Introduction
Lung cancer is the most frequently diagnosed
malignancy and the leading cause of cancer-associated mortality
worldwide (1). Non-small cell lung
cancer (NSCLC) accounts for 85–90% of all lung cancer cases
(2). Patients diagnosed with
symptomatic lung cancer have a poor prognosis, with an overall
5-year survival rate of 16% in the United States (3).
Lung carcinogenesis is a multistep process
associated with the activation of oncogenes and the inactivation of
tumor-suppressing genes (4). However,
the molecular mechanisms underlying the progression of NSCLC remain
poorly understood. Therefore, there is an urgent requirement for
further investigation of the mechanisms that lead to the
development and progression of lung cancer in order to identify
novel biomarkers and therapeutic targets.
MicroRNAs (miRNAs) are small, non-coding RNAs ~22
nucleotides in length that regulate gene expression by either
degradation or repression of mRNA translation (5). MicroRNAs serve essential roles in a
variety of biological processes, including cell death,
differentiation, proliferation and metabolism (5,6). Altered
miRNA expression occurs in numerous types of human cancer (7) and is associated with the initiation and
progression of cancer (8). The
expression of miRNAs may be controlled through epigenetic
mechanisms, with ~10% of miRNAs being regulated by DNA methylation
(9).
MicroRNA-137 (miR-137) is located on human
chromosome 1p21.3 and is embedded in a CpG island (10,11).
miR-137 is frequently downregulated in several types of cancer,
including colorectal cancer, gastric cancer, glioblastoma and NSCLC
(12–16). Previous studies have suggested that
miR-137 silencing may be the result of hypermethylation of the
miR-137 gene promoter (11,15,16).
miR-137 promoter methylation is associated with poor prognosis in
certain types of cancer, including gastric cancer (17) and squamous cell carcinoma of the head
and neck (18). A previous study by
Kang et al demonstrated that the level of miR-137 promoter
methylation was significantly higher in lung tumors than in the
adjacent non-tumor tissues (19).
However, the aforementioned study did not assess whether miR-137
promoter methylation has prognostic value for recurrence or overall
survival in lung cancer (19).
The principal aim of the present study was to
evaluate whether methylation of the miR-137 promoter represents a
prognostic biomarker for overall and disease-free survival in
NSCLC. The overall objective of the present study was to provide an
experimental and theoretical basis for further study of the
associations between miR-137 promoter methylation and prognosis in
NSCLC.
Materials and methods
Ethical approval
The present study was approved by the Ethics
Committee of Subei People's Hospital (Yangzhou, China) and written
informed consent was obtained from all participants.
Study population
A total of 10 pairs of 4% formalin-fixed for 24 h at
room temperature, paraffin-embedded (FFPE) NSCLC tissues and
corresponding matched non-tumor lung tissues were collected (from 4
patients with squamous cell carcinoma and 6 patients with
adenocarcinoma) between May 2012 and October 2012, at Subei
People's Hospital. A further 56 FFPE NSCLC tissues (from 31
patients with squamous cell carcinoma and 25 patients with
adenocarcinoma) were obtained between February 2008 and December
2009. All tissue samples were collected prior to treatment with
chemoradiotherapy. Each sample was confirmed by histopathological
evaluation using hematoxylin and eosin staining. Clinical data were
recorded at the time of resection and patients were prospectively
followed-up to ascertain vital status (the last follow-up date was
April 30, 2012).
Cell culture
Human lung cancer A549 and H1299 cells and normal
bronchial epithelial BEAS-2B cells were purchased from the Cell
Resource Center, Shanghai Institute of Biochemistry, China.
(http://www.sibcb.ac.cn/) BEAS-2B cells were
derived by transforming human bronchial epithelial cells with an
adenovirus 12-simian virus 40 construct, as previously described
(20). The cell lines were maintained
in RPMI-1640 medium supplemented with 10% fetal bovine serum (both
from Wisent, Inc., St. Bruno, QC, Canada) at 37°C in a humidified
air atmosphere containing 5% carbon dioxide.
Treatment with
5-aza-2′-deoxycytidine
A549 and H1299 cells were seeded onto 24-well plates
on day 0, exposed to the DNA methylation inhibitor
5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at a final concentration of 5 µmol/l between
day 1 and day 3 at 37°C in a humidified air atmosphere containing
5% CO2 and were harvested under Trypsin-EDTA digestion
harvested for RT-qPCR analysis of miR-137 expression on day 4.
