Introduction
Glioma is the most common type of brain tumor in
children and adults (1). Stage IV
glioblastoma (GBM), graded according to the World Health
Organization tumor classification system, as having a duration of
12–15 months (2). The 5-year survival
rate of patients with stage IV GBM is <5%, despite the
administration of chemotherapy, radiotherapy, surgical resection
and other intensive treatment modalities. As a result, it is
necessary to develop novel efficacious therapies to support the
continuous improvement of the prognosis of patients with GBM
(3).
MicroRNAs (miRNAs/miRs) are non-coding, small,
endogenous RNAs that regulate the expression of a number of genes
by specific antisense complementarity with target mRNAs. These
molecules can act as oncogenes and tumor suppressors (4). Previous studies have indicated that
miRNAs regulate a number of different biological processes,
including the invasion, apoptosis, proliferation and
differentiation of cells (1,5–7). Recent
studies have identified and implicated the miRNAs involved in the
progression of various cancer types as novel targets for anticancer
therapies (4–7).
The epithelial-mesenchymal-transition (EMT) involves
the transdifferentiation of epithelial cells into mesenchymal
cells. This process has been implicated in the progression of
cancer, including metastasis and invasion (8,9). The
present study demonstrated that EMT is closely associated with
malignant progression and clinical outcome in patients with glioma,
and identified the EMT biological processes associated with the
miRNA profile of GBM, which may provide potential novel targets for
GBM therapy.
Materials and methods
Microarray data and bioinformatics
analysis
Microarray data from the GSE16011 and Rembrandt
datasets were collected (10,11). The Rembrandt dataset (Affymetrix
GeneChip Human Genome U133 Plus 2.0 Array) and the CEL files for
GSE16011 were used, and the data were separately merged using
Matlab software R2012a (Mathworks, Inc., Natick, MA, USA). The
expression data were normalized according to the robust multi-array
average normalization. The array data from The Cancer Genome Atlas
(TCGA) GBM Agilent miRNAs (gene expression level 3) and HG-U133A
gene expression mRNAs (gene expression level 3) were downloaded
from TCGA Data Portal (12).
Gene expression signatures were used in the present
study to define the process in which epithelial cells transition to
the mesenchymal cells, based on a meta-analysis of gene expression
studies (GES) (13). A total of 130
downregulated or upregulated genes from the EMT-core-gene list,
with >10 GES (EMT_up and EMT_down genes) were used in the
present study. EMT_up and EMT_down gene set enrichment scores in
the gene expression microarray were assessed using gene set
variation analysis (GSVA) (14).
Oligonucleotide transfection and cell
culture
Human glioma U251 and LN229 cell lines were obtained
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China) and cultured in Dulbecco's modified Eagle's
medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
culture medium supplemented with 10% fetal bovine serum (FBS;
Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C and
5% CO2. The cells were regularly passaged at 2–3 day
intervals. Cells at passages 2–4 were used in the present
study.
The miR-95 and miR-223 mimic
(5′-UCAAUAAAUGUCUGUUGAAUU-3′ for miR-95 and
5′-CGUGUAUUUGACAAGCUGAGUU-3′ for miR-223) and control
(5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by Shanghai
GenePharma Co., Ltd., (Shanghai, China). Oligonucleotide
transfection was performed using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) in U251 and LN229 cells at 70–90% confluence. On the basis of
the manufacturer's instructions, the transfection complexes were
prepared and subsequently added to the glioma cells to obtain a
10-nmol/l final oligonucleotide concentration. The transfection
medium was replaced at 8 h post-transfection.
Invasion assays
Cell invasion assays were performed using Transwell
membranes coated with Matrigel (BD Biosciences, Franklin Lakes, NJ,
USA). According to the manufacturer's instructions, 500 µl DMEM
without serum was used to prehydrate the 24-well invasion chambers
(8.0 µm; BD Biosciences) for 2 h at 37°C with 5% CO2. In
DMEM without serum, the cells were seeded at a density of
5×104 cells/well in the upper chamber. The lower chamber
was filled with 500 µl 20% FBS as a chemo-attractant. The
non-migrating cells were removed from the top well using a cotton
swab following incubation for 24 h. The cells in the bottom chamber
were then fixed using 75% alcohol (37°C for 5 min) and subsequently
stained the cells with 0.1% crystal violet (37°C for 1 min). The
cells in each field of view migrating towards the bottom side
across the filter were counted at a magnification of ×100 (light
microscope) to quantify glioma cell migration. A total five fields
of view were counted in this experiment. Three independent
experiments were conducted.
