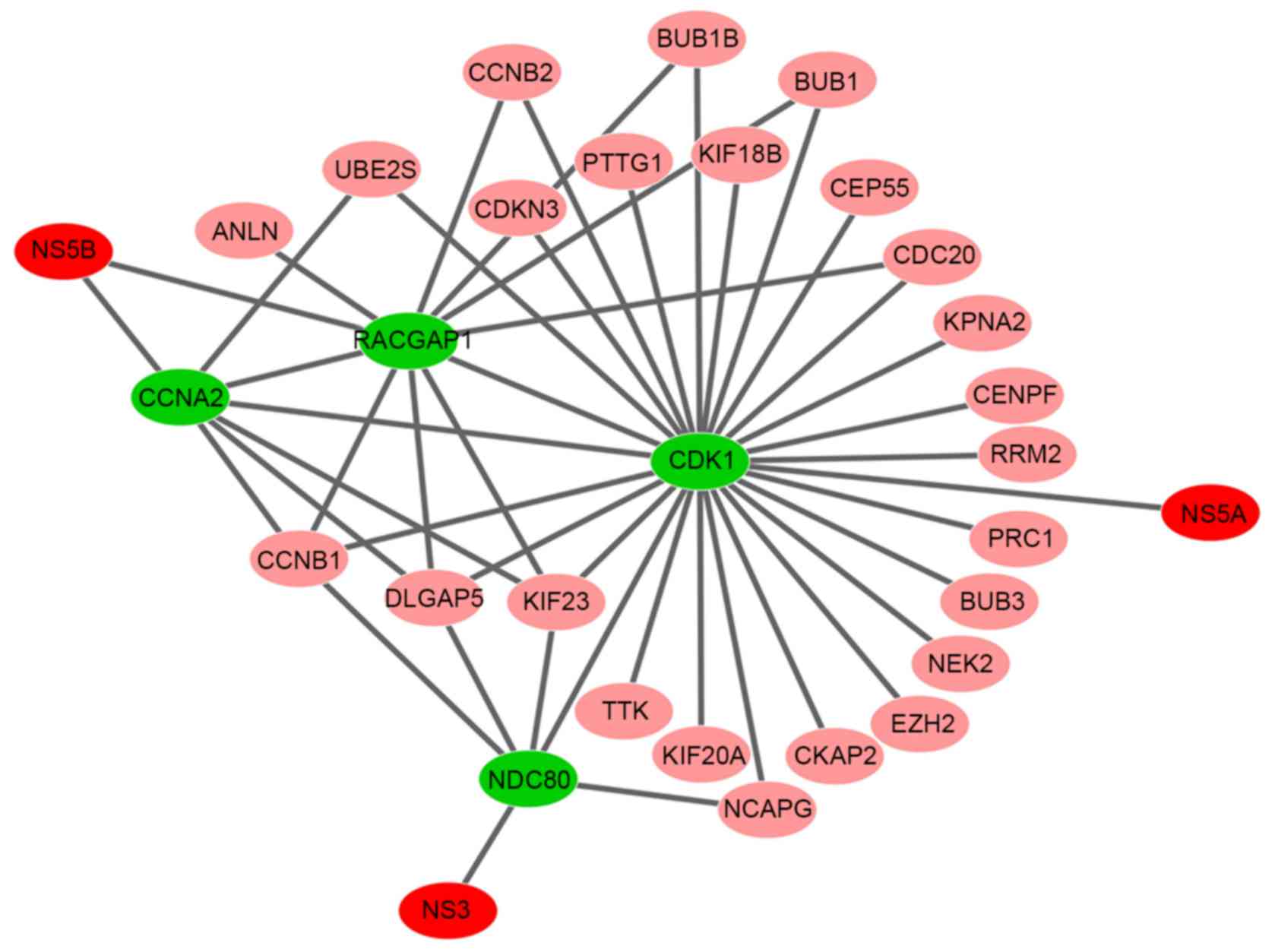
Pham LV, Ngo HT, Lim YS and Hwang SB:
Hepatitis C virus non-structural 5B protein interacts with cyclin
A2 and regulates viral propagation. J Hepatol. 57:960–966. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu MJ, Ke PY and Horng JT:
RacGTPase-activating protein 1 interacts with hepatitis C virus
polymerase NS5B to regulate viral replication. Biochem Biophys Res
Commun. 454:19–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koike K, Moriya K and Kimura S: Role of
hepatitis C virus in the development of hepatocellular carcinoma:
Transgenic approach to viral hepatocarcinogenesis. J Gastroenterol
Hepatol. 17:394–400. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chan KS, Koh CG and Li HY:
Mitosis-targeted anti-cancer therapies: Where they stand. Cell
Death Dis. 3:e4112012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kuo TC, Chang PY, Huang SF, Chou CK and
Chao CC: Knockdown of HURP inhibits the proliferation of
hepacellular carcinoma cells via downregulation of gankyrin and
accumulation of p53. Biochem Pharmacol. 83:758–768. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liao W, Liu W, Yuan Q, Liu X, Ou Y, He S,
Yuan S, Qin L, Chen Q, Nong K, et al: Silencing of DLGAP5 by siRNA
significantly inhibits the proliferation and invasion of
hepatocellular carcinoma cells. PLoS One. 8:e807892013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen JM, Chiu SC, Wei TY, Lin SY, Chong
CM, Wu CC, Huang JY, Yang ST, Ku CF, Hsia JY and Yu CT: The
involvement of nuclear factor-kappaB in the nuclear targeting and
cyclin E1 upregulating activities of hepatoma upregulated protein.
