Introduction
Melanoma is the most common malignancy of the eye,
with an incidence of 5–7 cases per million according to global
statistics in 1988 (1). Melanoma has
a high rate of metastasis, which primarily spreads to the liver
through the blood stream (2,3). Despite advances in the technology used
to treat melanoma, the mortality rate of metastatic disease remains
unchanged, and ~50% of all patients with melanoma eventually
succumb to mortality due to metastatic disease (4). As the molecular mechanisms of melanoma
aggressiveness are yet to be clearly understood, metastatic
melanoma cannot be effectively treated (5,6).
Understanding the crucial signals that contribute to the invasive
and metastatic potential of melanoma is necessary in order to
identify novel therapeutic targets.
Mono-ADP-ribosylation is a post-translational
protein modification involved in the transfer of the ADP-ribose
moiety from NAD+ to a specific amino acid in a target protein,
resulting in the alteration of the functional properties of the
target protein (7). This enzymatic
reaction was originally identified as the pathogenic mechanism of
bacterial toxins, including cholera, pertussis and diphtheria
toxins (8). ADP-ribosyltransferases
(ARTs) regulate endogenous protein functions, including DNA repair,
cell differentiation and cell cycle progression by attaching
ADP-ribose to specific amino acid residues in membrane proteins
(9). ART3 is a member of the ART
family, and is involved in cell division and the regulation of
inflammatory responses (10). Shi
et al (11) revealed that ART3
may contribute to the pathophysiological and biochemical
progression of a neural lesion. In one previous study, it was
identified that genetic variation of ART3 may result in a
functional defect in the process of spermatogenesis (12).
ARTs have been reported to be involved in
tumorigenesis. Xiao et al (13) confirmed that the knockdown of ART1
increased the apoptosis of CT26 cells in transplanted tumor types.
However, the biological function of ART3 in melanoma progression
has not previously been studied, to the best of our knowledge. In
the present study, it was revealed that ART3 was abnormally
expressed in melanoma tissues and melanoma cells. Then, following
the silencing of ART3 by small interfering RNA (siRNA) and short
hairpin RNA (shRNA) in order to study its function in melanoma, the
results revealed that ART3 knockdown may inhibit the migration
ability of melanoma cells.
Materials and methods
Cell lines and cell culture
Human melanoma cell lines OCM1, OM431 and OCM1A were
provided by Professor John F. Marshall (Tumor Biology Laboratory,
Cancer Research UK Clinical Center, John Vane Science Centre,
London, UK). A human retinal pigment epithelium (RPE-19) cell line
was provided by the Department of Ophthalmology, Ruijin Hospital,
Shanghai Jiao Tong University School of Medicine (Shanghai, China).
The melanoma and RPE cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.; cat. no. 10099141), 100 U
penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.; cat. no. 10378016). The cultures were maintained
in a humidified incubator at 37°C with 5% CO2.
Patients
A set of melanoma tissues (n=18) paired with
adjacent normal tissue (n=18) were obtained from patients diagnosed
with melanoma from Shanghai Ninth People's Hospital, Shanghai Jiao
Tong University, School of Medicine (Shanghai, China) from 2010 to
2015. There were 10 males and 8 females, their age ranged from
23–63, with a mean age of 41±8.23, after surgical resection of the
tumor, the tissues were put into the cryopreservation tube and
immediately preserved in the liquid nitrogen. Ethical approval
obtained from the Independent Ethics Committee of Shanghai Ninth
People's Hospital, Shanghai Jiao Tong University, School of
Medicine and written informed consent was obtained from all
patients involved.
ART3 siRNA oligonucleotides
ART3 siRNA oligonucleotides were as follows:
Si-ART3-1, GACAUGGCAGAUAUGCAUdTdT, si-ART3-2,
CACAGUUUGGGAUGGUCAUdTdT and si-ART3-3, CUGUAUUGAGAACCUAGAAdTdT. A
random homologous sequence of all human genes was set as si-NC:
UUCUCCGAACGUGUCACGUTTdTdT.
siRNA transfection
OCM1 and OM431 cells were seeded in 6-well plates at
a density of 2×105 cells/well. When the cells reached
70% confluency, cells were transfected with 50 nM of each siRNA
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
After 6–8 h, the transfection medium was replaced with DMEM with
10% FBS. After 48 h of transfection, cells were collected for RNA
extraction and protein extraction.
shRNA-expressing plasmid
construction
The two shRNA sequences (shART3-1:
5′-CACCGACATGGCAGATATGCATCGAAATGCATATCTGCCATGTC-3′; shART3-3:
5′-CACCGCTGTATTGAGAACCTAGAACGAATTCTAGGTTCTCAATACAG-3′) that target
sh-ART3 were cloned into pGIPZ lentivirus vector (System
Biosciences, Palo Alto, CA, USA).
