Introduction
Colorectal cancer (CRC) is the most predominant
malignant digestive tumor and fourth leading cause of
cancer-associated mortality globally (1). The rate of recurrence and metastasis
varies among patients with CRC, even between those at the same
stages (2,3). The lack of an ideal biological marker
for the prognosis of patients with advanced colorectal cancer makes
individualized treatment difficult (4). Therefore, the identification of novel
markers and therapeutic targets for CRC is urgently required.
Transcription factor AP4 (TFAP4; also termed AP4 or
AP-4) is a ubiquitously-expressed transcription factor belonging to
the basic helix-loop-helix (bHLH)/leucine zipper subgroup of bHLH
proteins, which exclusively form homodimers that bind to the CAGCTG
E-box motif (5–7). It has been suggested that TFAP4 is
involved in multiple essential cell processes, including cell
proliferation, mitotic division, cell cycle progression and
apoptosis (8–10). TFAP4 target genes also include stem
cell markers including LGR5, and regulators of
epithelial-mesenchymal transition including Snail Family
Transcriptional Repressor 1 and Epithelial cadherin (E-cadherin)
(11,12). In addition, TFAP4 is encoded by a
c-MYC target gene and is concomitantly upregulated with c-MYC in
CRC and breast cancer (11).
Previous studies have demonstrated that TFAP4 is
overexpressed in breast cancer, CRC and hepatocellular carcinoma,
and that it is implicated in tumor prognosis (13–15).
However, few studies have analyzed the clinical and functional
roles of TFAP4 in CRC. The present study investigated TFAP4
expression in CRC and evaluated its prognostic significance in 187
CRC samples. Changes in CRC cell phenotype were studied by
knockdown and overexpression of TFAP4. The results indicated that
TFAP4 is involved in CRC progression, and as a result, targeted
inhibition of TFAP4 may represent a novel approach for the
treatment of CRC.
Materials and methods
Patients
The present study included 187 patients with CRC who
underwent surgery between January 2007 and January 2008 in the
Department of Gastrointestinal Surgery at the First Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China). Patients
were included in the present study if they had undergone a curative
operation and had histologically proven colorectal cancer, and if
they had not received preoperative chemotherapy and/or
radiotherapy. Patients who succumbed to the disease within 3 months
of surgical resection were not included. Information on
clinicopathological characteristics, including age, sex, tumor
size, differentiation, lymph node metastasis and liver metastasis,
were retrieved from an inpatient database and are presented in
Table I. Clinical and pathological
classification and staging were determined according to the
American Joint Committee on Cancer tumor-node-metastasis (TNM)
staging system (16). Oncological
outcome data were collected by telephone and the outpatient
database every 3 months for the first year, every 6 months for the
second to fifth year and every 12 months after 5 years. Only
mortalities resulting specifically from CRC were defined as
clinical endpoints.
 | Table I.Association between TFAP4 expression
and the clinicopathological characteristics of patients with
colorectal cancer. |
Table I.
Association between TFAP4 expression
and the clinicopathological characteristics of patients with
colorectal cancer.
|
|
| TFAP4 expression, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variable | Total, n | Low or absent | High | P-valuea |
|---|
| Sex |
|
|
| 0.544 |
| Male | 109 | 58 (56.3) | 51 (60.7) |
|
|
Female | 78 | 45 (43.7) | 33 (39.3) |
|
| Age, years |
|
|
| 0.383 |
|
<60 | 80 | 47 (45.6) | 33 (39.3) |
|
| ≥60 | 107 | 56 (54.7) | 51 (60.7) |
|
| Location of primary
tumor |
|
|
| 0.047 |
|
Colon | 104 | 64 (62.1) | 40 (47.6) |
|
|
Rectal | 83 | 39 (37.8) | 44 (52.4) |
|
| Tumor size, cm |
|
|
| 0.632 |
|
<5 | 106 | 60 (58.3) | 46 (54.8) |
|
| ≥5 | 81 | 43 (41.7) | 38 (45.2) |
|
| T
classification |
|
|
| 0.276 |
| T1 +
T2 | 33 | 21 (20.4) | 12 (14.3) |
|
| T3 +
T4 | 154 | 82 (79.6) | 72 (85.7) |
|
| Lymph node
invasion |
|
|
| <0.001 |
| No | 116 | 77 (74.8) | 39 (46.4) |
|
|
Yes | 71 | 26 (25.2) | 45 (53.6) |
|
| Metastasis |
|
|
| <0.001 |
| M0 | 163 | 97 (94.2) | 64 (76.2) |
|
| M1 | 26 | 6 (5.8) | 20 (23.8) |
|
| TNM stage |
|
|
| <0.001 |
|
I–II | 107 | 74 (71.8) | 33 (39.3) |
|
|
III–IV | 80 | 29 (28.2) | 51 (60.7) |
|
| Pathological
differentiation |
|
|
| 0.024 |
|
Well | 14 | 12 (11.7) | 2 (2.4) |
|
|
Moderate | 138 | 76 (73.8) | 62 (73.8) |
|
|
Poor | 35 | 15 (14.6) | 20 (23.8) |
|
| Therapeutic
strategy |
|
|
| 0.392 |
| Surgery
only | 111 | 64 (62.1) | 47 (56.0) |
|
| Surgery
+ chemotherapy | 76 | 39 (37.9) | 37 (44.0) |
|
| Vital status (as
followed up) |
|
|
| <0.001 |
|
Alive | 124 | 83 (80.6) | 41 (48.8) |
|
|
Dead | 63 | 20 (19.4) | 43 (51.2) |
|
All 187 patient specimens selected for
immunohistochemical analysis were fixed in 10% formalin at room
temperature for 24–48 h and embedded in paraffin. In addition, 30
pairs of fresh tumor tissues with matched adjacent normal tissues
were stored in liquid nitrogen (at-196°C) until further use.
Written informed consent was obtained from all patients for the use
of their tissue samples. The Ethics Committee of the First
Affiliated Hospital of Sun Yat-sen University approved the protocol
of the present study.
Immunohistochemistry (IHC)
Immunohistochemical staining was used to determine
the expression levels of TFAP4 in CRC samples. Briefly, serial 4-mm
sections were cut from paraffin-embedded blocks. For antigen
retrieval, following deparaffinization and rehydration for 30 min
at 50°C using a descending alcohol series (100 for 5 min, 100 for 5
min, 95 for 5 min, 80 for 5 min, 70% for 5 min, then two rinses
with distilled water for 5 min each), sections were heated in 10
mmol/l citrate buffer (pH 6.0) at 125°C for 5 min and then cooled
at room temperature. To block endogenous peroxidase, sections were
treated in 0.3% hydrogen peroxide solution for 15 min at room
temperature. Following incubation with goat serum (Maxim Biotech,
Inc., Rockville, MD, USA) for 30 min at 37°C, polyclonal rabbit
TFAP4 antibodies (dilution, 1:100; cat no. SAB1404461; Aviva
Systems Biology Corporation, San Diego, CA, USA) were added and
incubated overnight at 4°C in a moist chamber. Phosphate buffer
saline (PBS) was used as a negative control. The following day,
sections were stained with a goat anti-rabbit IgG horseradish
peroxidase-conjugated secondary antibody for 30 min (dilution,
1:50; cat no. 4412S; Cell Signaling Technology, Inc., Danvers, MA,
USA) at room temperature and visualized using 3,3′-diaminobenzidine
hydrochloride for 10–15 sec at room temperature. Finally, the
sections were counterstained with Mayer's hematoxylin for 1 min at
room temperature, dehydrated, and images were captured.