RNA isolation
Total RNA was extracted from cultured cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Total tissue RNA was extracted from FFPE tissue
sections using the miRNeasy FFPE kit (Qiagen, Inc., Valencia, CA,
USA) according to the manufacturer's protocol. Paraffin was removed
from freshly cut FFPE tissue sections each up to 10-µm thick using
deparaffinization solution using the miRNeasy FFPE kit (Qiagen,
Inc.), and samples under went protease digestion at room
temperature to release RNA from the sections, then short incubation
(at 56°C for 15 min, then at 80°C for 15 min) to reverse formalin
cross-linking of the released nucleic acids and DNase digestion to
remove DNA. Total RNA (including miRNAs) was dissolved in 20 µl
RNase-free water. RNA concentrations were measured using a
NanoDrop-1000 (Thermo Fisher Scientific, Inc.) and RNA integrity
was determined by 1.5% agarose gel electrophoresis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miRNA-137
expression
RNA was reverse transcribed using the RevertAid
First Strand cDNA kit (Thermo Fisher Scientific, Inc.) according to
the manufacture's protocolin combination with a stem-loop primer
for miRNA-137U6 small nuclear RNA was used as an internal control
to normalize the expression levels of miRNA-137. The primer
sequences are presented in Table I.
Briefly, 1.0 µg total RNA was combined with 4.0 µl Ribo Lock RNase
inhibitor, 2.0 µl dNTP mix (10 mM each) and 1.0 µl RevertAid M-MuLV
reverse transcriptase in a total reaction volume of 20 µl, which
was incubated on an ABI Prism 7900HT Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) at 25°C for 5
min, 42°C for 60 min and 70°C for 5 min.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequence (5′-3′) |
|---|
| miRNA reverse
transcription primer sequence |
|
|
miRNA-137 |
TTATTGCTTAAGAATACGCGTAG |
| U6
snRNA |
AAAATATGGAACGCTTCACGAATTTG |
| qPCR primer
sequence |
|
| miRNA-137
forward |
CAAGGCTTGTTAACACTGTAAC |
| miRNA-137
reverse |
TCTGTCAATGTCTGAATAAATG |
| U6 snRNA
forward |
CTCGCTTCGGCAGCACATATACT |
| U6 snRNA
reverse |
ACGCTTCACGAATTTGCGTGTC |
| MS-qPCR primer
sequence |
|
|
Methylated alleles
forward |
5′-GCGGTAGTAGTAGCGGTAGC-3′ |
|
Methylated alleles
reverse |
5′-ACCCGTCACCGAAAAAAA-3′ |
|
Unmethylated alleles
forward |
5′-GGTGGTAGTAGTAGTGGTAGT-3′ |
|
Unmethylated alleles
reverse |
5′-TACCCATCACCAAAAAAAA-3′ |
RT-qPCR was performed using LightCycler®
480 SYBR-Green I Master mix on a LightCycler® 480
Real-Time PCR system (both Roche Diagnostics, Basel, Switzerland).
Each 10 µl PCR mixture contained 1 µl 20-fold diluted reverse
transcription product, 5 µl SYBR-Green Master mix, 2 µl RNase-free
water, and 1 µl forward and reverse primers. The reactions were
incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15
sec and 65°C for 60 sec. Relative miRNA-137 expression was
calculated using the 2−ΔΔCq method (21), where ΔCq is the difference in
threshold cycles (Cq) for the target and reference =
Cq(miRNA-137)-Cq(U6). RT and PCR primers were synthesized by
Shanghai Sheng Gong Biology Engineering Technology Service, Ltd.
(Shanghai, China).
Analysis of miR-137 promoter lesion
and sodium bisulfate conversion
The miR-137 CpG is lands were identified using
EMBOSS Software Version 6.3.1 (Institut Pasteur, Paris, France) and
EMBOSS (http://gensoft.pasteur.fr/docs/EMBOSS/6.3.1/). Total
tissue DNA was extracted from FFPE tissue scrolls using the Qiagen
EpiTect Plus FFPE Bisulfite kit (Qiagen, Inc.) according to the
manufacturer's protocol. FFPE tissue scrolls were deparaffinized,
followed by proteinase digestion and de-cross-linking as
aforementioned in RNA isolation paragraph. The DNA bisulfite
reaction was then set up and performed using an ABI Prism 7900HT
Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the Qiagen EpiTect Plus FFPE Bisulfite kit
(Qiagen, Inc.) according to the manufacturer's protocol. Upon
completion of the bisulfite conversion, modified DNA was purified
and eluted, the DNA concentrations were measured using a
NanoDrop-1000 (Thermo Fisher Scientific, Inc.) and the samples were
stored at −20°C for further analysis.
Methylation-specific (MS) qPCR
The methylation status of the miR-137 promoter in
the FFPE tissue sample was determined by MS-qPCR, as previously
described (19). Modified DNA (10 ng)
was subjected to PCR amplification on an ABI Prism 7900HT Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) at 90°C for 5 min, followed by 40 cycles of 95°C for 15 sec,
60°C for 30 sec and 72°C for 15 sec, then a 10 min final extension
at 72°C. The PCR products were diluted 500-fold with water, and 1
µl aliquots were subjected to MS-qPCR using a
LightCycler® 480 SYBR-Green I Master (Roche Diagnostics)
at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and
51°C for 60 sec, calculated with the formula
2−ΔCq[Cq(methylated)-Cq (unmethylated)] (21), and expressed as a percentages. qPCR
primer sequences are listed in Table
I.