Lentivirus package
The 293 T cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% (vol/vol) fetal bovine serum and maintained at 37°C at a
concentration of 6,000,000 cells and transfected using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) with 3 µg GIPZ-shART3, 3 µg pMD2.D, and 6.0 µg PsPax. After
incubation overnight with 293 T cells, the media was replaced with
5 ml fresh medium. The virus-containing supernatants were collected
at 48 and 72 h after transfection and then mixed and filtered
through a 0.45 µm cellulose acetate filter (Sartorius AG,
Göttingen, Germany). The viral supernatants were concentrated with
Amicon Ultra-15 Centrifugal Filter Units (EMD Millipore,
Schwalbach, Germany) at 4°C and spun at 3,913 × g for 30 min. viral
supernatants were added into OCM1 and OM431 cells for 48 h and then
replaced by fresh cell-culture medium. Then the colonies were
selected for subsequent culture after incubation with 4 g/ml
puromycin for 2 weeks.
Western blotting
Western blotting was performed as previously
described (14). Western blotting
results were repeated three times and quantitative analysis of
western blotting results were performed with Image J1.47 (National
Institutes of Health, Bethesda, MD, USA). Protein was extracted
from cells by RIPA Lysis and Extraction Buffer (Thermo Fisher
Scientific, Inc.; cat. no. 89900). Antibodies used were as follows:
Anti-ART3 polyclonal antibody (Wuhan Sanying Biotechnology, Wuhan,
China; cat. no. 15930-1-AP; dilution, 1:1,000), GAPDH antibody
(Wuhan Sanying Biotechnology; cat. no. 10494-1-AP; dilution,
1:5,000) and horseradish peroxidase-conjugated goat anti-rabbit
secondary antibodies was used for western blot (Rabbit IgG, Abcam,
Cambridge, UK; cat. no. ab6721; dilution, 1:5,000).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and cDNA was synthesized using the
PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). qPCR
analysis was performed using the RT qPCR Power SYBR™ Green Master
mix (Applied Biosystems; Thermo Fisher Scientific, Inc; cat.no.
4367659) using Applied Biosystems 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.), and amplified
qPCR products were quantified and normalized using GAPDH as a
control (15). GAPDH and ART3 primers
were as follows: GAPDH forward, 5′-ACTTCAACAGCGACACCCACTC-3′ and
reverse', 5′-GTCCACCACCCTGTTGCTGTAG-3′; ART3 forward,
5′GCAACCATGATTCTAGTGGAC-3′ and reverse, 5′-CTTTAGCAGTTGGGGAACG-3′.
Thermocycling conditions for ART3 expression were as follows: 45
cycles of denaturation at 95°C for 30 sec, 60°C for 30 sec,
extension at 72°C for 30 sec and a final extension at 72°C for 5
min.
Migration assay
The migration assay was based on the migration of
melanoma cells seeded in an upper chamber of transwell insert
through a membrane with an 8 µm pore size (EMD Millipore). Cells
were seeded into 6-well plates at a density of 2×105
cells/well, cultured for 18–24 h and then transfected with 50 nM of
each siRNA Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). After 6–8 h, the medium was replaced with fresh
medium.
A total of 24 h after transfection, cells were
detached and collected. The upper chamber was seeded with
1×105 cells of OCM1 and OM431 treated with siART3-1,
siART3-3, the cells stably expressing shART3-1 and shART3-3 were
contained in DMEM with 1% FBS (200 µl). The lower chamber contained
800 µl DMEM and 20% FBS was added to each well of the 24-well
plate. Following incubation at 37°C for 48 h, the upper chamber was
fixed with 100% methanol and stained with 0.1% crystal violet for 1
h as described previously (14).
Then, the stained chambers were observed and photographing in
microscope using a CCD camera (Visitron Systems GmbH, Puchheim,
Germany) on a Zeiss Axioplan 2 imaging microscope (Zeiss AG,
Oberkochen, Germany). Magnification: ×4.
Immunofluorescence staining
assays
Cells were grown on glass coverslips for 24 h, then
fixed with 4% paraformaldehyde in room temperature for 30 min and
permeabilized with 0.5% Triton X-100 for 5 min. Following blocking
with 10% FBS in DMEM in room temperature for 30 min, cells were
incubated with the following antibodies in 4°C overnight according
to the manufacturer's protocol: Anti-human ART3 (Wuhan Sanying
Biotechnology; cat. no. 15930-1-AP; dilution, 1:1,000), GAPDH
Rabbit mAb (Cell Signaling Technology; cat. no. 5174; dilution,
1:1,000) and Cy2-labeled goat anti-rabbit secondary antibodies were
used to visualize primary antibodies (Jackson Immuno Research
Laboratories, Inc., West Grove, PA, USA; cat. no. 111-225-006
dilution, 1:5,000). Cell micrographs were obtained using a CCD
camera (Visitron Systems GmbH) on a Zeiss Axioplan 2 imaging
microscope (Zeiss AG, Oberkochen, Germany). Magnification: ×20.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA). Results were expressed as the mean ± standard deviation.