To evaluate TFAP4 expression levels, two independent
pathologists assessed the images using a semi-quantitative scoring
method. In detail, the intensity of immunoreactivity was scored as:
0, negative staining; 1, weak staining; 2, moderate staining and 3,
strong staining. The extent of immunoreactivity was scored
according to the percentage of positive staining tumor cells in
each microscopic view (5 fields of view for every slice), as
follows: 0, 0–25% staining of tumor cells; 1, 26–50% staining of
tumor cells; 2, 51–75% staining of tumor cells; and 3, 76–100%
staining of tumor cells. The total IHC score was achieved by
multiplying the scores of extent and intensity. High TFAP4
expression was defined as a total score ≥4, and a score <4 was
defined as low TFAP4 expression.
Cell culture
The human colorectal cancer DLD-1, CaCo-2, HT-29,
HCT-116, LS174T, LoVo and SW480 cell lines were obtained from the
Cell Bank of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). The immortalized colorectal epithelial NCM460
cell line was obtained from Sun Yat-sen University. All cells were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and incubated in a
humidified incubator with 5% CO2 at 37°C.
Cell transfection with plasmids and
small interfering RNAs (siRNAs)
A total of three different TFAP4-specific siRNA
sequences and a negative control siRNA (a scramble control produced
by Guangzhou RiboBio Co., Ltd., Guangzhou, China) were used to
knockdown the expression of TFAP4 in CRC cell lines. The sequences
of the three siRNAs were as follows: si-001,
5′-GACGCATGCAGAGCATCAA-3′; si-002, 5′-GGACAAGGACGAAGGCATA-3′; and
si-003, 5′-CCTCGGTCATCAACTCTGT-3′. The expression plasmids for
TFAP-4 (EX-P0109-Lv201) and the control vector (EX-NEG-Lv201) were
obtained from GeneCopoeia, Inc. (Rockville, MD, USA). When cells
reached ~50% confluency, 50 µM siRNA per well was transfected,
performed using Lipofectamine 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) following the
manufacturer's protocol. Transfection efficiency was confirmed by
detecting the expression change of TFAP4 using western blotting and
quantitative polymerase chain reaction (qPCR) as described below.
Following transfection for 48 h, the cells were harvested and moved
into the 96-well culture plates, 6-well plates and transwell
chambers, for use in the proliferation, migration and invasion
assays, respectively.
Cell proliferation assay
Cell proliferation was examined using Cell Counting
Kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
according to the manufacturer's protocol. Briefly, transfected and
control cells (cells transfected with the control vector and cells
transfected with the control siRNA) were seeded onto 96-well
culture plates (3×103 cells/well) and cultured for 4
days. At each 24-h interval, 10 µl CCK-8 solution was added to each
well and incubated for 2 h at 37°C. Using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA), the absorbance was
measured at 450 nm.
Wound healing assay
Cells were seeded onto a 6-well plate and grown to
90% confluence. Confluent cells were then scratched using a 200-ml
pipette tip. Following rinsing three times with PBS to remove
detached cells, the remaining cells were incubated in
serum-deprived medium (RPMI-1640 medium with 1% FBS; Gibco; Thermo
Fisher Scientific, Inc.) at 37°C for 48 h. The wound status was
recorded using an inverted microscope at a magnification of ×40
(Olympus Corporation, Tokyo, Japan) at 0, 24 and 48 h after
scratching. Distances covered by migrated cells were quantified via
ImageJ software (version 1.48; National Institutes of Health,
Bethesda, MD, USA). The experiment was performed three times.
Migration and invasion assay
Cell invasion and migratory potential were evaluated
in 24-well plates using Transwell chambers (8 µm pore membrane; BD
Biosciences, Franklin Lakes, NJ, USA). For the migration assay,
cells (5×104) were seeded into the upper chamber. For
the invasion assay, 1×105 cells were seeded into the
upper chamber, where the membrane was covered with matrix gel (BD
Biosciences). To create a chemoattractant, cells were suspended in
100 µl serum-free RPMI-1640 medium in the upper chamber, whereas
the lower chamber contained 500 µl RPMI-1640 with 10% FBS. After 20
h of incubation at 37°C for the migration assay and 24 h for the
invasion assay, the membranes were fixed with 4% paraformaldehyde
for 20 min at room temperature and stained with 0.1% crystal violet
at room temperature for 20 min. The cells on the upper surface were
removed using a cotton swab. The number of invaded and migrated
cells was quantified by counting five different fields of view
under a light microscope at a magnification of ×200.
Western blot analysis
Human CRC tissues and cells were lysed in ice-cold
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) with added protease and phosphatase
inhibitors (Beyotime Institute of Biotechnology). Following removal
of the cell debris by centrifugation (13,000 × g at 4°C for 20
min), the protein concentration was measured using the
Bicinchoninic Acid Protein Assay kit (Beyotime Institute of
Biotechnology). Equal amounts of protein (20 µg) were separated on
10% SDS-PAGE gels and transferred to polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA). Following blocking
with 5% bovine serum albumin (Beyotime Institute of Biotechnology)
for 1 h at room temperature, the membranes were incubated overnight
at 4°C with the following antibodies: TFAP4 (dilution, 1:1,000; cat
no. SAB1404461; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and
β-tubulin (dilution, 1:10,000; cat no. 86298; Cell Signaling
Technology, Inc.). Following washing with TBS containing Tween-20,
the membranes were incubated with secondary antibodies [dilution,
1:5,000; goat anti-rabbit horseradish peroxidase (HRP)-conjugated
secondary antibody, cat no. 7074 and goat anti-mouse HRP-conjugated
secondary antibody, cat no. 7076; Cell Signaling Technology] at
room temperature for 1 h. Finally, proteins on the membranes were
visualized using enhanced chemiluminescence reagents (EMD
Millipore) in the ImageQuant Las4000mini detection system (GE
Healthcare Bio-Sciences, Pittsburgh, PA, USA).
