Introduction
Breast cancer is among the most common types of
cancer, which affects women of all age groups worldwide (1). The mortality rate of breast cancer has
remained ~20% for 5 years (1). This
is likely due to the fact that the complex molecular mechanisms of
breast cancer pathology remain to be fully understood. The
elucidation of these mechanism is an important requirement to
improve the treatment available for breast cancer (2).
Mitofusin 2 (Mfn2), also named hyperperplasia
suppressor gene (HSG), is expressed in numerous human
tissues and serves a pivotal role in the proliferation and
apoptosis of vascular smooth muscle cells (VSMCs) (3,4). However,
Mfn2 has been demonstrated to be hypoexpressed in various
types of human malignant tumors, including gastric cancer,
hepatocellular cancer, colorectal cancer, urinary bladder cancer
and breast cancer (5–9). Therefore, Mfn2 is considered a
cancer suppressor gene, and elucidation of its anti-tumor mechanism
may contribute to cancer therapy.
DNA methylation is an essential epigenetic mechanism
required for control of gene expression. Methylation usually occurs
at CpG sites in gene promoters mediated by DNA methyltransferases
(DNMTs) (10,11). Generally, 70–80% CpG sites in the
human genome are methylated (12).
Abnormal DNA methylation is a common event in human cancers,
including hypermethylation of a tumor suppressor gene,
hypomethylation of an oncogene and global hypomethylation of the
genome (13–15). DNA methylation is a reversible
process, making it a promising target for cancer therapy.
It is well established that the structure of a
protein determines its biological function. It has been
demonstrated that rat Mfn2 protein contains a p21Ras
signature motif (amino acids 77–92) and a PKA phosphorylation site
at Ser442 (3). The p21Ras
motif and PKA phosphorylation site are necessary for the
anti-proliferative effect of Mfn2 on rat vascular smooth muscle
cells (VSMCs) (3,16). The human homolog shares 95.2%
similarity with the rat Mfn2 amino acid sequence (3). In the present study, the DNA methylation
status of the Mfn2 promoter was analyzed in breast cancer
cells and tissues, as well as the effect of demethylation on
Mfn2 expression. Furthermore, the importance of the
p21Ras motif and PKA phosphorylation site at Ser 442 for
the anti-proliferation and anti-invasion effects of Mfn2 in breast
cancer was also demonstrated. The present study will provide a
novel viewpoint for understanding the anti-tumor function of Mfn2
in human breast cancer.
Materials and methods
Tissue specimens
The 7 pairs of fresh breast cancer tissue and
adjacent non-tumor tissue were obtained from patients (25–55 years
old) who underwent breast surgery in the Tangshan People's Hospital
(Tangshan, China) between March 2011 and March 2012. A distance of
~5 cm was required between the adjacent non-tumor tissues and the
tumor tissue boundary. The selected tumor tissues were confirmed by
pathologists to contain >80% tumor cells. None of the patients
had undergone preoperative chemotherapy, radiotherapy and/or other
treatments. The protocol and use of the specimens in the present
study was approved by the Institutional Ethics Committee of
Tangshan People's Hospital (Tangshan, China), and written consent
was obtained from all participants.
Cell lines
The human breast cancer cell line, MCF-7, was
purchased from the Cell Resource Center of the Institute of Basic
Medicine, Chinese Academy of Medical Sciences (Beijing, China). The
cells were cultured in DMEM medium (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Hyclone, GE Healthcare Life Sciences, Logan, UT, USA) at 37°C with
5% CO2. To demethylate cell DNA, the MCF-7 cells were
treated with 0, 5, 10 and 15 µM 5-aza-2′-deoxycytidine (5-aza-CdR)
for 5 consecutive days. The 5-aza-CdR is a cytidine analog that can
incorporate into DNA during S phase and disrupt the interaction
between DNA and DNMT. The culture medium was replaced every 48 h.
Untreated cells were used as a control.