Statistical analysis
Data were analyzed using the SPSS 18.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Values presented as the
mean ± standard deviation and were analyzed using one-way analysis
of variance with post hoc analysis by least significant difference
(LSD) test. The correlation between miR-137 expression and tissue
methylation levels was evaluated using Pearson's correlation
analysis. Descriptive statistics were used to compare the
demographic and clinicopathological characteristics (sex, age,
pathology, smoking, status, tumor size, histologic grade, T
category, lymph node metastasis and clinical stage) (22) of the study population stratified by
the miR-137 promoter methylation status, and categorical variables
were compared using analysis of variance with post hoc analysis by
LSD test. The univariate Kaplan-Meier method was used to estimate
disease-free survival and overall survival rates, and survival
differences were compared using the log-rank test. In analysis of
disease-free and overall survival rates, patients who succumbed
prior to recurrence were considered censored at the point of
mortality. A multivariable Cox proportional hazards model was used
to assess the prognostic value of the level of miR-137 promoter
methylation for disease-free and overall survival rates, following
adjustment for sex, age, histological grade, T category, age and
smoking status. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-137 is down regulated in lung
cancer cell lines and human tumor tissues
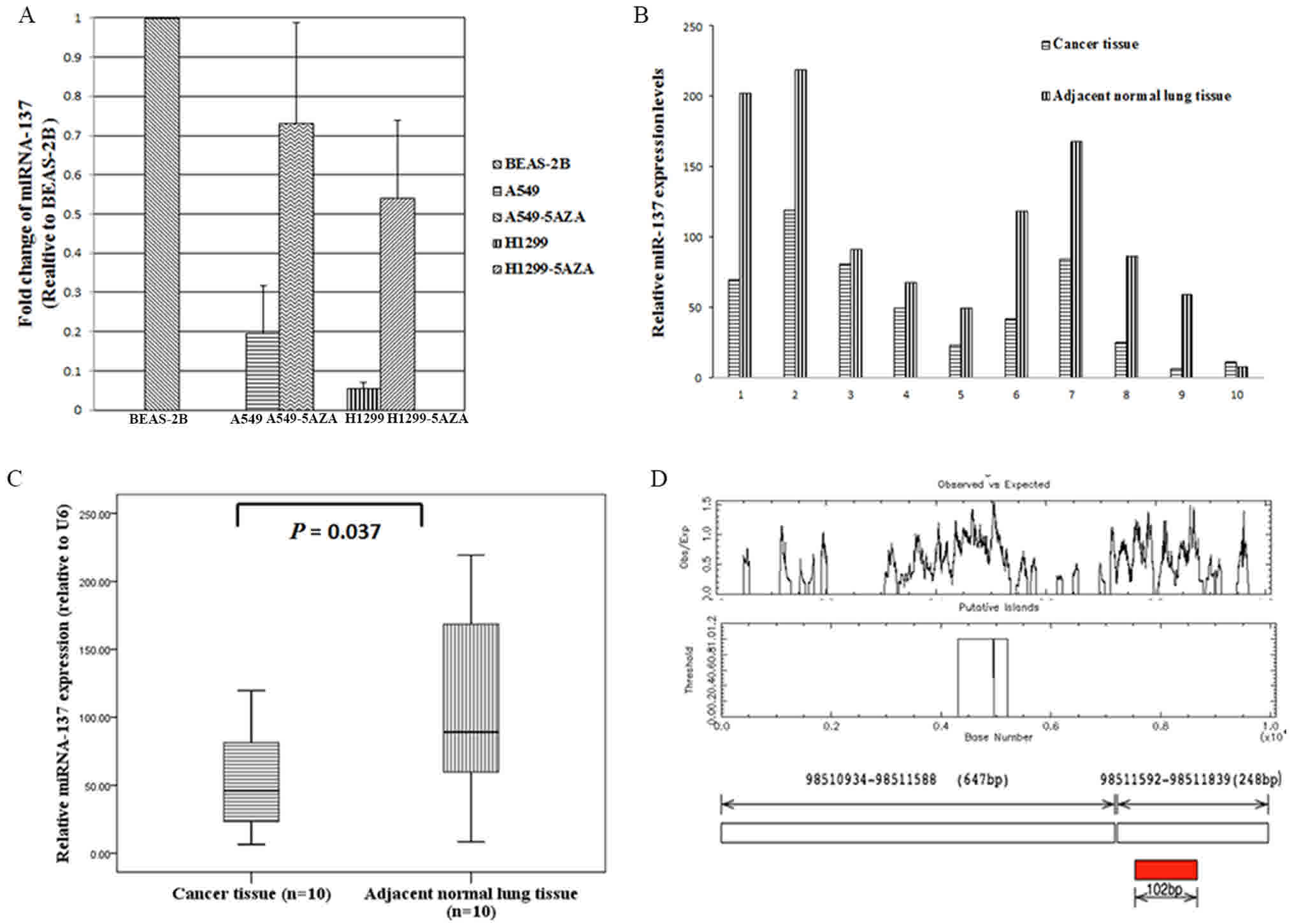
The expression of miR-137 was determined by qRT-PCR
(Fig. 1). miR-137 was markedly
downregulated in lung cancer A549 and H1299 cells compared with
that in normal lung bronchial epithelial BEAS-2B cells (Fig. 1A). In line with the cell line
analyses, the expression of miR-137 was significantly downregulated
in the 10 FFPE lung cancer tissue specimens compared with that in
the paired adjacent normal lung tissues (P=0.037; Fig. 1B and C).