RNA extraction and reverse
transcription-qPCR (RT-qPCR)
Total RNA was isolated from the cells or tissues
using TRIzol reagent (Takara Bio, Inc., Otsu, Japan) according to
the manufacturer's protocol, and 500 ng RNA was reverse-transcribed
into cDNA using the Prime Script RT Master Mix (Takara Bio, Inc.)
according to the manufacturer's protocol. RT-qPCR was performed
with the resultant cDNA as a template using the SYBR Premix Ex
Taq kit (RNaseH Plus; Takara Bio, Inc.) according to the
manufacturer's protocol in the Bio-Rad CFX96 cycler (Bio-Rad
Laboratories, Inc.) under the following conditions: 95°C for 30
sec, 40 cycles of 95°C for 3 sec, 60°C for 30 sec and 72°C for 50
sec. PCR reactions were performed in triplicate and repeated three
times. The relative level of TFAP4 mRNA was calculated using the
2−ΔΔCq method (17) with
normalization to GAPDH. Primers were designed as follows: TFAP4
forward, 5′-GAGCCAGCCTGGGATTGTC-3′; TFAP4 reverse,
5′-GTGCTTAAAGGAGAAAGAAGAAAACC-3′; GAPDH forward,
5′-TGTTGCCATCAATGACCCCTT-3′; and GAPDH reverse,
5′-CTCCACGACGTACTCAGCG-3′.
Statistical analysis
All data were analyzed using SPSS 19.0 statistical
software (IBM SPSS, Armonk, NY, USA). The data were presented as
the mean ± the standard deviation. The association between TFAP4
expression and clinicopathological parameters was analyzed using
the χ2 test. Survival analysis was performed using the
Kaplan-Meier method and the log-rank test. The significant
prognostic factors in the univariate survival analysis were
subsequently investigated with Cox's proportional hazards model for
multivariate survival analysis. Other comparisons between two
groups were made using Student's t-tests, with one-way analysis of
variance (followed by Tukey's post hoc test) used for comparisons
between more than two groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
TFAP4 expression and prognosis in
CRC
To investigate the function of TFAP4 in CRC, TFAP4
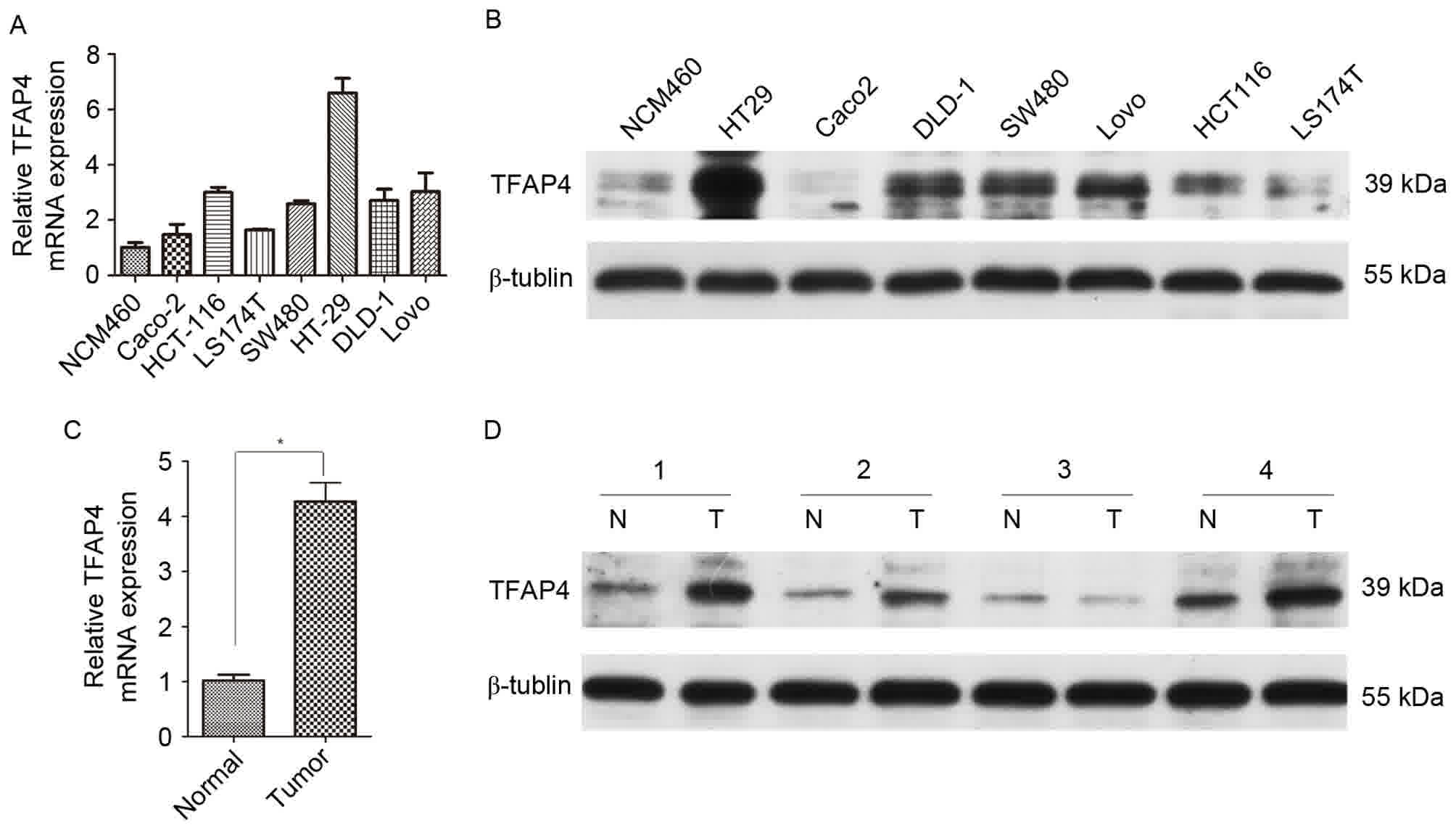
expression was examined in human CRC tissue and cell lines. RT-qPCR
experiments revealed that TFAP4 mRNA expression was increased in
all seven CRC cell lines compared with the immortalized colorectal
epithelial NCM460 cell line (Fig.
1A). Western blotting revealed similar results in terms of
protein TFAP4 expression (Fig. 1B).
In CRC tissues, the TFAP4 mRNA expression level was 4.3-fold higher
than that of adjacent normal tissues (Fig. 1C). Western blotting results also
revealed that with the exception of in lane number 3, TFAP4 protein
expression levels were markedly increased in the tumor tissues
compared with matched adjacent normal tissues (Fig. 1D).