Plasmids and transfection
pEGFP-N1 plasmids expressing the complete
Mfn2 open reading frame, the Mfn2 with
p21ras signature motif deletion mutant
(Mfn2ΔRas) and protein kinase A (PKA)
phosphorylation site deletion (Mfn2ΔPKA) were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The
pEGFP-N1 plasmid (Promega Corporation, Madison, WI, USA) was used
as control. The MCF-7 cells were seeded in 6-well plates
(2×105/well) and cultured for 24 h. Subsequently, the
cells were respectively transfected with the pEGFP-N1,
Mfn2ΔRas and Mfn2ΔPKA plasmids
(4 µg/2 ml) using Lipofectamine® 2000 Reagent (Thermo
Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from human tissue samples
and cultured cells (48 h post-transfection) using TRIzol (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions, and concentration was determined by NanoDrop 2000
(Thermo Fisher Scientific, Inc.). Reverse transcription was
performed using 2 µg total RNA with a Reverse Transcriptase First
Chain cDNA kit (Takara Bio, Inc., Otsu, Japan). The Mfn2
mRNA expression level was determined by RT-qPCR using a SYBR-Green
Master Mix kit and PikoReal 96 RT-PCR system (Thermo Fisher
Scientific, Inc.). PCR was performed in triplicate and each
experiment was repeated 3 times. The primer sequences used are as
follows: Mfn2, forward, 5′-TTCCACAAGGTGAGTGAGC-3′, and
reverse, 5′-TTAGCAGACACAAAGAAGATGC-3′; Cyclin D1, forward,
5′-CCGTCCATGCGGAAGATC-3′, and reverse, 5′-ATGGCCAGCGGGAAGAC-3′;
GADPH, forward, 5′-GAGAGGGAAATCGTGCGTGAC-3′, and reverse,
5′-CATCTGCTGGAAGGTGGACA-3′. GAPDH was used as an internal control.
The PCR condition was set to 94°C for 3 min; followed by 30 cycles
of 94°C for 30 sec, 60°C for 30 sec and 70°C for 60 sec; and a
final extension step at 72°C for 5 min. The 2−ΔΔCq
method (17) was used to analyze the
relative changes in Mfn2 expression.
Methylation-specific PCR (MSPCR)
The genomic DNAs of cells and human tissue samples
were extracted with 10 mg/ml proteinase K (Amresco, LLC, Solon, OH,
USA) digestion at 55°C for 15 min (cells) and 3 h (human tissue).
Subsequently, the purification was performed. Phenol: Chloroform:
Isoamyl alcohol (25:24:1; Tianjing Baishi Chemical Industry Co.,
Ltd., Tianjin, China) was added and vortexed thoroughly for 20 sec
followed by centrifugation at room temperature for 5 min at 16,000
× g. After upper aqueous phase was removed, the layer was
transferred to a fresh tube and was used for isopropyl alcohol
(Tianjin Fuyu Fine Chemicals Co., Ltd., Tianjin, China)
precipitation. The extracted DNA samples were stored in TE buffer
at −20°C until use. Bisulfite-based DNA modification, which
converts all unmethylated cytosine bases to uracil, was performed
using a Methylcode Bisulfite Conversion kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
HotStar Taq DNA polymerase (Fermentas; Thermo Fisher Scientific,
Inc.) was used for the PCR reaction. The modified DNA was used as a
template for MSPCR with primers specific for either the
modified-methylated or unmethylated Mfn2 gene promoter
sequences. PCR amplification was performed with the following
primer sets, which include the CpG island of Mfn2:
methylated forward, 5′-TGGTTTTGAATTTTCGATGTATTC-3′, methylated
reverse, 5′-CAAAACAATAAACACTAACCCGTA-3′; unmethylated forward,
5′-GGTTTTGAATTTTTGATGTATTTGT-3′, unmethylated reverse,
5′-CAAAACAATAAACACTAACCACATA-3′. The PCR amplification program
consisted of 10 min at 95°C hen followed by 40 cycles of
denaturation for 30 sec at, annealing for 30 sec at 60°C, extension
for 30 sec at 72°C, and a final extension at 72°C for 10 min. The
PCR products were separated on 1.5% agarose gels with DuRed Nucleic
acid dye (Beijing Fanbo Biochemicals Co., Ltd., Beijing, China) and
visualized under ultraviolet illumination.
Western blot analysis
Total protein was collected using protein lysis
buffer containing 1 mM phenylmethylsulfonyl fluoride protease
inhibitor and centrifugation at 14,000 × g for 30 min at 4°C.