Promoter hypermethylation
downregulated miR-137 in lung cancer cell lines and human tumor
tissues
CpG plot EMBOSS Software version 6.3.1 software
(http://gensoft.pasteur.fr/docs/EMBOSS/6.3.1/; Institut
Pasteur, Paris, France) identified 2 CpG islands located close to
the miR-137 gene (Fig. 1D) and
promoter hypermethylation has previously been reported to be
responsible for the repression of miR-137 (14). Therefore, miR-137 expression was
analyzed in a lung cell line treated with the methyltransferase
inhibitor 5-aza-CdR (5 µmol/l). Untreated control cells expressed
lower levels of miR-137, whereas 5-aza-CdR-treated cells expressed
higher levels of miR-137 (Fig. 1A).
The methylation status of the CpG sites was consistent with the
levels of miR-137 expressed in the lung cancer cell lines.
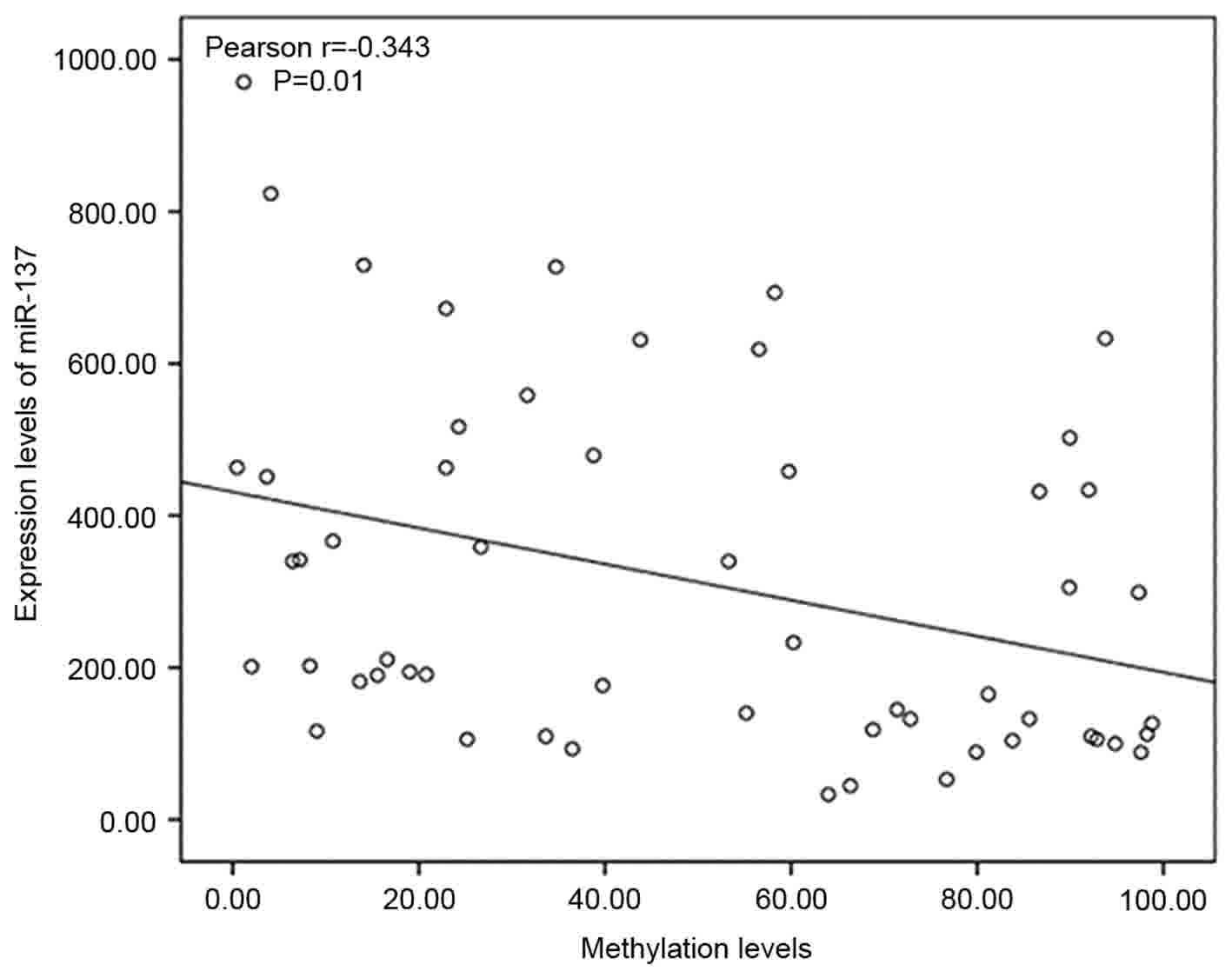
Subsequently, miR-137 expression and the promoter
methylation status in the 56 FFPE lung cancer tissues were compared
using RT-qPCR and MS-qPCR. A significant negative correlation
between the expression level of miR-137 and miR-137 promoter
methylation was observed in the lung cancer tissues (Pearson's
correlation, r=−0.343, P=0.01; Fig.