Parametric and non-parametric Kolmogorov-Smirnov tests were applied
to assess normality, and the expression of ART3 in melanoma tissues
was analyzed using a paired Student's t-test. Data were evaluated
using a one-way analysis of variance with a Tukey's honest
significant difference post-hoc test for comparisons between
groups. P<0.05, P<0.01 and P<0.001 were considered to
indicate a statistically significant, markedly significant and very
significant difference, respectively. Western blotting results were
analyzed using gray scale with Image J1.47 (National Institutes of
Health, Bethesda, MD, USA).
Results
ART3 is overexpressed in melanoma
tissues and melanoma cells
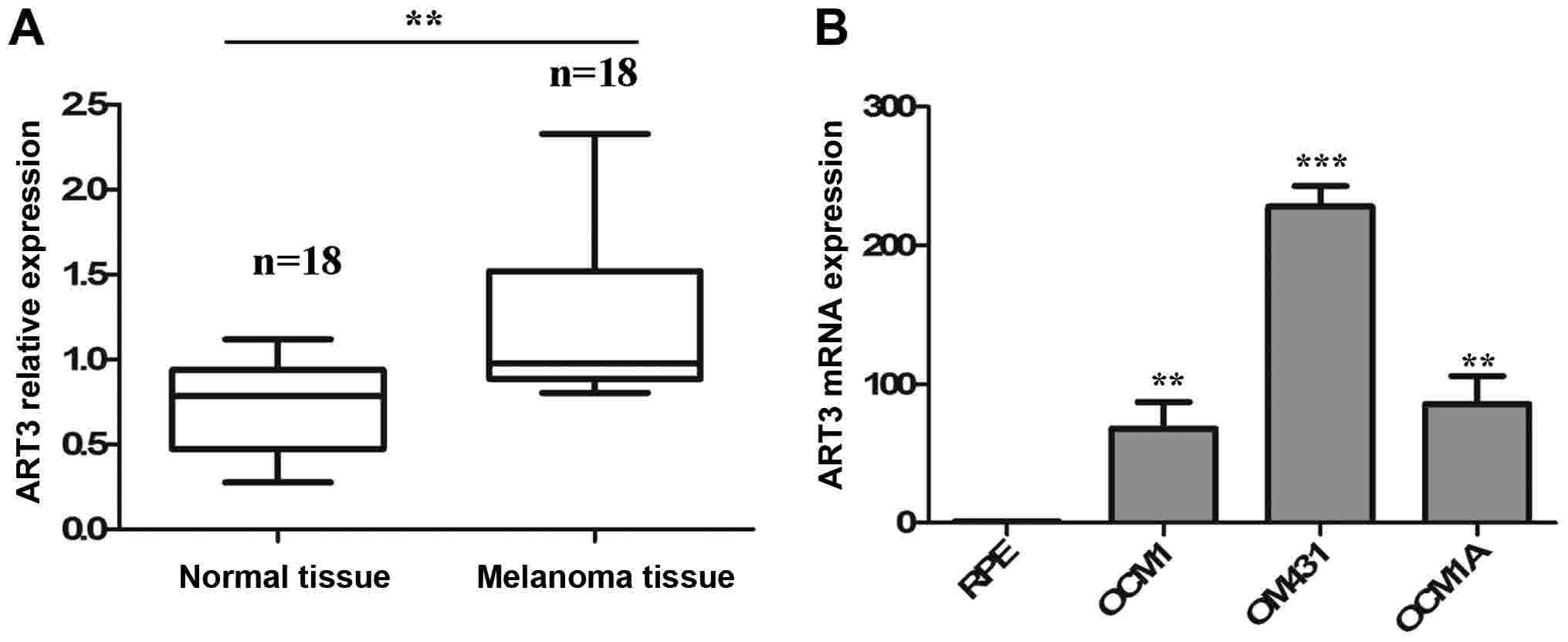
To investigate the association between the abnormal
expression of ART3 and the occurrence of melanoma, a set of
melanoma tissues (n=18) paired with adjacent normal tissue (n=18)
were obtained from diagnosed patients and used to examine the
expression of ART3. It was revealed that the expression levels of
ART3 were significantly increased in melanoma tissues compared with
adjacent normal tissues (P<0.01; Fig.
1A). To further verify the clinical significance of ART3, ART3
expression in three melanoma cell lines (OCM1, OM431 and OCM1A) and
one normal cell line (RPE) was examined using RT-qPCR. Consistent
with the results in melanoma tissues, the melanoma cell lines
expressed significantly higher levels of ART3 compared with the RPE
cells (P<0.01; Fig. 1B). These
data indicate the clinical importance of ART3 in melanoma.