Protein concentration in the supernatant was determined by BCA
assay (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China), according to the manufacturer's protocol. A total
of 45 µM protein was dissolved in loading buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China), and denatured
by heating at 100°C for 5 min. The proteins were then separated by
10% SDS-PAGE Following separation, the proteins were transferred
onto an immunoblot polyvinylidene difluoride membrane (Thermo
Fisher Scientific, Inc.). The protein bands were confirmed by 0.2%
Ponceau S staining for 5 min at room temperature. Membranes were
blocked with 5% BSA (Sigma-Aldrich; Merck KGaA; Darmstadt, Germany)
for 2 h at 37°C, then incubated with the following primary
antibodies overnight at 4°C: Anti-Mfn2 (0.5 µg/ml; cat. no.
ab50838), anti-Cyclin D1 (1:100; cat. no. ab16663), anti-p-ERK
(1:50; cat. no. ab223500), anti-ERK (1:500; cat. no. ab17942),
anti-GAPDH (1:1,000; cat. no. ab9485) and anti-β-Actin (1:1,000;
cat. no. ab8227; all Epitomics; Abcam, Cambridge, UK). The
membranes were washed 3 times in Tris-buffered saline solution with
Tween-20 (1X TBST) and incubated with horseradish
peroxidase-conjugated goat-anti-rabbit antibody (Beijing Solarbio
Science & Technology Co., Ltd.; 1:500; cat. no. SE134) for 1 h
at 37°C. Following a final wash with 1X TBST, immunoreactive bands
were detected using the ChampGel automatic gel imaging analyzer
(Beijing Sage Creation Science Co., Ltd., Beijing, China). Optical
band density was quantified using ImageJ (version 1.44p; National
Institutes of Health, Bethesda, MD, USA).
Cell proliferation assays
Cell proliferation assays were performed using Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), according to the manufacturer's instructions. The
cells were seeded into 96-well microtiter plates at a density of
7.0×103 cells/well and cultured for 24 h. The cells were
then treated with increasing concentrations of 5-aza-CdR (0, 5, 10,
15 µM) for 5 days. Subsequently, 10 µl CCK-8 solution was added to
each well. The absorbance was measured at 450 nm using a microplate
reader, and a calibration curve was prepared using the data
obtained from standardized wells that contained known numbers of
viable cells. Each experiment was performed in 5 replicate wells
and repeated 3 times independently.
Cell cycle analysis
Flow cytometry was performed for cell cycle
analysis. The cells were seeded into 6-well plates and treated with
increasing concentrations of 5-aza-CdR (0, 5, 10, 15 µM) for 1, 2
and 3 days. Subsequently, the treated and untreated cells were
washed with PBS followed and fixed in ice-cold 70% ethanol in PBS
for 24 h at 4°C. Subsequent to another wash, the fixed cells were
treated with 0.01% RNase (Sigma-Aldrich; Merck KGaA) for 10 min at
37°C, and then stained with 0.05% propidium iodide for 20 min at
4°C in the dark. The cell cycle distribution was determined using a
FACSCanto flow cytometry system (Becton Dickinson; BD Biosciences,
Franklin Lakes, NJ, USA), and analyzed with Modfit 3.2 software
(Verity Software House, Inc., Topsham, ME, USA; http://www.vsh.com). The experiment was repeated 3
times.
Scratch wound assay
MCF-7 cells were cultured in 6-well plates until
~95% confluent. Then, a scratch wound was created using a 200-µl
pipette tip. The wounded cells were washed twice with culture
medium to remove the detached cells, then replaced in DMEM medium
supplemented with 10% FBS at 37°C with 5% CO2. Images of
the wounds were acquired using a digital camera 24 h after the
wounds were made. Wound width (µm) was measured using a standard
caliper, and the experiment was performed in triplicate, repeated
≥3 times.
Transwell invasion assay
The cell invasion capability was detected using
transwell chamber culture systems (pore diameter, 8-µm; Corning
Incorporated, Corning, NY, USA). A total of 2×104 cells
were placed onto a Matrigel-coated transwell chamber with
serum-free opti-MEM medium (Thermo Fisher Scientific, Inc.). The
DMEM medium containing 10% FBS was added to the lower chamber as a
chemoattractant. After 24 h, the cells attached to the lower
surface of the insert filter were counted using 0.1% crystal violet
staining for 20 min at room temperature. The images were captured
using an Olympus IX71 fluorescent microscope equipped with an
Olympus DP73 digital camera (both Olympus Corporation, Tokyo,
Japan).
Statistical analysis
All statistical analysis was performed using SPSS
software version 13.0 (SPSS, Inc., Chicago, IL, USA). The data are
presented as the mean ± standard deviation. One-way analysis of
variance was used to analyze differences between groups. Scheffe
post hoc testing was used to determine pairwise differences between
means. P<0.05 was considered to indicate a statistically
significant difference.