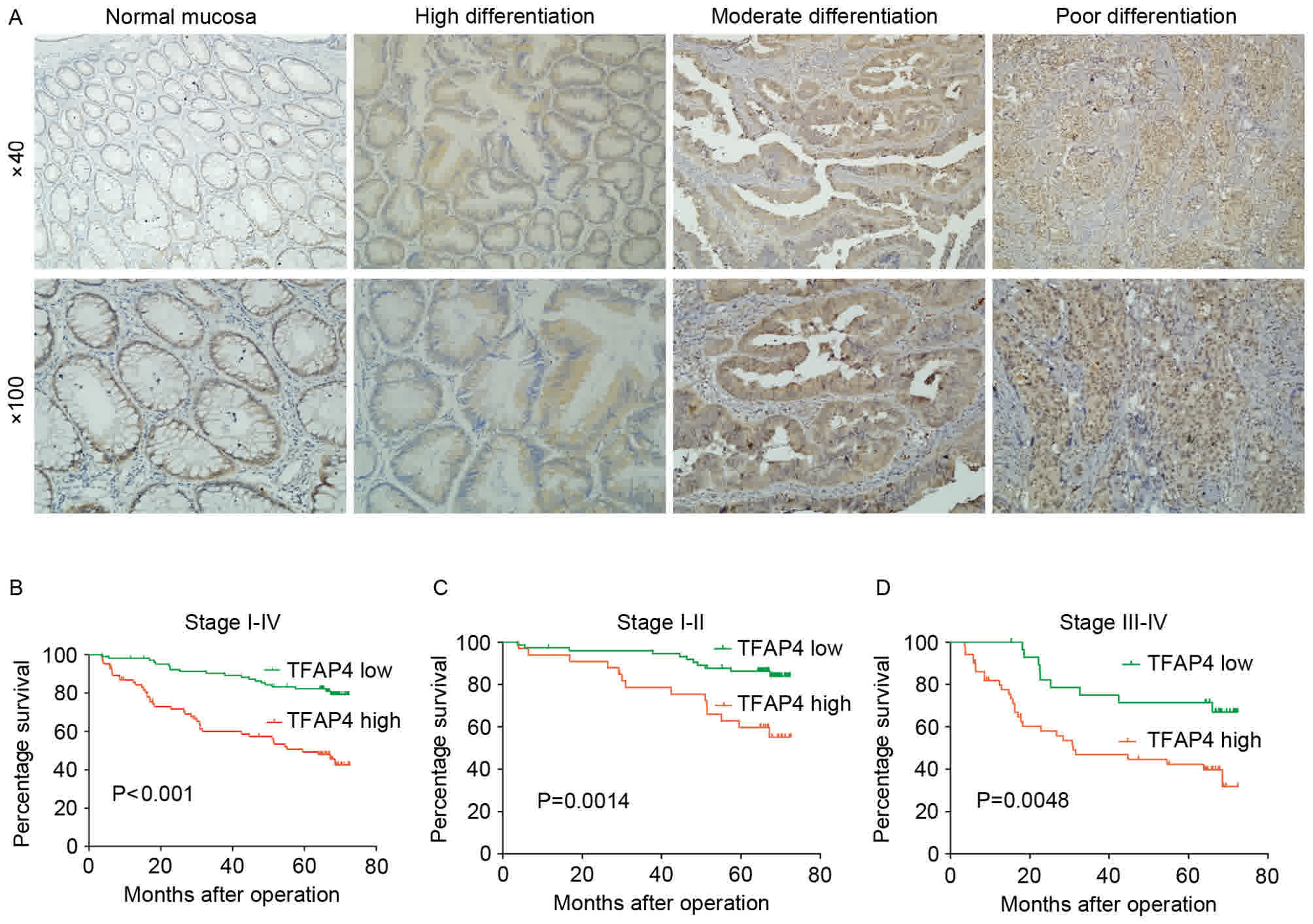
Next, TFAP4 protein expression was examined in 187
CRC tissues by IHC. Representative images of cytoplasmic and
nuclear TFAP4 protein expression in the tumor cells are presented
in Fig. 2A. The positive expression
rate was 76.5% (143 of 187). To evaluate the association between
TFAP4 expression and clinicopathological parameters, patients were
divided into two groups based on the aforementioned IHC score
assessment system. The results revealed that high TFAP4 expression
was positively associated with TNM stage (P<0.001), lymph node
invasion (P<0.001), liver metastasis (P<0.001) and
differentiation (P=0.024). More details are presented in Table I.
Since TFAP4 overexpression was associated with CRC
progression, the present study investigated whether high TFAP4
expression was a prognostic factor in patients with CRC.
Kaplan-Meier survival analysis demonstrated that high TFAP4
expression was significantly associated with poor overall survival
(OS), and was also a significant prognostic factor within stage
I–II and stage III–IV patients (P<0.001; Fig. 2B-D). In addition, univariate and
multivariate Cox regression analysis was performed to examine
whether TFAP4 expression was an independent prognostic factor.
Univariate analysis revealed that high TFAP4 expression, degree of
differentiation, lymph node metastasis, liver metastasis and TNM
stage were prognostic factors for OS time. However, in the
multivariate Cox regression analysis, only liver metastasis and
high TFAP4 expression were associated with poor OS (Table II). The present data demonstrated
that high TFAP4 expression was an independent prognostic factor for
patients with CRC (hazard ratio=2.607; 95% confidence interval,
1.469–4.627; P<0.001).
 | Table II.Univariate and multivariate analyses
of various potential prognostic factors in patients with colorectal
cancer. |
Table II.