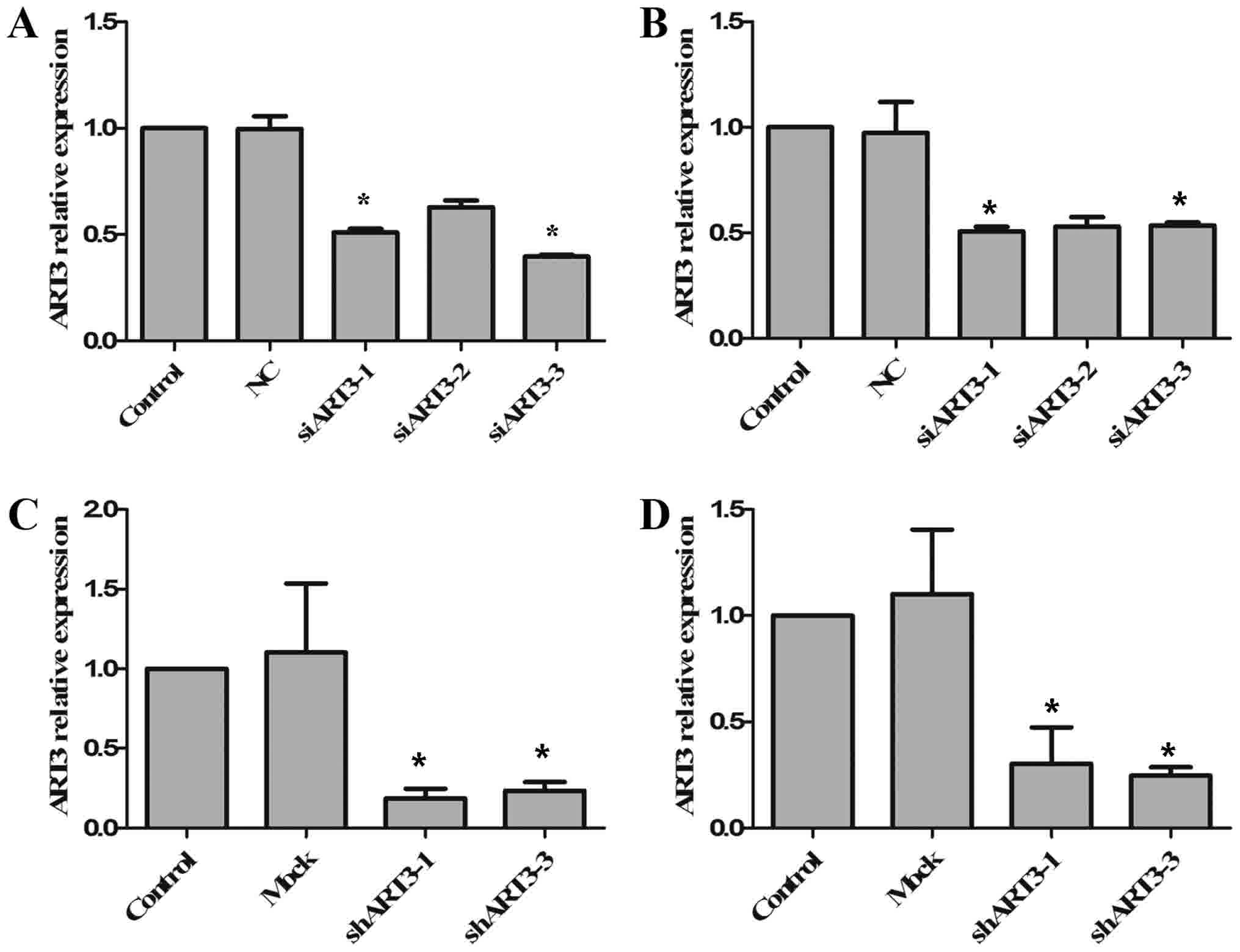
Targeted gene interference of ART3
with siRNA and shRNA
As ART3 is highly expressed in melanoma cells and
melanoma tissues, it was speculated that ART3 may serve a function
in the occurrence of melanoma. siRNA-mediated interference of ART3
expression was performed in OCM1 and OM431 cells, in order to
exclude off-target effects. OCM1 and OM431 cells were transfected
with three si-ART3 and si-negative control (NC). The RNA
interference efficiency was tested using RT-qPCR, among which the
interference efficiency of si-ART3-1 and si-ART3-3 reached >50%,
revealing a significant difference compared with the NC (P<0.05;
Fig. 2A and B). Next, one siRNA
(si-ART3-1) was selected for the construction of pGIPZ ART3-shRNA
plasmids. The pGIPZ ART3-shRNA vectors with an empty vector (mock)
were then packaged into lentiviruses, and transduced into OCM1 and
OM431 cells. Following transduction, the cells were screened using
puromycin for the stable expression of sh-ART3. The expression
levels of ART3 in stable cell clones were then detected using
RT-qPCR (Fig. 2C and D). It was
revealed that ART3 expression levels were significantly knocked
down in the two shART3-expressing cell lines, OCM1 (P<0.05;
Fig. 2C) and OM431 (P<0.05;
Fig. 2D).