Cell Signal. 27:26–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang XD, Dai YC, Li ZY, Gan MF, Zhang SR,
Yin-Pan, Lu HS, Cao XQ, Zheng BJ, Bao LF, et al: Expression and
function analysis of mitotic checkpoint genes identifies TTK as a
potential therapeutic target for human hepatocellular carcinoma.
PLoS One. 9:e977392014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xing C, Xie H, Zhou L, Zhou W, Zhang W,
Ding S, Wei B, Yu X, Su R and Zheng S: Cyclin-dependent kinase
inhibitor 3 is overexpressed in hepatocellular carcinoma and
promotes tumor cell proliferation. Biochem Biophys Res Commun.
420:29–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geng J, Li X, Zhou Z, Wu CL, Dai M and Bai
X: EZH2 promotes tumor progression via regulating VEGF-A/AKT
signaling in non-small cell lung cancer. Cancer Lett. 359:275–287.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Collett K, Eide GE, Arnes J, Stefansson
IM, Eide J, Braaten A, Aas T, Otte AP and Akslen LA: Expression of
enhancer of zeste homologue 2 is significantly associated with
increased tumor cell proliferation and is a marker of aggressive
breast cancer. Clin Cancer Res. 12:1168–1174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi L, Zhang SL, Li K, Hong Y, Wang Q, Li
Y, Guo J, Fan WH, Zhang L and Cheng J: NS5ATP9, a gene up-regulated
by HCV NS5A protein. Cancer Lett. 259:192–197. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Quan M, Liu S, Li G, Wang Q, Zhang J,
Zhang M, Li M, Gao P, Feng S and Cheng J: A functional role for
NS5ATP9 in the induction of HCV NS5A-mediated autophagy. J Viral
Hepat. 21:405–415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Brendle A, Brandt A, Johansson R, Enquist
K, Hallmans G, Hemminki K, Lenner P and Försti A: Single nucleotide
polymorphisms in chromosomal instability genes and risk and
clinical outcome of breast cancer: A Swedish prospective
case-control study. Eur J Cancer. 45:435–442. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Waseem A, Ali M, Odell EW, Fortune F and
Teh MT: Downstream targets of FOXM1: CEP55 and HELLS are cancer
progression markers of head and neck squamous cell carcinoma. Oral
Oncol. 46:536–542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li KK, Ng IO, Fan ST, Albrecht JH,
Yamashita K and Poon RY: Activation of cyclin-dependent kinases
CDC2 and CDK2 in hepatocellular carcinoma. Liver. 22:259–268. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ito Y, Takeda T, Sakon M, Monden M,
Tsujimoto M and Matsuura N: Expression and prognostic role of
cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma.
Oncology. 59:68–74. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Y, Riley DJ, Zheng L, Chen PL and Lee
WH: Phosphorylation of the mitotic regulator protein Hec1 by Nek2
kinase is essential for faithful chromosome segregation. J Biol
Chem. 277:49408–49416. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qu Y, Li J, Cai Q and Liu B: Hec1/Ndc80 is
overexpressed in human gastric cancer and regulates cell growth. J
Gastroenterol. 49:408–418. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bièche I, Vacher S, Lallemand F,
Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E,
Lerebours F, Ripoche H, et al: Expression analysis of mitotic
spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in
early breast tumorigenicity, and a two-gene signature for
aneuploidy. Mol Cancer. 10:232011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen Y, Riley DJ, Chen PL and Lee WH: HEC,
a novel nuclear protein rich in leucine heptad repeats specifically
involved in mitosis. Mol Cell Biol. 17:6049–6056. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Glinsky GV, Berezovska O and Glinskii AB:
Microarray analysis identifies a death-from-cancer signature
predicting therapy failure in patients with multiple types of
cancer. J Clin Invest. 115:1503–1521. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liang Y, Liu M, Wang P, Ding X and Cao Y:
Analysis of 20 genes at chromosome band 12q13: RACGAP1 and MCRS1
overexpression in nonsmall-cell lung cancer. Genes Chromosomes
Cancer. 52:305–315. 2013. View Article : Google Scholar : PubMed/NCBI
|