Univariate and multivariate analyses
of various potential prognostic factors in patients with colorectal
cancer.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Factors | Cases, n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years |
| 1.668
(0.981–2.836) | 0.059 | 1.552
(0.875–2.783) | 0.140 |
|
<60 | 80 |
|
|
|
|
|
≥60 | 107 |
|
|
|
|
| Sex |
| 1.373
(0.818–2.304) | 0.231 | 1.486
(0.864–2.557) | 0.152 |
|
Male | 109 |
|
|
|
|
|
Female | 78 |
|
|
|
|
| Tumor size, cm |
| 1.409
(0.860–2.311) | 0.174 | 1.105
(0.658–1.858) | 0.705 |
|
<5 | 106 |
|
|
|
|
| ≥5 | 81 |
|
|
|
|
| Therapeutic
strategy |
| 0.942
(0.568–1.560) | 0.816 | 0.770
(0.433–1.370) | 0.374 |
|
Sur | 111 |
|
|
|
|
| Sur +
chemo | 76 |
|
|
|
|
|
Differentiation |
| 0.161
(0.022–1.161) | 0.070 | 0.334
(0.044–2.528) | 0.288 |
|
Well | 14 |
|
|
|
|
|
Moderate/poor | 173 |
|
|
|
|
| Depth of
invasion |
| 1.581
(0.753–3.320) | 0.226 | 1.121
(0.511–2.460) | 0.775 |
|
T1-T2 | 33 |
|
|
|
|
|
T3-T4 | 154 |
|
|
|
|
| Lymph node
invasion |
| 2.343
(1.427–3.848) | 0.001 | 1.594
(0.526–4.828) | 0.410 |
|
Present | 71 |
|
|
|
|
|
Absent | 116 |
|
|
|
|
| Metastasis |
| 5.957
(3.425–10.360) | <0.001 | 3.833
(1.868–7.863) | <0.001 |
|
Present | 26 |
|
|
|
|
|
Absent | 163 |
|
|
|
|
| TNM stage |
| 2.880
(1.734–4.782) | <0.001 | 1.060
(0.315–3.569) | 0.925 |
|
I–II | 107 |
|
|
|
|
|
III–IV | 80 |
|
|
|
|
| TFAP4
expression |
| 3.703
(2.174–6.306) | <0.001 | 2.607
(1.469–4.627) | 0.001 |
|
High | 84 |
|
|
|
|
|
Low | 103 |
|
|
|
|
TFAP4 promotes colorectal cancer cell
proliferation, migration and invasion in vitro
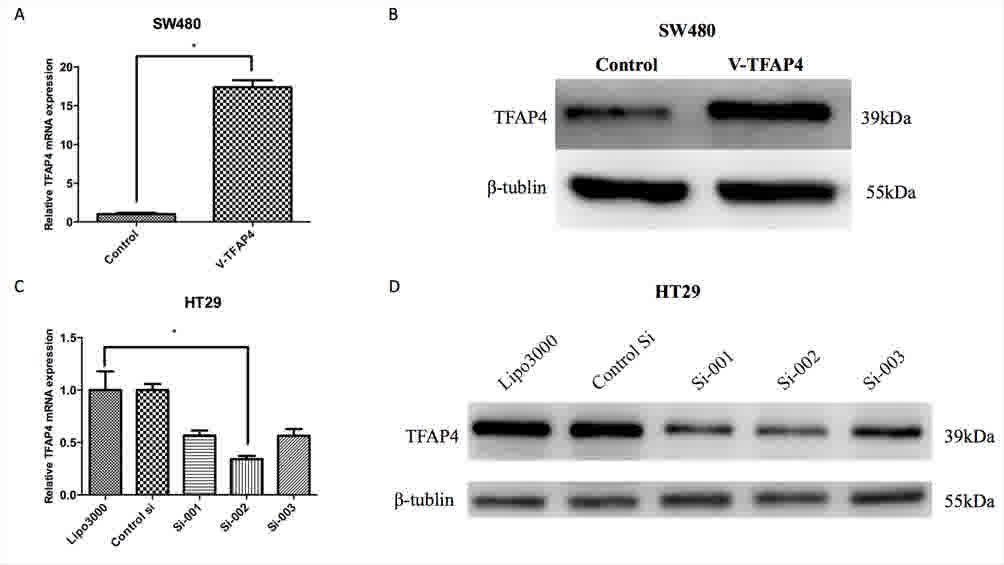
To determine the function of TFAP4 in CRC cells,
cell proliferation, motility and invasion assays were performed to
evaluate the effect of TFAP4 overexpression or knockdown on CRC
cell lines in vitro. The SW480 cell line, which exhibits
relatively low TFAP4 expression, was transfected with a
TFAP4-expressing plasmid (V-TFAP4). To knockdown TFAP4, the HT29
cell line, which expresses a relatively high level of TFAP4, was
transfected with specific siRNAs. RT-PCR and western blot analysis
demonstrated that TFAP4 expression was reduced by siRNA and
increased by cDNA transfection, respectively (Fig. 3). The results indicated that si-002
was the most effective sequence in terms of TFAP4 inhibition.
Therefore, si-002 was used for subsequent experiments, and the
negative control siRNA (50 nM) was used as a control.
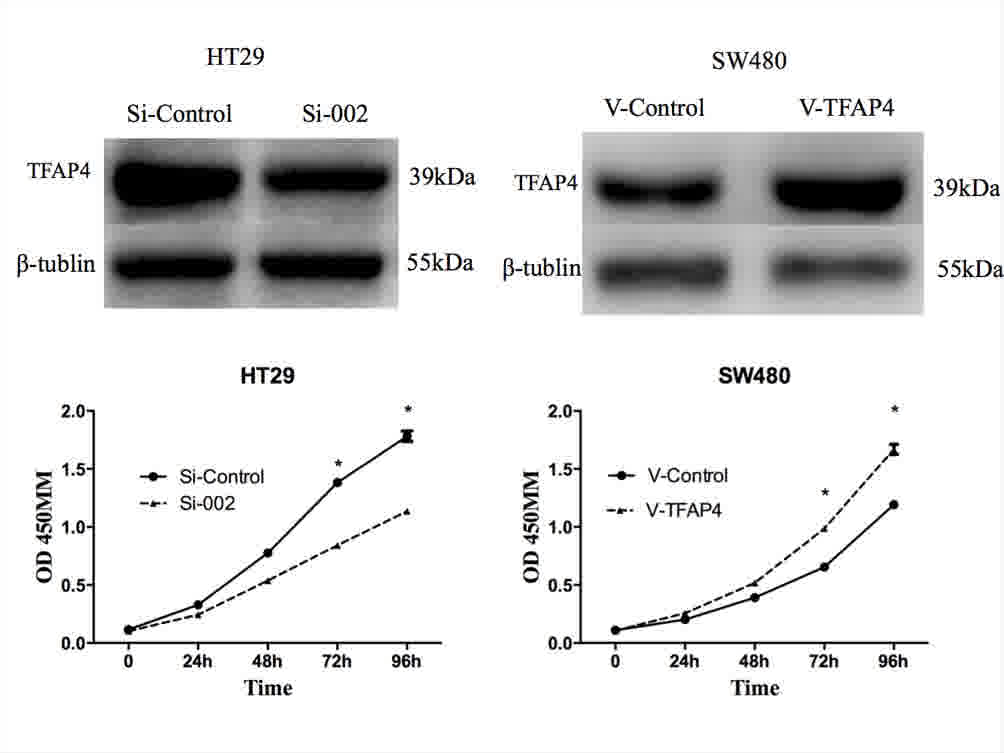
In cell proliferation assays, overexpression of
TFAP4 was revealed to significantly promote proliferation, while
downregulation of TFAP4 inhibited CRC cell proliferation
(P<0.05; Fig. 4). Compared with
control cells, overexpression of TFAP4 stimulated increased
migration and invasion, whereas knockdown had an inhibitory effect
on these phenomena (P<0.05; Figs.