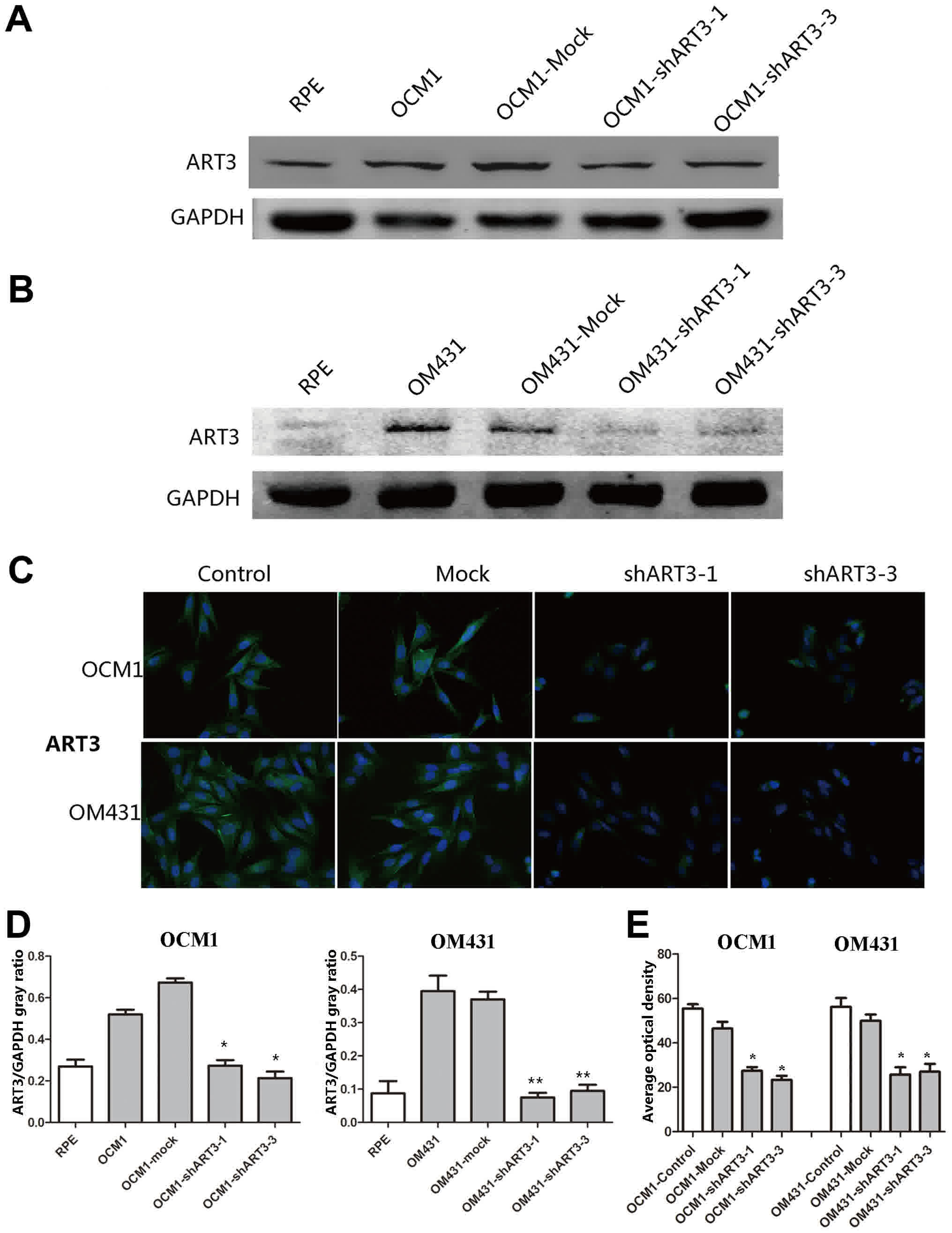
Detecting the knockdown of ART3 in
melanoma cells by western blotting and immunofluorescence
staining
To further verify the knockdown of ART3 in melanoma
cells, western blotting (Fig. 3A and
B) and immunofluorescence staining (Fig. 3C) was used to examine the ART3
expression in shART3 expressing cells. As expected, the quantified
results of the western blotting and immunofluorescence revealed
that the expression levels of ART3 in stable cell clones was
significantly knocked down compared with the empty vector
transfected cells (mock; Fig. 3D and
E).