Reverse transcription quantitative PCR
(RT-qPCR)
RNA was extracted from U251 and LN229 glioma cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Total RNA was converted
into a cDNA template using PrimeScript™ RT reagent kit (Takara Bio,
Inc., Otsu, Japan). N-cadherin mRNA level was analyzed by RT-qPCR
on the ABI 7300 HT Sequence Detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using the SYBR®
PrimeScript™ RT-PCR kit (Takara Bio, Inc.). N-cadherin were
amplified with the primer: 5′-GGTGGAGGAGAAGAAGACCAG-3′ (Sense) and
5′-GGCATCAGGCTCCACAGT-3′ (Antisense). The amplification of β-actin
with primer: 5′-AAGACCTGTACGCCAACACAGT-3′ (Sense) and
5-AGAAGCATTTGCGGTGGACGAT-3′ (Antisense) was taken as an internal
control. Relative gene expression was calculated via the
2−ΔΔCq method (15).
Statistical analysis
Significant differences were calculated using
one-way analysis of variance followed by the Student-Newman-Keuls
method for multi-group comparisons; Student's t-test was used to
perform two-group comparisons. Data are presented as the mean ±
standard deviation. SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was
used to perform statistical analysis, other than Pearson's
correlation analysis, which was conducted using Matlab, and the
log-rank test of Kaplan-survival curves for survival analysis,
which was performed using GraphPad Prism 6 (GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
EMT is closely associated with
malignant progression and clinical outcomes in glioma
In the present study, the association of EMT with
malignant progression and clinical outcome was investigated. The
EMT_up and EMT_down gene set enrichment scores in the gene
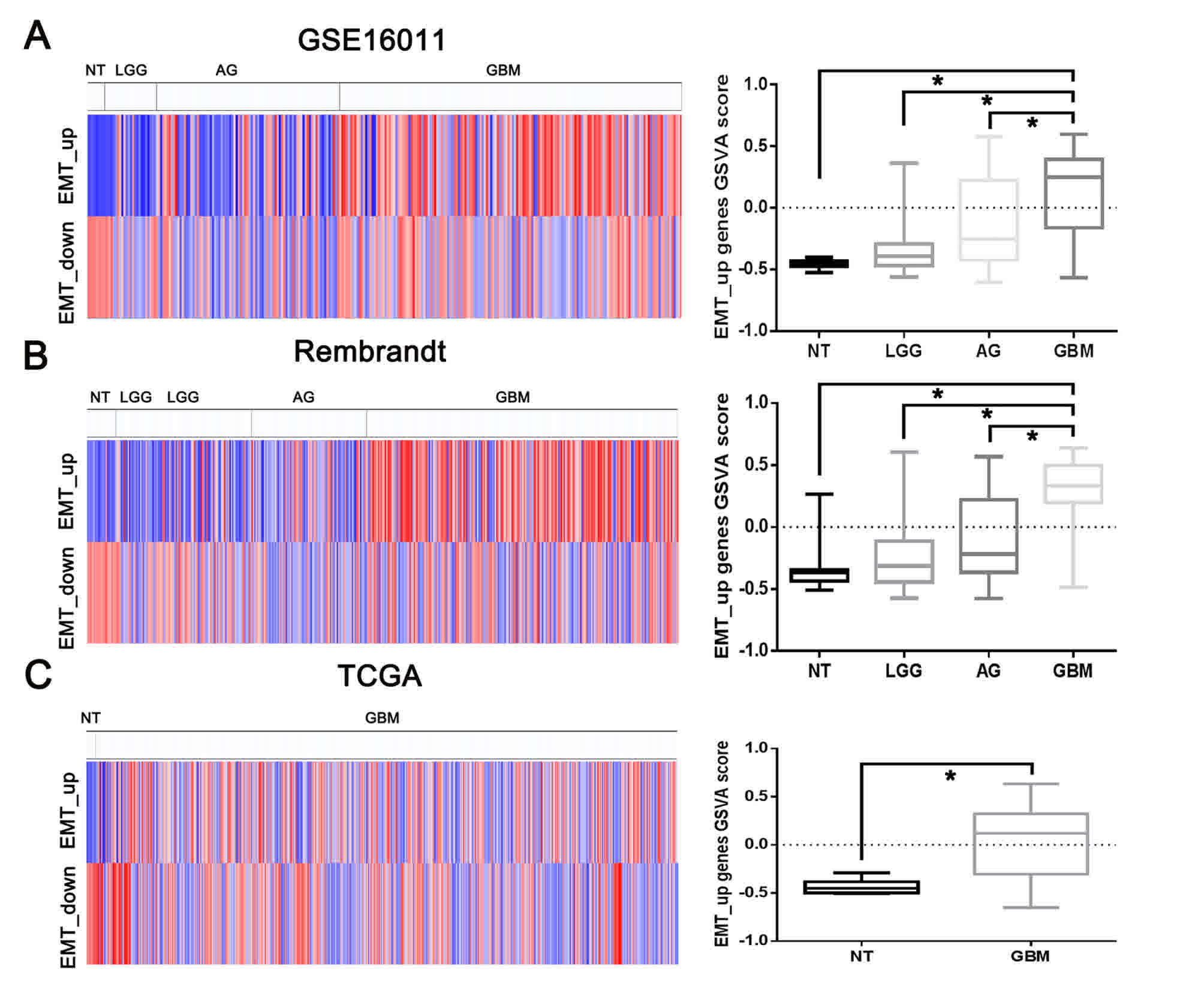