2), suggesting that promoter methylation silences miR-137 in
lung cancer.
miR-137 promoter methylation status is
associated with lymph node metastasis and advanced clinical
stage
To determine whether the miR-137 promoter
methylation status is associated with lung cancer, the association
between the miR-137 promoter methylation status and the
clinicopathological characteristics of lung cancer were further
analyzed. Low levels of miR-137 promoter methylation were
significantly associated with smoking, positive lymph node
metastasis and advanced clinical stage (P=0.027, P=0.004 and
P=0.021, respectively) (Table II).
There was no significant association between miR-137 promoter
methylation and sex, age, pathology, tumor size, histological grade
or T category (Table II).
 | Table II.Characteristics of 56 patients with
non-small cell lung cancer by microRNA-137 promoter methylation
levels. |
Table II.
Characteristics of 56 patients with
non-small cell lung cancer by microRNA-137 promoter methylation
levels.
| Variable | No. | Mean ± SD |
P-valuea |
|---|
| Sex |
|
| 0.773 |
|
Male | 43 | 49.2±30.1 |
|
|
Female | 13 | 51.8±25.6 |
|
| Age (years) |
|
| 0.508 |
|
<60 | 24 | 46.8±29.0 |
|
|
≥60 | 32 | 52.0±29.0 |
|
| Smoking status |
|
| 0.027 |
| 0 | 18 | 35.8±21.4 |
|
| <20
pack/year | 13 | 50.5±37.6 |
|
| ≥20
pack-y | 25 | 59.5±25.2 |
|
| Histiotype |
|
| 0.643 |
|
Adenocarcinoma | 25 | 48.1±27.6 |
|
|
Squamous cell | 31 | 51.8±30.8 |
|
| T-status |
|
| 0.610 |
| T1 | 30 | 46.9±28.5 |
|
| T2 | 21 | 51.4±31.9 |
|
| T3 | 5 | 60.2±17.0 |
|
| N-status |
|
| 0.004 |
| N0 | 31 | 38.9±26.5 |
|
| N1 | 14 | 60.6±29.9 |
|
| N2 | 11 | 66.8±21.5 |
|
| TNM |
|
| 0.021 |
| I | 26 | 38.8±28.1 |
|
| II | 17 | 57.8±27.1 |
|
|
III | 13 | 61.7±26.2 |
|
|
Differentiation |
|
| 0.621 |
|
Well | 11 | 50.6±27.1 |
|
|
Moderately | 34 | 51.9±29.6 |
|
|
Poorly | 11 | 42.1±29.7 |
|
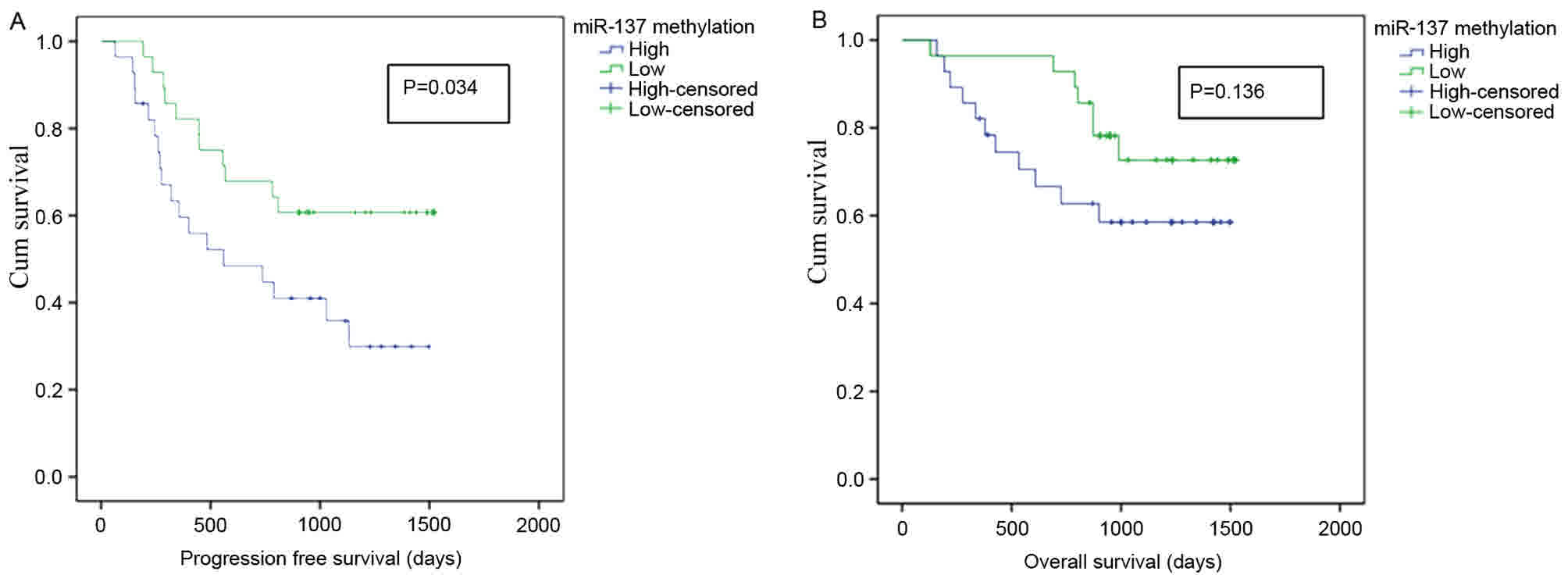
High levels of miR-137 promoter
methylation are associated with poor disease-free survival
Kaplan-Meier survival analysis and multivariate Cox
proportional hazards analysis were employed to evaluate the
association between miR-137 promoter methylation and prognosis in
NSCLC. In the present study, 1 patient succumbed to pneumonia
shortly after surgery and 8 patients were lost to follow-up.
Patients were stratified according to the median relative miR-137
promoter methylation level (43.79%) in the tumor specimens from the