Results
Hypoexpression of Mfn2 and
hypermethylation of its promoter in breast cancer
The mRNA expression level of Mfn2 was
analyzed in 7 pairs of human breast cancer and adjacent non-tumor
tissues using RT-qPCR. As demonstrated in Fig. 1A, Mfn2 was hypoexpressed in
breast cancer tissues compared with the corresponding adjacent
non-tumor tissues (P<0.05). Given that DNA methylation is
associated with gene expression, the methylation level of
Mfn2 promoter was analyzed by MSPCR, with the aim of
clarifying the mechanism behind Mfn2 silencing in breast
cancer. Using the MethPrimer tool, 1 typical CpG island was
identified in the upstream promoter of Mfn2. As demonstrated
in Fig. 1B, the Mfn2 promoter
was hypermethylated in the majority of breast tumor tissues (6/7)
compared with the corresponding adjacent non-tumor tissues. These
data indicate that silencing of Mfn2 by DNA hypermethylation
on its promoter may contribute to the tumorigenesis of breast
cancer. Mfn2 may be a tumor suppressor gene in breast
cancer.
Demethylation treatment of MCF-7
cells
DNA methylation is a reversible process, making it a
promising target for cancer therapy. 5-aza-CdR is widely used as an
inhibitor of DNMTs and can increase the expression of genes
silenced by DNA methylation (18–21). In
order to confirm whether the expression of Mfn2 in human
breast cancer cells is regulated by DNA methylation of its
promoter, the MCF-7 breast cancer cells were treated with 5-aza-CdR
for 1, 2 and 3 days at concentrations of 5.0 10 and 15 µM. The
methylation level of the Mfn2 promoter in the
5-aza-CdR-treated groups decreased in a dose-dependent manner
compared with the untreated group (Fig.
1C). Conversely, the mRNA expression levels of Mfn2 in
the 5-aza-CdR-treated groups increased in a dose-dependent manner
compared with that of the untreated group (P<0.05; Fig. 1D). These data indicate that
demethylation treatment could upregulate Mfn2 expression in
breast cancer.
Overexpression of Mfn2 inhibits
growth, migration and invasion of MCF-7 cells
To clarify the anti-tumor role of Mfn2 in breast
cancer, the MCF-7 cells were transfected with pEGFP-N1-Mfn2
plasmids for 48 h, producing ~85% GFP-positive cells (data not
shown). The overexpression of Mfn2 in MCF-7 cells
transfected with N1-Mfn2 were confirmed by RT-qPCR
(P<0.01; Fig. 2A) and immunoblot
analysis (P<0.01; Fig. 2B).