5–7). In detail, in the wound
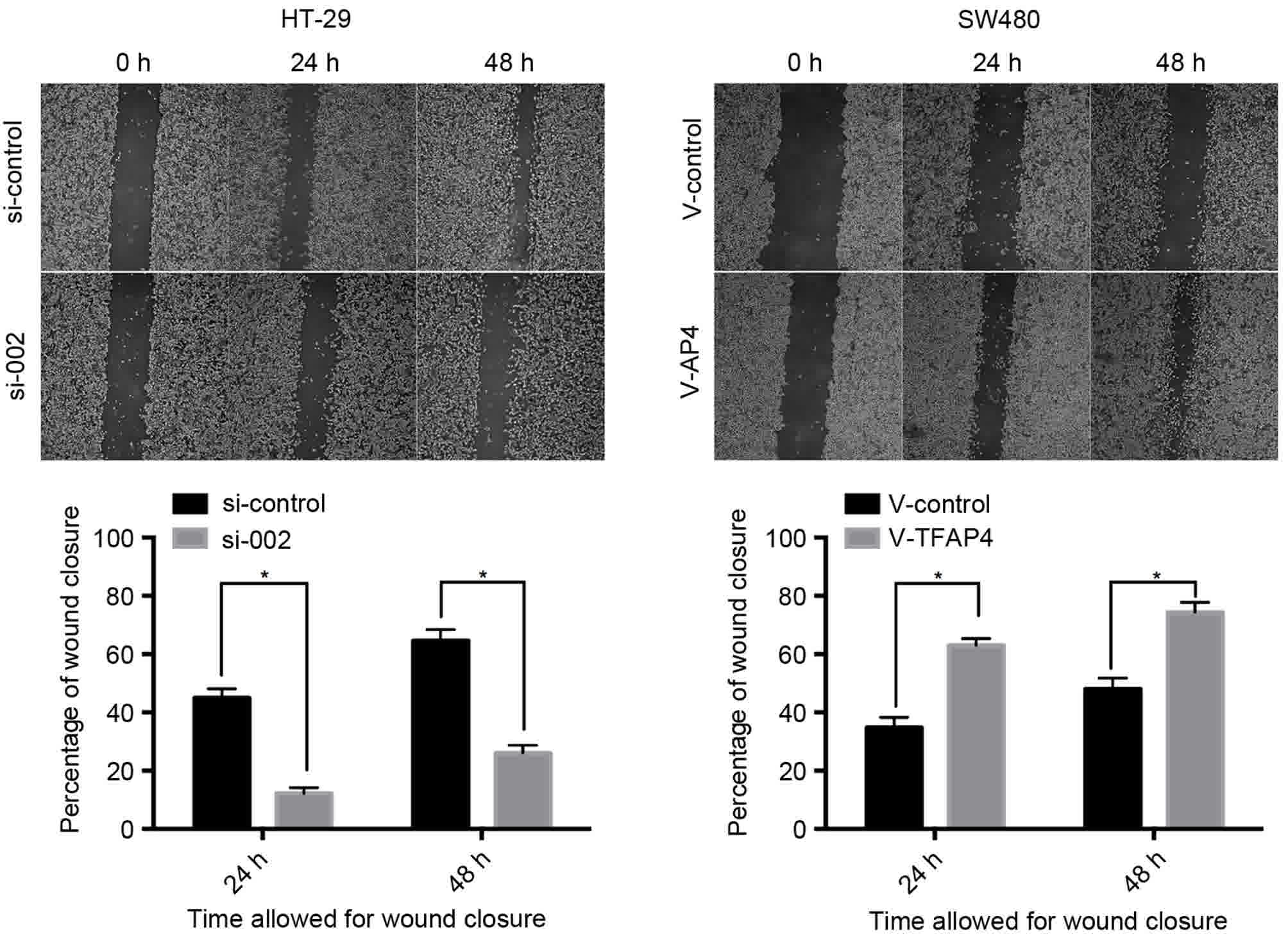
healing assays, knockdown of TFAP4 repressed the cell motility of
CRC cells, but overexpression of TFAP4 improved their motility
(Fig. 5). Transwell migration assays
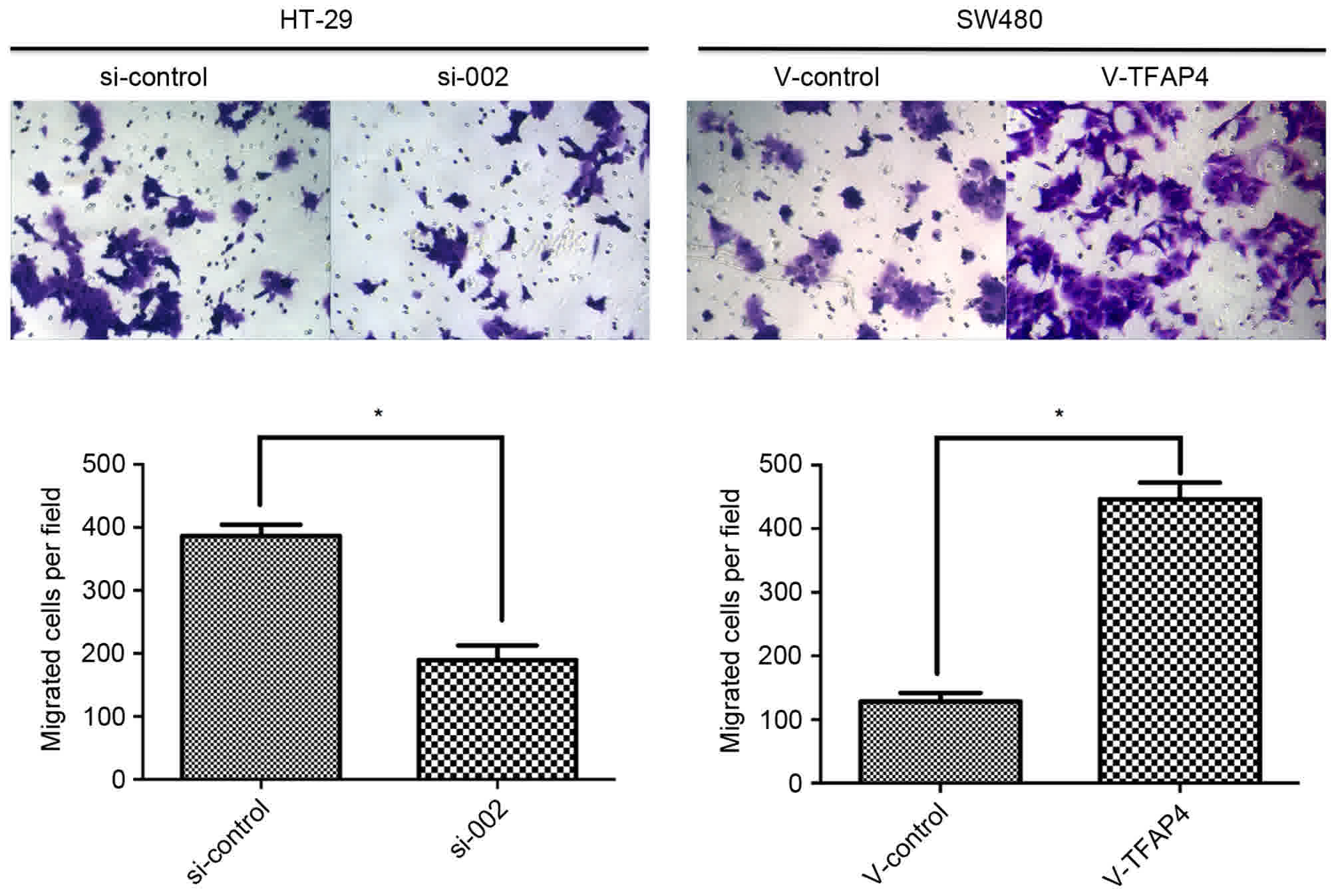
revealed that ectopic expression of TFAP4 stimulated tumor cell
migration, and that the number of migrated cells was reduced
following TFAP4 knockdown (Fig. 6).