remaining 47 tumor tissues; low miR-137 promoter methylation (n=24;
≤median) and high miR-137 promoter methylation group (n=23;
>median). In univariate Kaplan-Meier analysis, patients with
high levels of miR-137 promoter methylation exhibited significantly
poorer disease-free survival rates (P=0.034; Fig. 3A), but no significant difference was
observed in overall survival rates (P=0.136; Fig. 3B), compared with patients with low
levels of miR-137 promoter methylation.
In the multivariable Cox proportional hazards
analysis, adjusting for gender, age, histologic grade, smoking
status and T stage, the results showed that patients with high
levels of miR-137 promoter methylation had a higher risk of
disease-free death (hazard ratio, 3.333; 95% CI, 1.108–10.028,
P=0.032, Table III) compared with
the patients with low levels of miR-137 promoter methylation;
levels of miR-137 promoter methylation was not significantly
associated with overall survival (P=0.366).
 | Table III.Cox proportional hazards model
analysis of adjusted hazard ratios for progression-free and overall
survival rates according to miRNA-137 promoter methylation levels
in NSCLC patients. |
Table III.
Cox proportional hazards model
analysis of adjusted hazard ratios for progression-free and overall
survival rates according to miRNA-137 promoter methylation levels
in NSCLC patients.
| miR-137 promoter
methylation levels | HR | 95% CI | P-value |
|---|
| Progression-free
survival |
|
|
|
| Low
levels | 1.000 |
|
|
| High
levels | 3.333 | 1.108–10.029 | 0.032 |
| Sex | 0.298 | 0.061–1.453 | 0.134 |
|
Age | 1.081 | 0.302–3.866 | 0.904 |
| Smoking
status | 1.359 | 0.678–2.726 | 0.387 |
|
Histiotype | 2.255 | 0.554–9.172 | 0.256 |
|
T-stage | 0.044 | 1.021–4.919 | 0.044 |
| Overall
survival |
|
|
|
| Low
levels | 1.000 |
|
|
|
High | 2.537 | 0.934–6.890 | 0.366 |
| Sex | 0.341 | 0.070–1.655 | 0.182 |
|
Age | 1.503 | 0.407–5.548 | 0.541 |
| Smoking
status | 1.439 | 0.711–2.913 | 0.311 |
|
Histiotype | 2.497 | 0.572–10.983 | 0.572 |
|
T-stage | 1.554 | 0.781–3.091 | 0.209 |
Discussion
The aim of the present study was to investigate
whether miR-137 promoter hypermethylation is associated with
overall survival and disease-free survival in lung cancer. Although
miR-137 has been reported to exert tumor-suppressor activity in
various types of cancer (12–18), there is limited data on the function
of miR-137 in NSCLC. The present study demonstrated that miR-137 is
downregulated in NSCLC cell lines and tumor tissues.