Compared with the control cells (N1), the proliferation ability of
Mfn2-overexpressing MCF-7 cells was markedly decreased
(P<0.05; Fig. 2C), and the cell
cycle was mostly arrested in G0/G1 phase (P<0.05; Fig. 2D). This suggests that the growth
suppression effect of Mfn2 may be the result of inhibition
of the cell cycle. In addition, the migration and invasion
abilities of MCF-7 cells overexpressing Mfn2 were markedly
inhibited compared with the control cells, as detected by the wound
assay (P<0.05; Fig. 2E) and the
invasion assay (P<0.05; Fig. 2F).
These data suggest that overexpression of Mfn2 could
suppress the growth and metastasis of breast cancer.
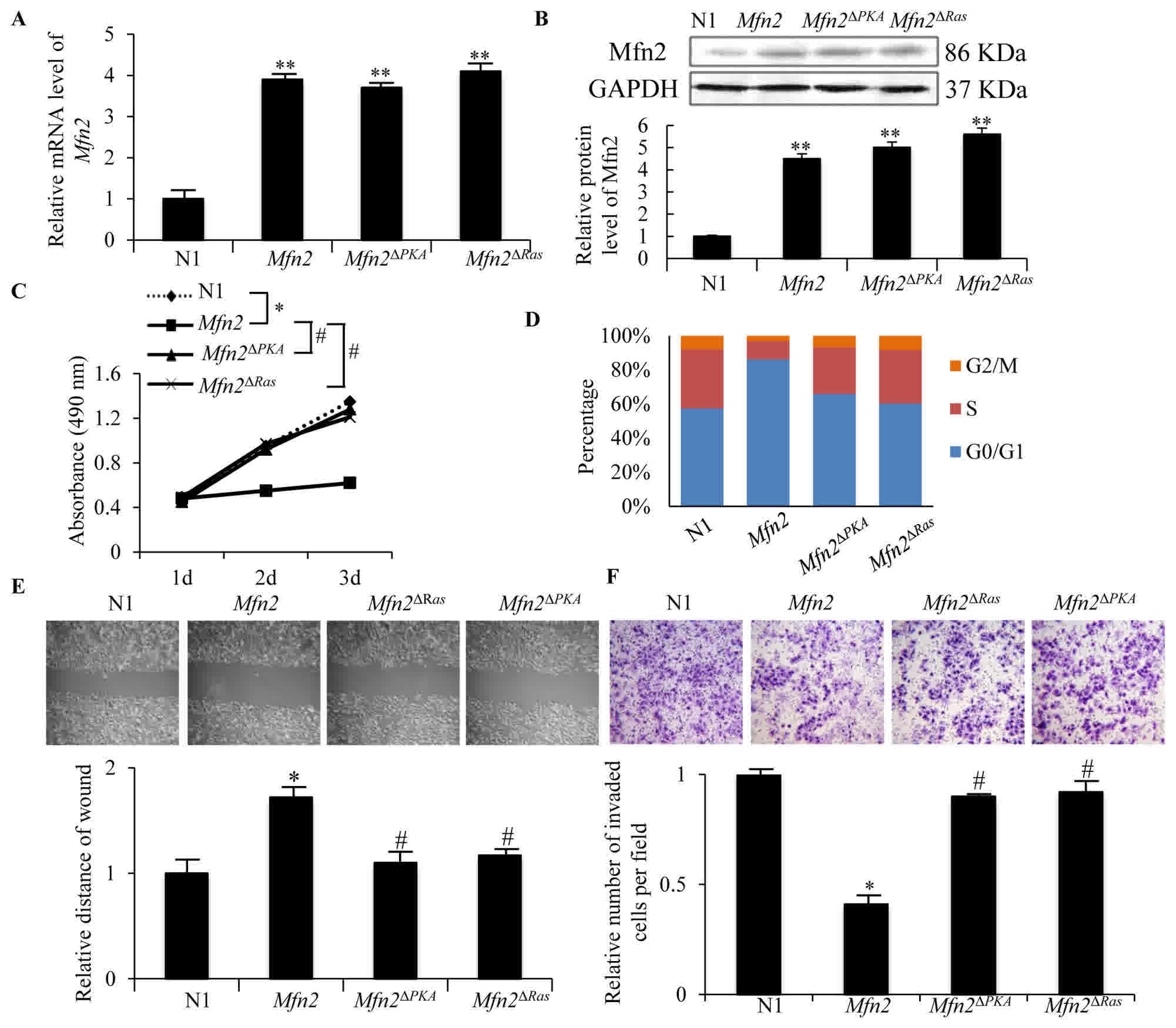 | Figure 2.Proliferation, migration and invasion
abilities of MCF-7 cells transfected with N1, Mfn2,
Mfn2ΔRas and Mfn2ΔPKA plasmids.
(A) The mRNA level of Mfn2 in each group was detected by
reverse transcription-quantitative polymerase chain reaction 24 h
post-transfection, using GAPDH as internal control. The data
are presented as expression relative to that of N1. (B) Mfn2
protein expression was detected by western blotting 48 h
post-transfection. Quantification of the protein band was analyzed
by Image Pro Plus software. (C) Cells of each group were seeded
into 96 plates at 7.0×103/well. Cell proliferation was
detected using a Cell counting kit-8 assay for 3 days. The data are
representative of 3 experiments. (D) Cells were seeded in a 25
cm2 tissue culture flask and harvested at 80% confluence
for cell cycle distribution analysis by flow cytometry. (E)
Migration activity was detected using a scratch wound assay. ×100,
magnification. The bar graph demonstrates the fold change of wound
distance relative to N1. (F) Invasion ability was analyzed using a
transwell assay with Matrigel. The cells attached to the lower
surface of the insert were stained with crystal violet, imaged and
counted in 5 random fields of view (×200, magnification). The data
represent the mean ± standard deviation of the number of cells
relative to the N1 group in 3 experiments. *P<0.05 and
**P<0.01 vs. N1 group; #P<0.05 vs. Mfn2
group. N1, EGFP-N1 plasmid; Mfn2, mitofusin 2;
Mfn2ΔRas, Mfn2 ORF lacking the
p21Ras coding sequence; Mfn2ΔPKA,
Mfn2 ORF lacking the protein kinase A phosphorylation site
coding sequence. |
The anti-proliferation, anti-migration
and anti-invasion effects of Mfn2 are mediated by the
Ras-Raf-ERK1/2 signaling pathway