expression microarray were assessed using GSVA. As depicted in
Fig. 1, the EMT_up geneset enrichment
score was significantly upregulated in gliomas, compared with
non-tumor tissues, and the increasing expression of EMT_up geneset
enrichment score was significantly associated with the grade of
glioma malignancy in the GSE16011 and Rembrandt datasets. In TCGA,
the EMT_up gene set enrichment score was also increased in GBMs,
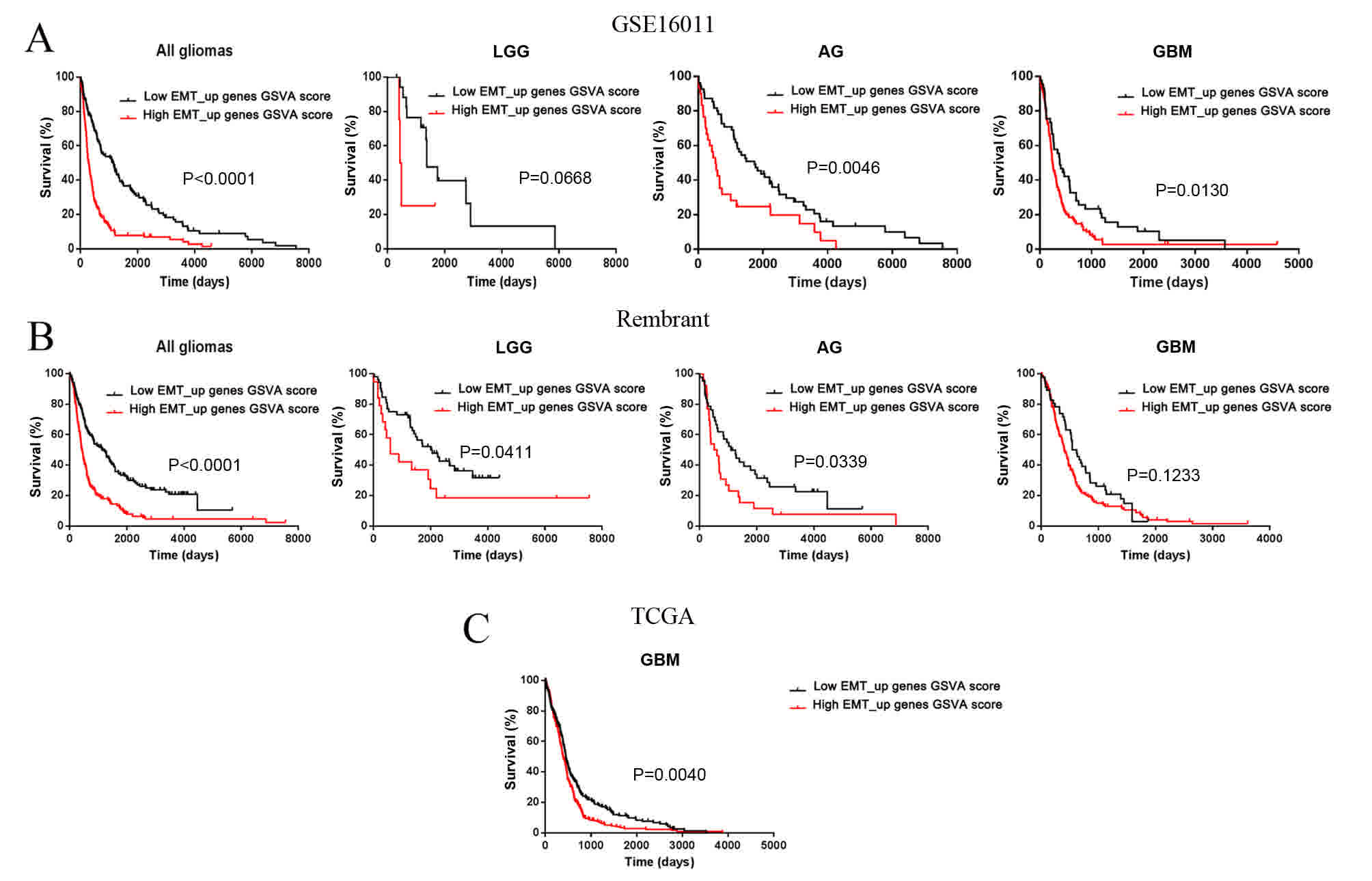
compared with non-tumor tissues. Furthermore, the EMT_up geneset
enrichment score could predict the clinical consequences in
patients with low-grade gliomas, GBMs and anaplastic gliomas
(Fig. 2).
Identification of EMT biological
process associated with miRNA profiles
To analyze aberrant gene expression during EMT, the
paired profiling data of miRNAs and mRNA profiling (level 3) were