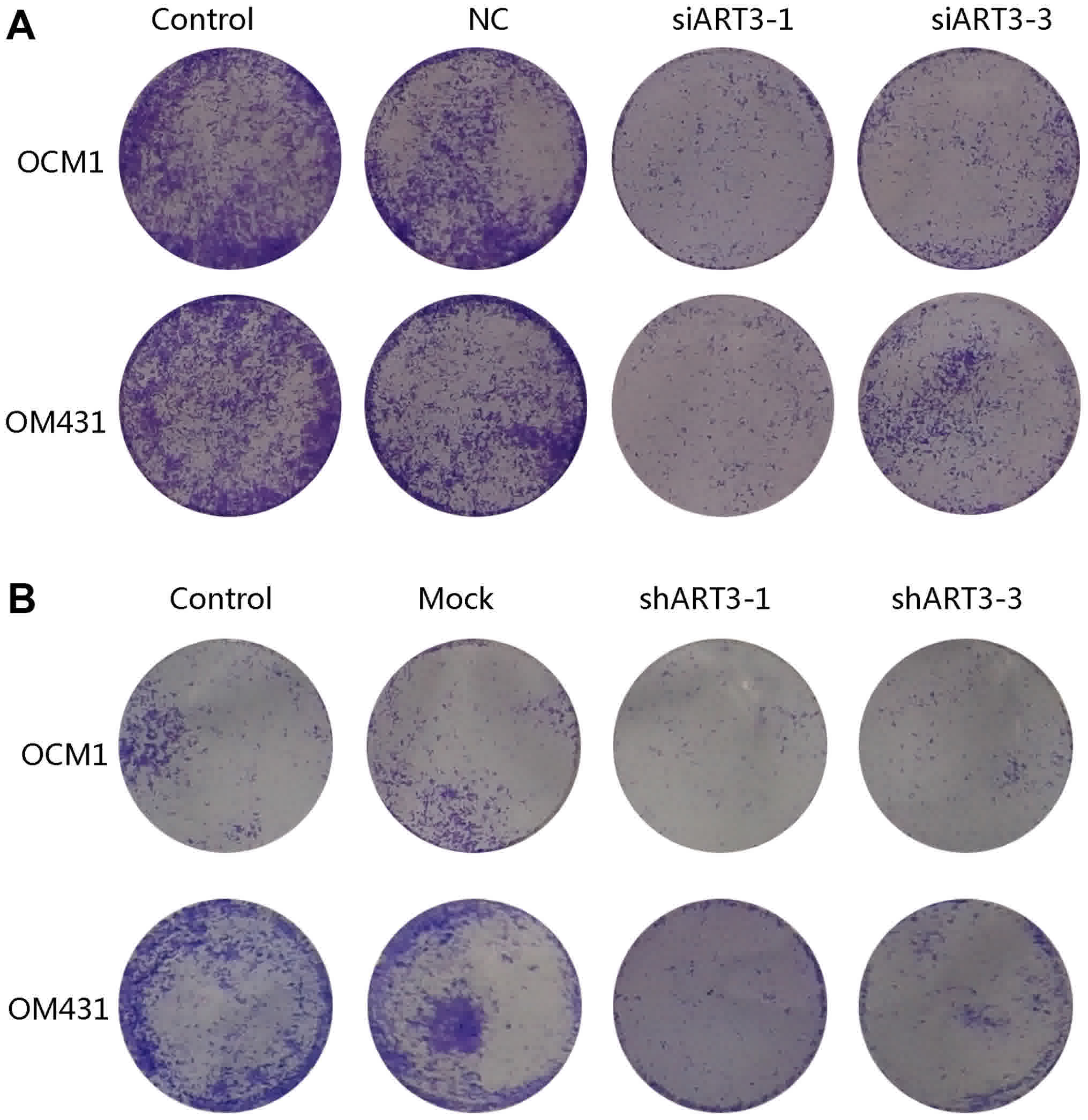
Knockdown of ART3 inhibits the
migration of melanoma cells
To study the effect of ART3 on migration, si-ART3-1
and si-ART3-3 transfected OCM1 and OM431 cells were harvested
subsequent to being cultured for 48 h, stained with 0.1% crystal
violet and imaged. The result of the Transwell chamber assay
indicated that the number of si-ART3-1 and si-ART3-3 cells
migrating through the filtration membrane was markedly lower
compared with that of the controls (Fig.
4A). This indicated that the interference of ART3 may inhibit
the migratory ability of melanoma cells. In addition, a markedly
decreased migratory rate in shART3-expressing OCM1 and OM431 cells
compared with that of the controls after 48 h was observed
(Fig. 4B). These data indicate that
ART3 serves a regulatory role in melanoma cell migration and may
serve as a novel therapeutic target.
Discussion
ADP ribosylation is a notable posttranslational
protein modification, regulating a host of critical cellular
processes, including tumorigenesis and cell apoptosis (16). Momii and Koide (17) suggested that mouse testicular cells
possess an ART that catalyzes the transfer of the ADP-ribose moiety
from NAD+ to an unknown acceptor protein. Genetic variation of ART3
may result in a functional defect in the process of
spermatogenesis. Okada et al (18) revealed that ART3 was identified as a
susceptible gene underlying non-obstructive azoospermia. The
biological function of ART3 varies in different tissues and organs
(19); however, the effect of ART3
function on melanoma tumorigenesis has yet to be reported.