It was reported that Mfn2 could bind to Ras protein
with its p21Ras motif in rat VSMCs (3). The anti-proliferative effect of Mfn2 on
VSMCs was mediated by inhibition of the Ras-Raf-ERK1/2 signaling
pathway. Deletion of the p21Ras motif abolished
Mfn2-induced growth arrest of VSMCs. Due to the 95% homology of the
amino acid sequence of Mfn2 in rats and humans, it was speculated
that Mfn2 may interact with Ras via the p21Ras
motif in human breast cancer cells. To test this hypothesis, MCF-7
cells were transfected with plasmids containing the Mfn2
open reading frame (ORF) lacking the p21Ras coding
sequence (Mfn2ΔRas). As demonstrated in Fig. 2A and B, the mRNA and protein
expression levels of Mfn2 in MCF-7 cells transfected with
Mfn2ΔRas plasmids were not significantly
different from cells transfected with Mfn2 full length
plasmids. However, the proliferation (P<0.05) (Fig. 2C), migration (Fig. 2E) and invasion (Fig. 2F) abilities of MCF-7 cells transfected
with Mfn2ΔRas plasmids were markedly increased compared
with those of MCF-7 cells transfected with Mfn2 full length
plasmids (P<0.05). The cell cycle arrest was also rescued
(Fig. 2D). Thus, the
anti-proliferation, anti-migration and anti-invasion functions of
Mfn2ΔRas were partially abolished, indicating that the
p21Ras motif of Mfn2 is required for it anti-tumor
effects in breast cancer and that Mfn2 probably serves a role in
regulating the Ras-Raf-MEK-ERK1/2 signaling cascade.
The protein expression level of ERK1/2, a major
downstream signaling protein of Ras, and activated phosphorylated
ERK1/2 (p-ERK1/2) were detected. As demonstrated in the Fig. 3A, the protein level of p-ERK1/2 was
markedly decreased by overexpression of Mfn2, but not
Mfn2ΔRas (P<0.05). Given that Ras-ERK1/2
signaling has been demonstrated to induce Cyclin D1
expression to modulate the G1-S phase transition (22), the expression of Cyclin D1 was
also analyzed. The mRNA and protein expression levels of Cyclin
D1 were significantly downregulated in MCF-7 cells transfected
with Mfn2 compared with control cells (P<0.05; Fig. 3B and C). This suggests that
Mfn2-induced cell-cycle arrest in the G0/G1 phases is attributable,
at least in part, to inhibition of the Ras-ERK1/2-cyclin D1
pathway. Deletion of the p21Ras motif partially
abolished Mfn2-induced inhibition of ERK1/2 phosphorylation
(Fig. 3A) and expression of Cyclin
D1 (Fig. 3B and C), indicating a