downloaded from TCGA. A total of 491 TCGA GBM samples were examined
in the present study. Matlab software was used to calculate the
Pearson's correlation to determine the association between the
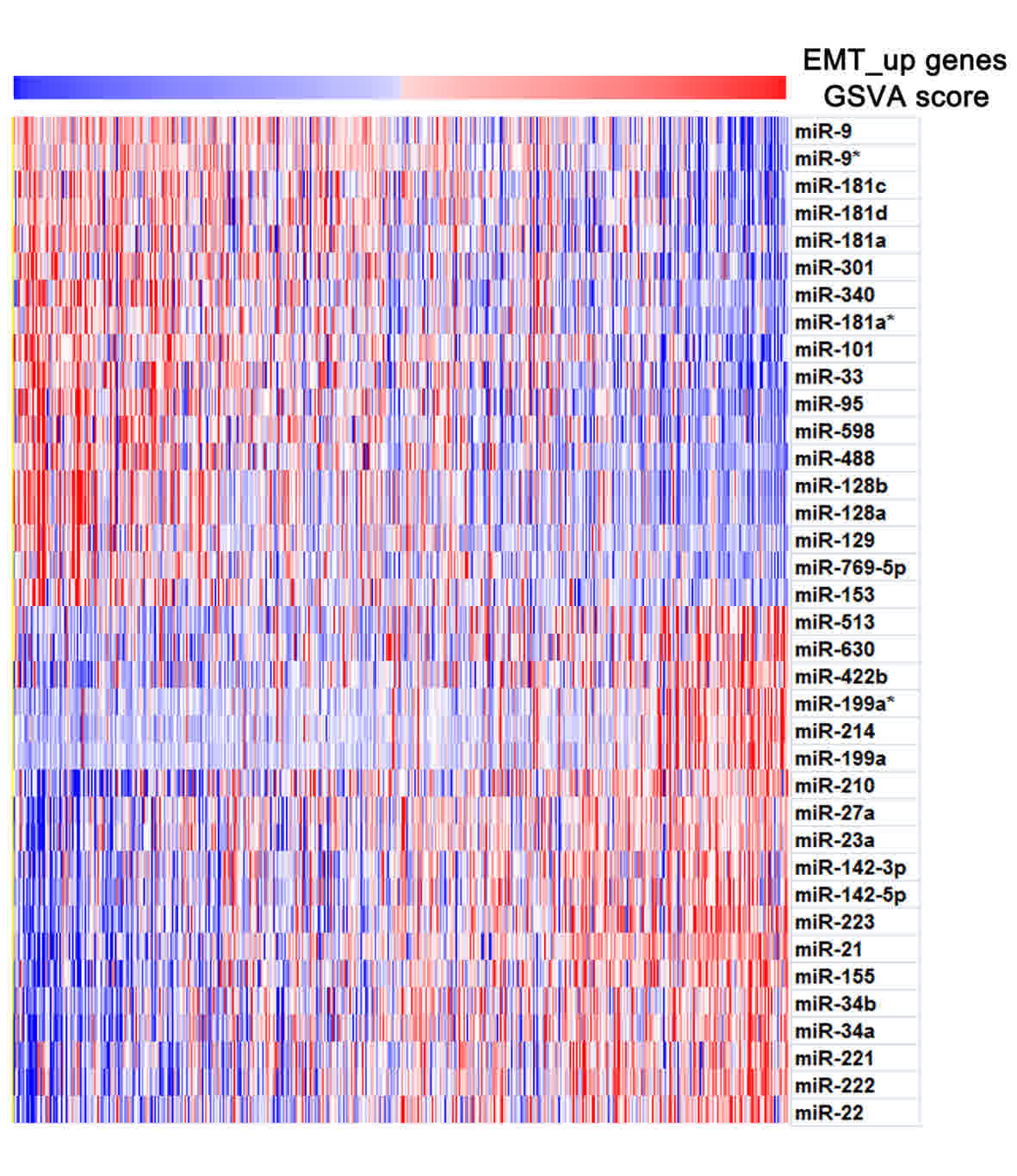
miRNAs and the EMT_up gene set enrichment score. miRNAs exhibited a
high correlation with the EMT_up gene set enrichment score
(P<0.01, r<-0.3 and r>0.3 correlated with EMT_up gene set
enrichment score, Table I and
Fig. 3), and were considered as the
EMT-specific miRNA signature. As depicted in Table I and Fig.
3, the EMT-specific miRNA signature included 18 and 19 miRNAs
negatively and positively correlated with EMT_up gene set
enrichment scores, respectively. In the present study, several
well-characterized tumorigenesis-associated miRNAs, including
miR-21-, miR-181- and miR-128-family members, were detected. During