In the present study, it was revealed that there was
an abnormal expression of ART3 in melanoma cells and melanoma
tissues, which indicated the potential function of ART3 in melanoma
tumorigenesis. To gain a better understanding of the biological
function of ART3, siRNA and shRNA were used to silence the ART3
gene in melanoma cell lines, the results revealed the efficient
inhibition of ART3. The effects of ART3 silencing on cell migration
were further examined. The results revealed that ART3 knockdown
notably inhibited melanoma migration.
Considering that melanoma is a highly metastatic
disease and ~1/2 of all patients with melanoma succumb to mortality
from metastases (20,21), the treatment of melanoma faces
numerous challenges. Single-agent therapies (mono-therapies) have
not yet demonstrated a clear clinical benefit (22). Selumitinib provides a significant
advantage in increased progression-free survival rates, but with an
insignificant impact on improved overall survival (23). The use of histone deacetylase
inhibitors to induce the differentiation of melanoma cells is being
explored clinically, but evidence of antitumor activity in
pre-clinical models remains limited (24). The combination of si-B-cell lymphoma 2
and H101 recombinant oncolytic adenovirus has proven to be
effective in the treatment of melanoma (25). Hu et al (26) revealed that the pharmacological
nuclear factor-κB inhibitor, BAY11-7082, induced cell apoptosis and
inhibited the migration of human melanoma cells. Therefore,
identifying effective targets that inhibit melanoma metastasis is
necessary and ART3 may be a potential therapeutic target in the
future.
The mechanism of the involvement of ART3 in melanoma
cell migration requires further study, as the ADP-ribose moiety
from NAD+ was transferred to the cysteine residues of the target
protein (11), suggesting that ART3
may be a cysteine-specific enzyme. Paraoan et al (27) revealed that the shift in the balance
between cathepsin S and cystatin C may be part of the deregulated
proteolytic pathways that contribute to the invasive phenotype of
melanoma. It has been suggested that ART3 may function as a
cysteine enzyme involved in melanoma metastasis. Previously, small
molecular inhibitors have been studied as novel promising drugs for
the treatment of melanoma (28). Bi
et al (14) revealed that a
chemical inhibitor of C-X-C chemokine receptor type 4, AMD3100, may
inhibit the proliferation and migration of melanoma cells. At
present, no small molecule inhibitors of ART3 have been identified,
thus the development of novel ART3 inhibitors is a promising area
of research.
In summary, the targeted silencing of ART3
expression inhibited the migration of melanoma cells. This suggests
that ART3 may be a metastasis-associated gene in melanoma. Future
research should focus on the mechanism of the ART3 anti-metastasis
function and examine its potential role as a molecular target for
the treatment of melanoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific
Research Program of the National Health and Family Planning
Commission of China (grant no. 201402014), the National Natural
Science Foundation of China (grant nos. 81372469, 81301952 and
U1432117), the Program for Professor of Special Appointment
(Eastern Scholar) at the Shanghai Institutions of Higher Learning
(grant no. 1410000159), the SMC-ChenXing Yong Scholar Program
(2014, Class B) and the Science and Technology Commission of
Shanghai (grant no. 17DZ2260100).
Availability of data and materials
Not applicable.
Authors' contributions
In this report, XQF and SFG designed and directed
the experiments, discussed, revised and wrote the manuscript; JH,
YYL, and YW designed and performed the experiments and drafted the
manuscript; JH and HZ were responsible for sample collection and
data analysis. All authors approved this manuscript.
Ethics approval and consent to
participate
The ethical approval was signed by ethics committee
of Shanghai Ninth People's Hospital affiliated to Shanghai Jiao
Tong University School of Medicine (Shanghai, China), and written
informed consent was obtained from all patients involved.
Consent for publication
Informed consent for the publication of their
clinical data was obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sato T, Han F and Yamamoto A: The biology
and management of uveal melanoma. Curr Oncol Rep. 10:431–438. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh AD, Turell ME and Topham AK: Uveal
melanoma: Trends in incidence, treatment, and survival.
Ophthalmology. 118:1881–1885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Jia R, Wang J, Xu X, Yao Y, Ge S
and Fan X: Targeted silencing of MART-1 gene expression by RNA
interference enhances the migration ability of uveal melanoma
cells. Int J Mol Sci. 14:15092–15104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yousef YA and Alkilany M:
Characterization, treatment, and outcome of uveal melanoma in the
first two years of life. Hematol Oncol Stem Cell Ther. 8:1–5. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sagoo MS, Harbour JW, Stebbing J and
Bowcock AM: Combined PKC and MEK inhibition for treating metastatic
uveal melanoma. Oncogene. 33:4722–4723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pereira PR, Odashiro AN, Lim LA, Miyamoto
C, Blanco PL, Odashiro M, Maloney S, De Souza DF and Burnier MN Jr:
Current and emerging treatment options for uveal melanoma. Clin
Ophthalmol. 7:1669–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okazaki IJ and Moss J:
Mono-ADP-ribosylation: A reversible posttranslational modification
of proteins. Adv Pharmacol. 35:247–280. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bourne HR: ADP-Ribosylating toxins and G
proteins. Insights into signal transduction. Joel Moss and Martha
Vaughan, Eds. American society for microbiology, Washington, DC,
1990. xviii, 567 pp., illus. $79; to ASM members, $69. Science.