crucial interaction between Mfn2 and Ras in breast cancer
cells.
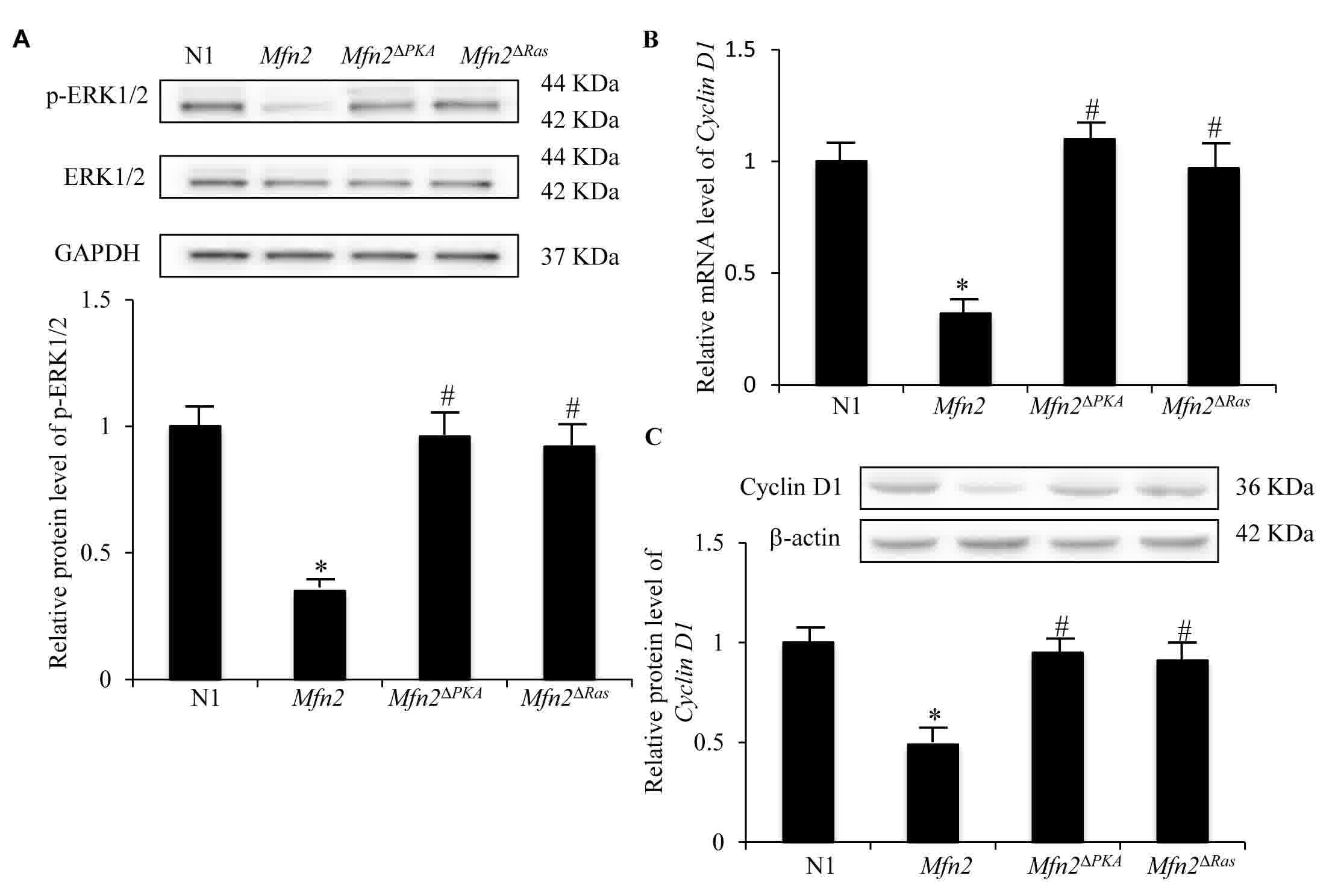 | Figure 3.Expression of p-ERK and Cyclin D1 in
MCF-7 cells transfected with N1, Mfn2, Mfn2ΔRas
and Mfn2ΔPKA plasmids. (A) Protein expression
levels of ERK and p-REK in determined by western blotting, using
GAPDH as a control. The results are expressed as the mean ±
standard deviation. *P<0.05 vs. N1 group; #P<0.05
vs. Mfn2 group. (B) mRNA expression of Cyclin D1
determined by reverse transcription-quantitative polymerase chain
reaction, relative to the N1 group, using GAPDH as an
internal control. (C) Relative protein expression levels of
Cyclin D1 determined by western blotting using β-actin as a
control. Showed. Results are expressed as the mean ± standard
deviation of 3 experiments, relative to the N1 group. *P<0.05
vs. N1 group; #P<0.05 vs. Mfn2 group. p-,
phosphorylated; ERK1/2, extracellular signal-regulated kinase 1/2;
N1, EGFP-N1 plasmid; Mfn2, mitofusin 2;
Mfn2ΔRas, Mfn2 ORF lacking the
p21Ras coding sequence; Mfn2ΔPKA,
Mfn2 ORF lacking the protein kinase A phosphorylation site
coding sequence. |
The PKA site phosphorylation of Mfn2
is also required for its anti-proliferation, anti-migration and
anti-invasion functions
It has been established that Ser442 of Mfn2 is a
protein kinase A (PKA) phosphorylation site (4). The PKA site of Mfn2 is essential for
Mfn2-mediated inhibition of ERK1/2 signaling, and the consequent
anti-proliferative effect, in rat VSMCs (16). In order to evaluate the importance of
the PKA site of Mfn2 in human breast cancer, MCF-7 cells were
transfected with plasmids containing the Mfn2 ORF with a PKA
phosphorylation site deletion (Mfn2ΔPKA). A total
of 48 h post-transfection, the mRNA and protein expression levels
of Mfn2 in MCF-7 cells transfected with
Mfn2ΔPKA plasmids were not significantly
different to those of MCF-7 cells transfected with Mfn2 full
length plasmids (Fig. 2A and B).
However, the proliferation (Fig. 2C),
migration (Fig. 2E) and invasion
(Fig. 2F) abilities of MCF-7 cells
transfected with Mfn2ΔPKA plasmids were markedly
increased compared with cells transfected with Mfn2 full
length plasmids (P<0.05). Cell cycle arrest was also rescued
(Fig. 2D). This indicates that the
PKA phosphorylation site was necessary for the anti-proliferation,
anti-migration and anti-invasion functions of the Mfn2 protein.