EMT, these molecules exhibited the same, decreased or increased
expression. Furthermore, a number of miRNAs with currently unknown
functions, including miR-95, miR-155 and miR-223, exhibited similar
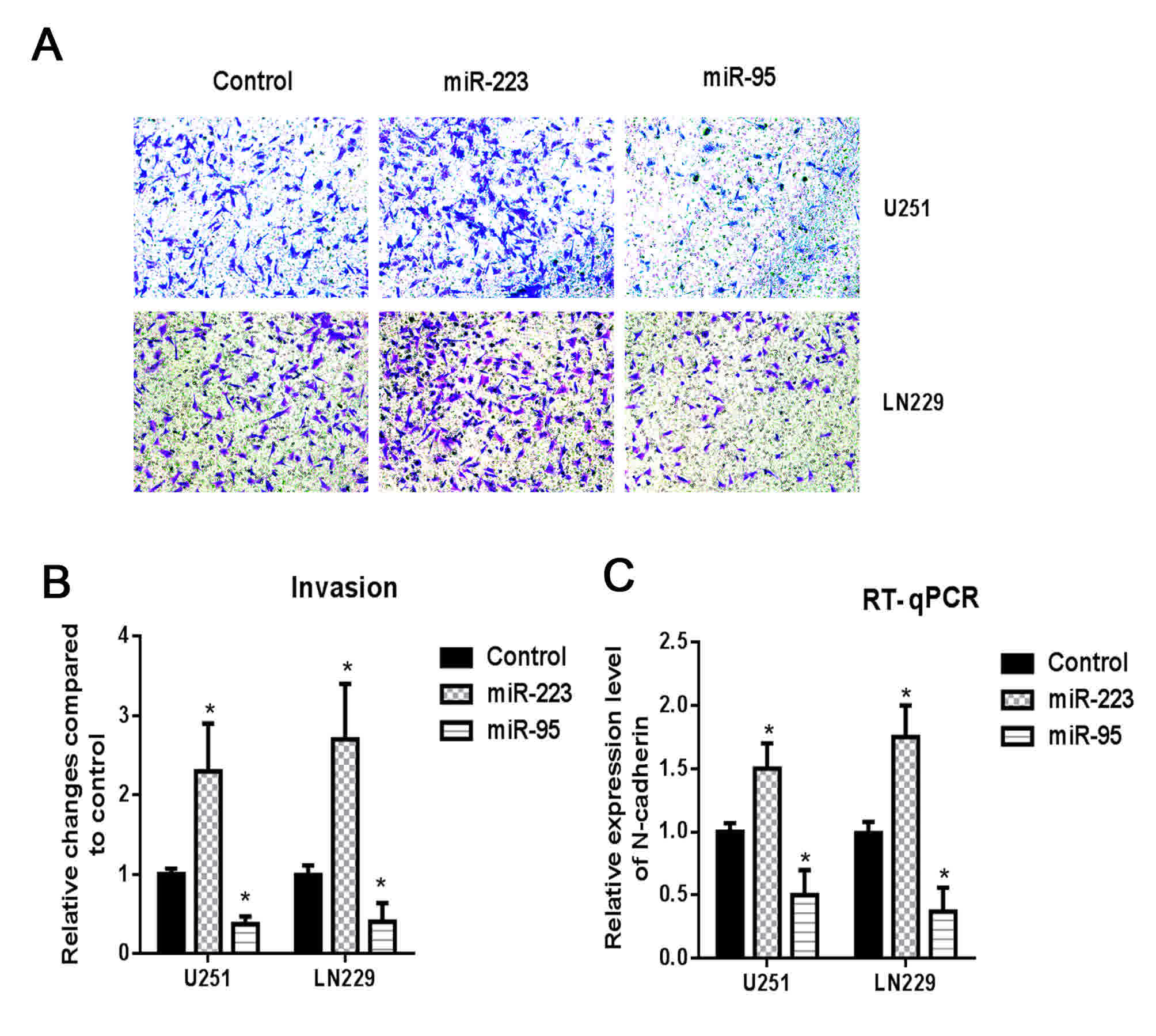
characteristics. Of these molecules, two miRNAs, miR-223 and
miR-95, exhibited a high positive and negative correlation with
EMT, respectively, and were selected for functional validation. The
upregulation of miR-95 was demonstrated to inhibit cellular
invasion in U251 and LN229 glioma cells and reduce the expression
of the mesenchymal marker N-catenin, whereas miR-223 was
demonstrated to have the opposite effect (Fig. 4).
 | Table I.Specific miRNA signature of the
epithelial-mesenchymal transition biological process. |
Table I.
Specific miRNA signature of the
epithelial-mesenchymal transition biological process.