Several factors may reduce the expression of miRNAs.
DNA methylation of CpG islands is an important regulatory mechanism
for gene expression, which has also been revealed to be responsible
for inactivating the expression of miRNAs, including that of
miRNA-137 (19). miR-137 is
downregulated in the tissues of several types of cancer compared
with normal tissues (11–16,23) and
functions as a tumor suppressor by targeting several genes,
including cyclooxygenase-2, cell division protein kinase 6, cell
division control protein 42 homolog, C-terminal-binding protein 1,
estrogen-related receptor, KIT proto-oncogene receptor tyrosine
kinase, glioma pathogenesis-related protein-1, paxillin and solute
carrier family 22 member 8 (12–15,24–29).
Treatment with the DNA methytransferase inhibitor 5-aza-dC
increased miR-137 expression in A549 and H1299 cells, suggesting
that promoter methylation may be one mechanism that leads to the
silencing of miR-137 in lung cancer, which is consistent with the
study of Kang et al (19). In
agreement with this hypothesis, the level of miR-137 promoter
methylation was significantly correlated with miR-137 expression in
the 56 human lung cancer tissues tested in the present study.
In line with the results of a study undertaken by
Zhang et al (12) on
non-small-cell lung cancer, low levels of miR-137 promoter
methylation were significantly associated with smoking, positive
lymph node status and advanced Tumor-Node-Metastasis stage in human
NSCLC, but not with tumor size, tumor status, sex, differentiation
or histological type. Furthermore, to the best of our knowledge,
the present study provides the first evidence that high levels of
miR-137 promoter methylation are associated with poor disease-free
survival, and multivariate Cox proportional hazards analysis
demonstrated that miR-137 promoter methylation was an independent
prognostic factor for disease-free survival.
The present study has a number of limitations. To
begin with, the role of miR-137 as a tumor-suppressor in lung
cancer was not confirmed. Secondly, further research is required to
investigate the environmental and personal risk factors associated
with miR-137 promoter methylation. Finally, due to the relatively
small sample size and short follow-up in the present study, further
research is required to confirm the association between miR-137
promoter methylation and survival outcomes in NSCLC.
In conclusion, aberrant miR-137 promoter methylation
is a common feature in NSCLC. Further studies should focus on the
quantitative assessment of miR-137 promoter methylation in tumor
tissues and specific types of lung cancer, with the aim of
developing etiological and prognostic markers to prolong survival
in lung cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81302016 and
81302015), the National Natural Science Foundation of Jiangsu
Province, China (grant nos. BK20130456 and BK20140101) and the Six
Talent Peaks Project in Jiangsu, China (grant no. WSN-106).
Availability of data and materials
The datasets used or analyzed during the present
study are available from the corresponding author on reasonable
request.
Author's contributions
Conceptualization, LM, FW and XX. Formal analysis,
LM. Funding acquisition, LM and YC. Methodology, SL, YC and JY.
Resources, LM and SH. Software, LM and SL. Supervision, XX and SH.
Analysis and interpretation of data, SH. Writing-original draft,
LM. Writing-review and editing, XX.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Subei People's Hospital (Yangzhou, China) and written
informed consent was obtained from all participants.
Consent for publication
Written informed consent was obtained from all
participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reck M, Popat S, Reinmuth N, De Ruysscher
D, Kerr KM and Peters S: ESMO Guidelines Working Group: Metastatic
non-small-cell lung cancer (NSCLC): ESMO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 25
Suppl 3:iii27–iii39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Field JK, Oudkerk M, Pedersen JH and Duffy
SW: Prospects for population screening and diagnosis of lung
cancer. Lancet. 382:732–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Minna JD, Roth JA and Gazdar AF: Focus on
lung cancer. Cancer Cell. 1:49–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han L, Witmer PD, Casey E, Valle D and
Sukumar S: DNA methylation regulates MicroRNA expression. Cancer
Biol Ther. 6:1284–1288. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Silber J, Lim DA, Petritsch C, Persson AI,
Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello
JF, et al: miR-124 and miR-137 inhibit proliferation of
glioblastoma multiforme cells and induce differentiation of brain
tumor stem cells. BMC Med. 6(14)2008.PubMed/NCBI
|
|
12
|
Zhang B, Liu T, Wu T, Wang Z, Rao Z and
Gao J: microRNA-137 functions as a tumor suppressor in human
non-small cell lung cancer by targeting SLC22A18. Int J Biol
Macromol. 74:111–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bier A, Giladi N, Kronfeld N, Lee HK,
Cazacu S, Finniss S, Xiang C, Poisson L, deCarvalho AC, Slavin S,
et al: MicroRNA-137 is downregulated in glioblastoma and inhibits
the stemness of glioma stem cells by targeting RTVP-1. Oncotarget.