250:841–842. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ludden PW: Reversible ADP-ribosylation as
a mechanism of enzyme regulation in procaryotes. Mol Cell Biochem.
138:123–129. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Isa M A, Abu-Rafea B, Al-Asiri S,
Al-Motawa J, Almady K and Alwaznah R: Ovarian stimulation
medications and patients' responses as prognostic factors in
IUI-treated infertile Saudi patients. Iran J Reprod Med.
12:493–498. 2014.PubMed/NCBI
|
|
11
|
Shi W, Gong P, Fan J, Yan YH, Ni L, Wu X,
Cui G, Wu X, Gu X and Chen J: The expression pattern of
ADP-ribosyltransferase 3 in rat traumatic brain injury. J Mol
Histol. 43:37–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rajender S, Avery K and Agarwal A:
Epigenetics, spermatogenesis and male infertility. Mutat Res.
727:62–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao M, Tang Y, Chen WW, Wang YL, Yang L,
Li X, Song GL and Kuang J: Tubb3 regulation by the Erk and Akt
signaling pathways: A mechanism involved in the effect of arginine
ADP-ribosyltransferase 1 (Art1) on apoptosis of colon carcinoma
CT26 cells. Tumour Biol. 37:2353–2363. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bi J, Li P, Li C, He J, Wang Y, Zhang H,
Fan X, Jia R and Ge S: The SDF-1/CXCR4 chemokine axis in uveal
melanoma cell proliferation and migration. Tumour Biol.
37:4175–4182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Chang W, Dynek JN and Smith S: NuMA is a
major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis.
Biochem J. 391:177–184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Momii A and Koide SS: Adenosine
diphosphate-ribosyltransferase activity in permeabilized mouse
testicular cells. Arch Biochem Biophys. 214:628–633. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okada H, Tajima A, Shichiri K, Tanaka A,
Tanaka K and Inoue I: Genome-wide expression of azoospermia testes
demonstrates a specific profile and implicates ART3 in genetic
susceptibility. PLoS Genet. 4:e262008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Friedrich M, Grahnert A, Klein C, Tschöp
K, Engeland K and Hauschildt S: Genomic organization and expression
of the human mono-ADP-ribosyltransferase ART3 gene. Biochim Biophys
Acta. 1759:270–280. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cools-Lartigue JJ, McCauley CS, Marshall
JC, Di Cesare S, Gregoire F, Antecka E, Logan P and Burnier MN:
Immunomagnetic isolation and in vitro expansion of human uveal
melanoma cell lines. Mol Vis. 14:50–55. 2008.PubMed/NCBI
|
|
21
|
Nareyeck G, Zeschnigk M, Prescher G,
Lohmann DR and Anastassiou G: Establishment and characterization of
two uveal melanoma cell lines derived from tumors with loss of one
chromosome 3. Exp Eye Res. 83:858–864. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng H, Terai M, Kageyama K, Ozaki S,
McCue PA, Sato T and Aplin AE: Paracrine effect of NRG1 and HGF
drives resistance to MEK inhibitors in metastatic uveal melanoma.
Cancer Res. 75:2737–2748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carvajal RD, Sosman JA, Quevedo JF, Milhem
MM, Joshua AM, Kudchadkar RR, Linette GP, Gajewski TF, Lutzky J,
Lawson DH, et al: Effect of selumetinib vs. chemotherapy on
progression-free survival in uveal melanoma: A randomized clinical
trial. JAMA. 311:2397–2405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Landreville S, Agapova OA, Matatall KA,
Kneass ZT, Onken MD, Lee RS, Bowcock AM and Harbour JW: Histone
deacetylase inhibitors induce growth arrest and differentiation in
uveal melanoma. Clin Cancer Res. 18:408–416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang H, Wang H, Zhang J, Qian G, Niu B,
Fan X, Lu J, Hoffman AR, Hu JF and Ge S: Enhanced therapeutic
efficacy by simultaneously targeting two genetic defects in tumors.
Mol Ther. 17:57–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu S, Luo Q, Cun B, Hu D, Ge S, Fan X and
Chen F: The pharmacological NF-κB inhibitor BAY11-7082 induces cell
apoptosis and inhibits the migration of human uveal melanoma cells.
Int J Mol Sci. 13:15653–15667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paraoan L, Gray D, Hiscott P,
Garcia-Finana M, Lane B, Damato B and Grierson I: Cathepsin S and
its inhibitor cystatin C: Imbalance in uveal melanoma. Front Biosci
(Landmark Ed). 14:2504–2513. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johansson Hertzman C and Brage Egyhazi S:
BRAF inhibitors in cancer therapy. Pharmacol Ther. 142:176–182.
2014. View Article : Google Scholar : PubMed/NCBI
|