| Negatively correlated
miRNAs | Positively correlated
miRNAs |
|---|
|
|
|---|
| ID | R-value | P-value | ID | R-value | P-value |
|---|
| miR-128b | −0.4205 |
1.85×10−22 | miR-21 | 0.4774 |
2.53×10−29 |
| miR-95 | −0.4185 |
3.09×10−22 | miR-223 | 0.4715 |
1.53×10−28 |
| miR-128a | −0.4167 |
4.78×10−22 | miR-155 | 0.4384 |
1.78×10−24 |
| miR-9 | −0.3815 |
1.85×10−18 | miR-222 | 0.4046 |
9.10×10−21 |
| miR-340 | −0.3596 |
1.96×10−16 | miR-34a | 0.3978 |
4.62×10−20 |
| miR-9* | −0.3581 |
2.67×10−16 | miR-199a | 0.3773 |
4.71×10−18 |
| miR-101 | −0.3555 |
4.47×10−16 | miR-22 | 0.3679 |
3.47×10−17 |
| miR-301 | −0.3534 |
6.93×10−16 | miR-214 | 0.3599 |
1.86×10−16 |
| miR-488 | −0.3523 |
8.54×10−16 | miR-210 | 0.3441 |
4.30×10−15 |
| miR-181c | −0.3495 |
1.48×10−15 | miR-199a* | 0.3400 |
9.44×10−15 |
| miR-33 | −0.3431 |
5.20×10−15 | miR-142-5p | 0.3356 |
2.17×10−14 |
| miR-181d | −0.3351 |
2.37×10−14 | miR-221 | 0.3284 |
8.22×10−14 |
| miR-598 | −0.3320 |
4.22×10−14 | miR-422b | 0.3279 |
9.07×10−14 |
| miR-181a | −0.3177 |
5.63×10−13 | miR-34b | 0.3271 |
1.04×10−13 |
| miR-181a* | −0.3166 |
6.82×10−13 | miR-23a | 0.3201 |
3.65×10−13 |
| miR-769-5p | −0.3068 |
3.68×10−12 | miR-27a | 0.3192 |
4.30×10−13 |
| miR-129 | −0.3021 |
8.08×10−12 | miR-142-3p | 0.3181 |
5.23×10−13 |
| miR-153 | −0.3009 |
9.83×10−12 | miR-630 | 0.3090 |
2.53×10−12 |
|
|
|
| miR-513 | 0.3011 |
9.46×10−12 |
Discussion
miRNAs exhibit different expression signatures that
exert an influence on the behaviors of a number of cancer cell
types (16). EMT involves the
conversion of cells to a mesenchymal phenotype from an epithelial
phenotype, which leads to increased chemo-resistance and high cell
mobility and therefore represents a notable event during the
dissemination and progression of cancer (13). However, the role of EMT and the
patterns of miRNA expression involved in the EMT of gliomas have
not sufficiently been investigated to date. The present study
observed that EMT was positively associated with malignant
progression and clinical outcome in three independent glioma
datasets, GSE16011, Rembrandt and TCGA. Furthermore, integrated
analysis of miRNAs and the profiling of mRNAs 491 GBM samples were
performed, which revealed that the EMT-associated miRNA profile may
provide potential novel targets for GBM therapy.
The progression of the majority of carcinomas
towards malignancy is associated with an increase in the
mesenchymal phenotype and a loss of epithelial differentiation,
accompanied by increased invasion and mobility (17). In the present study, the clinical
significance of EMT was analyzed in three independent glioma
datasets, GSE16011, Rembrandt and TCGA. The results demonstrated
that EMT was closely associated with malignant progression in
gliomas, as previously proposed (8,9). These
results indicated that EMT was also involved in the malignant
transformation of glioma.
miRNAs inhibit EMT-associated phenotypic changes in
a number of different cancer types, and can provide anticancer
therapies with a novel target (18).
In the present study, the EMT-associated miRNA profile was
identified through the integrated analysis of miRNAs and mRNA
profiling in 491 TCGA GBM samples. Among the miRNAs profiled in the
current study, a number of well-characterized
tumorigenesis-associated miRNAs, including miR-21, miR-181 and
miR-128 family members, and numerous miRNAs with presently unknown
functions, including miR-95, miR-155 and miR-223, also exhibited
similarly decreased or increased expression during EMT. A number of
previous studies have recognized EMT as an early event of
invasion/metastasis (8,9,13). The
upregulation of miR-95, which is negatively correlated with EMT,
was demonstrated to inhibit cellular invasion in U251 and LN229
glioma cells and reduce the levels of N-catenin expression, whereas
miR-223, which was positively correlated with EMT, produced the
opposite effects. However, only the mRNA expression level of
N-cadherin, and not that of the protein, was examined in the
present study, which is a limitation of the current study. The
protein expression of N-cadherin should therefore be studied
further in future. These results indicated that the EMT-specific
miRNA expression profile may provide potential targets for GBM
therapy.