4:665–676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen L, Wang X, Wang H, Li Y, Yan W, Han
L, Zhang K, Zhang J, Wang Y, Feng Y, et al: miR-137 is frequently
down-regulated in glioblastoma and is a negative regulator of
Cox-2. Eur J Cancer. 48:3104–3111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Q, Chen X, Zhang M, Fan Q, Luo S and
Cao X: miR-137 is frequently down-regulated in gastric cancer and
is a negative regulator of Cdc42. Dig Dis Sci. 56:2009–2016. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Balaguer F, Link A, Lozano JJ, Cuatrecasas
M, Nagasaka T, Boland CR and Goel A: Epigenetic silencing of
miR-137 is an early event in colorectal carcinogenesis. Cancer Res.
70:6609–6618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Steponaitiene R, Kupcinskas J, Langner C,
Balaguer F, Venclauskas L, Pauzas H, Tamelis A, Skieceviciene J,
Kupcinskas L, Malfertheiner P and Link A: Epigenetic silencing of
miR-137 is a frequent event in gastric carcinogenesis. Mol
Carcinog. 55:376–386. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Langevin SM, Stone RA, Bunker CH,
Lyons-Weiler MA, LaFramboise WA, Kelly L, Seethala RR, Grandis JR,
Sobol RW and Taioli E: MicroRNA-137 promoter methylation is
associated with poorer overall survival in patients with squamous
cell carcinoma of the head and neck. Cancer. 117:1454–1462. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kang N, Choi SY, Kim YK, Yoo IeR, Han DH,
Lee DS, Kim YS, Hong SH, Kang JH, Lee KY, et al: Silencing of
miR-137 by aberrant promoter hypermethylation in surgically
resected lung cancer. Lung Cancer. 89:99–103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reddel RR, Ke Y, Gerwin BI, McMenamin MG,
Lechner JF, Su RT, Brash DE, Park JB, Rhim JS and Harris CC:
Transformation of human bronchial epithelial cells by infection
with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via
strontium phosphate coprecipitation with a plasmid containing SV40
early region genes. Cancer Res. 48:1904–1909. 1988.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shepherd FA, Crowley J, Van Houtte P,
Postmus PE, Carney D, Chansky K, Shaikh Z and Goldstraw P;
International Association for the Study of Lung Cancer
International Staging Committee and Participating Institutions, .
The international association for the study of lung cancer lung
cancer staging project: Proposals regarding the clinical staging of
small cell lung cancer in the forthcoming (seventh) edition of the
tumor, node, metastasis classification for lung cancer. J Thorac
Oncol. 2:1067–1077. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dacic S, Kelly L, Shuai Y and Nikiforova
MN: miRNA expression profiling of lung adenocarcinomas: Correlation
with mutational status. Mod Pathol. 23:1577–1582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bemis LT, Chen R, Amato CM, Classen EH,
Robinson SE, Coffey DG, Erickson PF, Shellman YG and Robinson WA:
MicroRNA-137 targets microphthalmia-associated transcription factor
in melanoma cell lines. Cancer Res. 68:1362–1368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng Y, Deng H, Bi F, Liu J, Bemis LT,
Norris D, Wang XJ and Zhang Q: MicroRNA-137 targets
carboxyl-terminal binding protein 1 in melanoma cell lines. Int J
Biol Sci. 7:133–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y
and He F: miR-137 targets estrogen-related receptor alpha and
impairs the proliferative and migratory capacity of breast cancer
cells. PLoS One. 7:e391022012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu X, Li Y, Shen H, Li H, Long L, Hui L
and Xu W: miR-137 inhibits the proliferation of lung cancer cells
by targeting Cdc42 and Cdk6. FEBS Lett. 587:73–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bi Y, Han Y, Bi H, Gao F and Wang X:
miR-137 impairs the proliferative and migratory capacity of human
non-small cell lung cancer cells by targeting paxillin. Hum Cell.
27:95–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li P, Ma L, Zhang Y, Ji F and Jin F:
MicroRNA-137 down-regulates KIT and inhibits small cell lung cancer
cell proliferation. Biomed Pharmacother. 68:7–12. 2014. View Article : Google Scholar : PubMed/NCBI
|