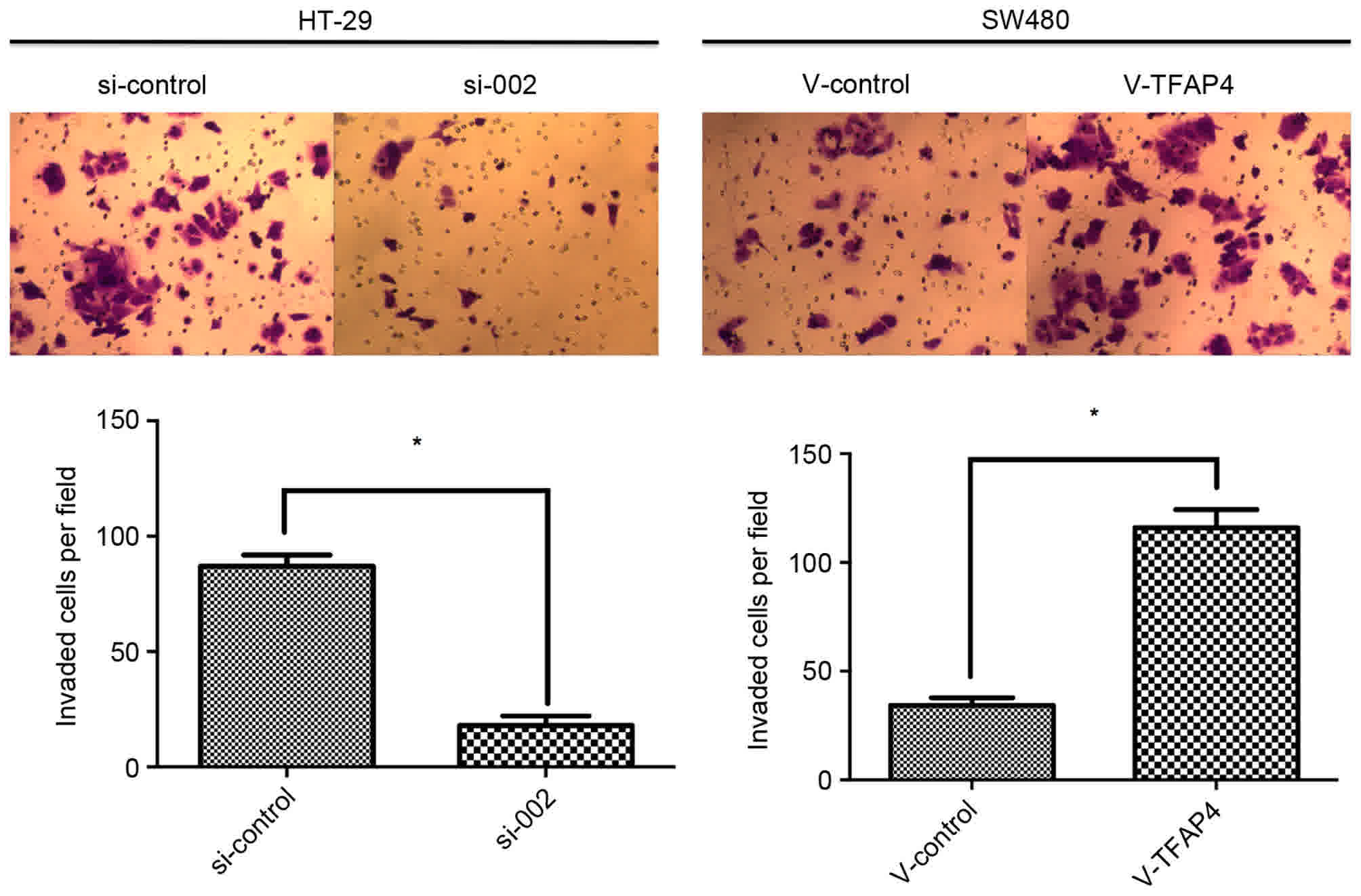
Similarly, the invasive ability was improved in TFAP4-overexpressed
CRC cells; however, TFAP4 knockdown in CRC cells indicated the
opposite effect (Fig. 7). Taken
together, the present data demonstrated that TFAP4 promoted CRC
proliferation, migration and invasion in vitro.
Discussion
In the present study, high expression of TFAP4 in
CRC cell lines and specimens compared with normal cells and tissue
was observed. Multivariate survival analysis revealed that the
TFAP4 expression level in CRC was an independent prognostic factor.
Functional analysis demonstrated that TFAP4 contributed to CRC cell
proliferation, migration and invasion.
TFAP4 was initially identified as a cellular protein
that binds to the simian virus 40 enhancer and activates viral late
gene transcription (18). TFAP4
controls transcriptional networks during cellular differentiation
through homodimer formation and binding to the symmetrical DNA
sequence, CAGCTG (5). High TFAP4
expression has been reported in CRC, breast cancer and
hepatocellular carcinoma (13–15). These
results also indicate that TFAP4 overexpression is associated with
a worse prognosis.
Previous studies have established a functional role
for TFAP4 in several crucial aspects of tumor progression,
including proliferation, apoptosis (9,11,12,19),
transformation (10), invasion,
metastasis (12) and chemoresistance
(13,20), indicating that TFAP4 is potentially
involved in tumorigenesis. Loss of p21 expression is associated
with the promotion of neoplasia, tumorigenesis and progression in
CRC (21). The p21 gene encodes a
cell cycle-dependent kinase inhibitor, which is induced by p53 and
numerous other anti-proliferative factors (22). Jackstadt et al (9,10) reported
that TFAP4 promotes cell proliferation via direct repression of p21
by occupying four CAGCTG motifs in the basic region of the p21
promoter. Since TFAP4 is a c-MYC target gene and is upregulated in
colorectal tumors, and this upregulation is a hallmark of CRC
(23), it was hypothesized that TFAP4
may be involved in tumor progression. In the present study,
knockdown of endogenous TFAP4 inhibited CRC cell proliferation,
whereas ectopic expression of TFAP4 enhanced these capacities.
These results are consistent with previous studies (12,14)
highlighting the involvement of TFAP4 in the progression of
CRC.