In summary, the present study demonstrated that EMT
was closely associated with malignant progression and clinical
outcome in glioma. To the best of our knowledge, the current study
provided the first evidence of an EMT-specific miRNA expression
profile, which may provide potential targets for GBM therapy. These
data can aid the identification of therapeutic and prognostic
markers. Additional studies are required to characterize the roles
of the miRNAs involved and to identify further miRNAs as molecular
targets of therapy for patients with GBM.
Acknowledgements
The authors would like to thank Dr. Lei Shi for
editing the grammar of the paper.
Funding
This work was financially supported by a grant from
the National Natural Science Foundation of China (grant no.
81472362).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and YY concieved and designed the study. YZ, AZ,
SL, XW and WY downloaded the gene expression data and performed the
bioinformatics analysis. RL, AZ and SL performed the in
vitro experiments. YZ, RL, XW and WY wrote the manuscript. All
authors gave the final approval of the version to be published.
Ethics approval and consent to
participate
All patients provided signed informed consent and
the study was approved by the institutional Review Board of Nanjing
Medical University.
Consent for publication
Written informed consent was obtained from all
participants for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yan W, Zhang W and Jiang T: Oncogene
addiction in gliomas: Implications for molecular targeted therapy.
J Exp Clin Cancer Res. 30:582011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taylor TE, Furnari FB and Cavenee WK:
Targeting EGFR for treatment of glioblastoma: Molecular basis to
overcome resistance. Curr Cancer Drug Targets. 12:197–209. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C, Feng Y, Coukos G and Zhang L:
Therapeutic microRNA strategies in human cancer. AAPS J.
11:747–757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu
Z and You Y: hsa-mir-181a and hsa-mir-181b function as tumor
suppressors in human glioma cells. Brain Res. 1236:185–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Zhang J, Hoadley K, Kushwaha D,
Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, et al:
miR-181d: A predictive glioblastoma biomarker that downregulates
MGMT expression. Neuro Oncol. 14:712–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang XF, Shi ZM, Wang XR, Cao L, Wang YY,
Zhang JX, Yin Y, Luo H, Kang CS, Liu N, et al: MiR-181d acts as a
tumor suppressor in glioma by targeting K-ras and Bcl-2. J Cancer
Res Clin Oncol. 138:573–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gabriely G, Wurdinger T, Kesari S, Esau
CC, Burchard J, Linsley PS and Krichevsky AM: MicroRNA 21 promotes
glioma invasion by targeting matrix metalloproteinase regulators.
Mol Cell Biol. 28:5369–5380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan W, Zhang W, Sun L, Liu Y, You G, Wang
Y, Kang C, You Y and Jiang T: Identification of MMP-9 specific
microRNA expression profile as potential targets of anti-invasion
therapy in glioblastoma multiforme. Brain Res. 1411:108–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gravendeel LA, Kouwenhoven MC, Gevaert O,
de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB,
Kloosterhof NK, et al: Intrinsic gene expression profiles of
gliomas are a better predictor of survival than histology. Cancer
Res. 69:9065–9072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Madhavan S, Zenklusen JC, Kotliarov Y,
Sahni H, Fine HA and Buetow K: Rembrandt: Helping personalized
medicine become a reality through integrative translational
research. Mol Cancer Res. 7:157–167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gröger CJ, Grubinger M, Waldhör T,
Vierlinger K and Mikulits W: Meta-analysis of gene expression
signatures defining the epithelial to mesenchymal transition during
cancer progression. PLoS One. 7:e511362012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kong D, Li Y, Wang Z and Sarkar FH: Cancer
stem cells and epithelial-to-mesenchymal transition
(EMT)-phenotypic cells: Are they cousins or twins? Cancers (Basel).
3:716–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Li Y, Ahmad A, Azmi AS, Kong D,
Banerjee S and Sarkar FH: Targeting miRNAs involved in cancer stem
cell and EMT regulation: An emerging concept in overcoming drug
resistance. Drug Resist Updat. 13:109–118. 2010. View Article : Google Scholar : PubMed/NCBI
|