In gastrointestinal adenocarcinoma, TFAP4 promotes
cell proliferation by affecting key regulators of the cell cycle
(9,20). In breast cancer, TFAP4 suppresses cell
proliferation by inducing the c-Jun N-terminal kinase/TFAP4/protein
tyrosine phosphatase, non-receptor type 6 signaling pathway
(13). The contradictory effect of
TFAP4 in colorectal cancer and breast cancer suggests that TFAP4
performs a differential role in cell proliferation and that
TFAP4-activated signaling pathways may be tumor-specific. The
present study demonstrated that TFAP4 promotes the proliferation of
CRC cells. The results were consistent with those of previous
studies, demonstrating that TFAP4 serves an essential function in
tumor cell proliferation (9,20).
Jackstadt et al (12) reported that TFAP4 is a novel regulator
of the epithelial-mesenchymal transition (EMT), which contributes
to the metastatic processes in colorectal cancer. TFAP4 directly
represses E-cadherin via a non-canonical TFAP4-binding motif and
induces neural-cadherin, the process of which is a hallmark of the
EMT (12). Consistent with the study
by Jackstadt et al (12), the
present study revealed that TFAP4 expression is also positively
associated with liver metastasis, and that TFAP4 overexpression
promotes cell migration and invasion in vitro. These results
indicated that TFAP4 activation may also induce EMT and enhance the
migration and invasion of CRC cells. However, additional studies on
the molecular mechanisms underlying these events are required.
In summary, consistent with the function of TFAP4 in
numerous different aspects of tumor malignancy, the present study
demonstrated that TFAP4 is overexpressed in colorectal cancer and
that TFAP4 is associated with poor clinical outcome in patients
with CRC. In addition, TFAP4 is an independent prognostic factor
for the OS of patients with CRC. Overexpression of TFAP4 promotes
cell proliferation, migration and invasion. Future studies should
focus on the identification of TFAP4-interacting proteins and the
validation of the potential effectiveness of TFAP4 as a target for
intervening with CRC progression.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Planning Project of Guangdong Province Daya Bay (grant
no. 2013A01015).
Availability of data and materials
The datasets generated and analyzed in the present
study are included in this published article.
Authors' contributions
JY designed the procedure and study, analyzed the
data and wrote the report. JPM and XHZ assisted design of study
protocol and critically revised the report. JBX, CQC, SRC and YLH
performed the operations and perioperative care of the patient and
revised the report. SX provided assistance during the experiments
of RT-PCR and the revision of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients for the use of their tissue samples. The Ethics Committee
of the First Affiliated Hospital of Sun Yat-sen University approved
the protocol of the present study.
Consent for publication
The study participants provided consent for the data
to be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pritchard CC and Grady WM: Colorectal
cancer molecular biology moves into clinical practice. Gut.
60:116–129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Augestad KM, Bakaki PM, Rose J, Crawshaw
BP, Lindsetmo RO, Dørum LM, Koroukian SM and Delaney CP: Metastatic
spread pattern after curative colorectal cancer surgery. A
retrospective, longitudinal analysis. Cancer Epidemiol. 39:734–744.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo HY and Xu RH: Predictive and
prognostic biomarkers with therapeutic targets in advanced
colorectal cancer. World J Gastroenterol. 20:3858–3874. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu YF, Lüscher B, Admon A, Mermod N and
Tjian R: Transcription factor AP-4 contains multiple dimerization
domains that regulate dimer specificity. Genes Dev. 4:1741–1752.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Atchley WR and Fitch WM: A natural
classification of the basic helix-loop-helix class of transcription
factors. Proc Natl Acad Sci USA. 94:5172–5176. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jung P and Hermeking H: The c-MYC-AP4-p21
cascade. Cell Cycle. 8:982–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim MY, Jeong BC, Lee JH, Kee HJ, Kook H,
Kim NS, Kim YH, Kim JK, Ahn KY and Kim KK: A repressor complex, AP4
transcription factor and geminin, negatively regulates expression
of target genes in nonneuronal cells. Proc Natl Acad Sci USA.
103:13074–13079. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jackstadt R and Hermeking H: AP4 is
required for mitogen- and c-MYC-induced cell cycle progression.
Oncotarget. 5:7316–7327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jackstadt R, Jung P and Hermeking H: AP4
directly downregulates p16 and p21 to suppress senescence and
mediate transformation. Cell Death Dis. 4:e7752013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jung P, Menssen A, Mayr D and Hermeking H:
AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci
USA. 105:15046–15051. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A
and Hermeking H: AP4 is a mediator of epithelial-mesenchymal
transition and metastasis in colorectal cancer. J Exp Med.
210:1331–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amin S, Kumar A, Nilchi L, Wright K and
Kozlowski M: Breast cancer cells proliferation is regulated by
tyrosine phosphatase SHP1 through c-jun N-terminal kinase and
cooperative induction of RFX-1 and AP-4 transcription factors. Mol
Cancer Res. 9:1112–1115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xinghua L, Bo Z, Yan G, Lei W, Changyao W,
Qi L, Lin Y, Kaixiong T, Guobin W and Jianying C: The
overexpression of AP-4 as a prognostic indicator for colorectal
carcinoma. Med Oncol. 29:871–877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hu BS, Zhao G, Yu HF, Chen K, Dong JH and
Tan JW: High expression of AP-4 predicts poor prognosis for
hepatocellular carcinoma after curative hepatectomy. Tumour Biol.
34:271–276. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Edge SB, Byrd SR, Compton CC, et al: AJCC
cancer staging manual. 7th edition. Springer-Verlag; New York (NY):
pp. 143–164. 2010
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mermod N, Williams TJ and Tjian R:
Enhancer binding factors AP-4 and AP-1 act in concert to activate
SV40 late transcription in vitro. Nature. 332:557–561. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsujimoto K, Ono T, Sato M, Nishida T,
Oguma T and Tadakuma T: Regulation of the expression of caspase-9
by the transcription factor activator protein-4 in
glucocorticoid-induced apoptosis. J Biol Chem. 280:27638–27644.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Zhang B, Guo Y, Liang Q, Wu C, Wu
L, Tao K, Wang G and Chen J: Down-regulation of AP-4 inhibits
proliferation, induces cell cycle arrest and promotes apoptosis in
human colorectal cancer cells. PLoS One. 7:e370962012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zirbes TK, Baldus SE, Moenig SP, Nolden S,
Kunze D, Shafizadeh ST, Schneider PM, Thiele J, Hoelscher AH and
Dienes HP: Prognostic impact of p21/waf1/cip1 in colorectal cancer.
Int J Cancer. 89:14–18. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bunz F, Dutriaux A, Lengauer C, Waldman T,
Zhou S, Brown JP, Sedivy JM, Kinzler KW and Vogelstein B:
Requirement for p53 and p21 to sustain G2 arrest after DNA damage.
Science. 282:1497–1501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cancer Genome Atlas Network, . Muzny DM,
Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL,
Lewis L, Morgan MB, et al: Comprehensive molecular characterization
of human colon and rectal cancer. Nature. 487:330–337. 2012.
View Article : Google Scholar : PubMed/NCBI
|