Furthermore, the expression levels of p-ERK1/2 (Fig. 3A) and Cyclin D1 (Fig. 3B and C) were significantly increased
in MCF-7 cell transfected with Mfn2ΔPKA plasmids
compared with those in MCF-7 cells transfected with Mfn2 full
length plasmids (P<0.05). These data indicate a crucial role of
the PKA phosphorylation site of Mfn2 in the inhibition of the
Ras-ERK1/2-Cyclin D1 pathway.
Discussion
The present study confirmed that Mfn2 was
hypoexpressed, and its promoter was hypermethylated, in the MCF-7
cell line and in human breast cancer specimens. However, Sorianello
et al (23) reported that the
promoter of Mfn2 was unmethylated in rat insulinoma and
hepatoma cells, as analyzed by bisulfite sequencing. It is
speculated that Mfn2 expression is regulated via different
mechanisms which vary between species and tissues. In addition,
demethylation treatment of MCF-7 cells with 5-aza-CdR resulted in
upregulation of Mfn2 expression in a dose-dependent manner.
This suggests that Mfn2 hypoexpression in breast cancer is
at least partially attributable to hypermethylaton of its promoter.
Although 5-aza-CdR can rescue the expression of tumor suppressor
genes by demethylating their promoter CpG sites (24–31), its
clinical anti-tumor applications are limited due to lack of gene
specificity. Furthermore, it was reported that Mfn2 is a
direct target of miR-761 and upregulation of Mfn2 expression
by inhibiting miR-761 repressed hepatocarcinoma growth and
metastasis (32). These results
indicate that epigenetic mechanisms serve an important role in
regulating Mfn2 expression at a transcriptional and
post-transcriptional level.
It has been established that the drosophila, rat and
human homologues of HSG serve a critical role in mitochondrial
fusion (33–36). In the present study, the
anti-proliferation, anti-migration and anti-invasion effects of
Mfn2 were confirmed in breast cancer cells. The results are
consistent with previous studies in VSMCs (3) and in various types of cancer, including
breast cancer (18,37) gastric cancer (18), urinary bladder carcinoma (38) and hepatocellular carcinoma (20,21,23) and
lung adenocarcinoma (5,8,24,38–41).
However, tumor-promoting functions of Mfn2 in lung adenocarcinoma
have also been reported (42,43). These conflicting results may be due to
differing experimental methods, but suggest that the roles of Mfn2
in different types of cancer are more complicated than
expected.
Furthermore, the effect of Mfn2 protein structure of
on its function in breast cancer was analyzed. In rat VSMCs, the
p21Ras motif of Mfn2 serves an essential role in
Mfn2-mediated inhibition of ERK1/2 signaling and growth arrest
(33–36). In the present study, it was
demonstrated that the p21Ras motif of Mfn2 is also
necessary for its anti-proliferation function in breast cancer
cells. Mfn2-Ras binding negatively regulated the Ras-ERK1/2-cyclin
D1 signaling pathway and resulted in cell cycle arrest in the G0/G1
phases of breast cancer cells. This suggests that Mfn2 is an
important protein in the Ras-signaling pathway. Furthermore, it is
known that Mfn2 contains a PKA phosphorylation site at Ser442
(33–36) and the cAMP-PKA signaling pathway is a
versatile signal pathway involved in regulation of cellular
functions through phosphorylation (44–48). The
present study demonstrated that the PKA phosphorylation site at
Ser442 of Mfn2 was essential for Mfn2-mediated inhibition of ERK1/2
signaling and reduced proliferation of breast cancer cells. These
results support those demonstrated in VSMCs (16,33–36). It
was speculated that deletion of the PKA phosphorylation site at
Ser442 of Mfn2 affects its tertiary structure and, consequently,
its interaction with Ras.
Although the absence of the p21Ras motif
or the PKA phosphorylation site at Ser442 of Mfn2 has no effect on
its mitochondrial membrane localization (3), altered mitochondrial function in breast
cancer has been observed (49). In
breast cancer cells, with the overexpression of Mfn2, the
vesicular endocytosis-associated protein, SH3 domain-containing
GRB2-like protein 2 (SH3GL2), was demonstrated to translocate to
mitochondria, and induce the production of superoxide and the
release of cytochrome C from the mitochondria to the cytoplasm
(49). This was accompanied by
decreased lung and liver metastases and primary tumor growth
(49). SH3GL2 depletion reversed this
phenotype (49). This indicates that
the anti-tumor functions of Mfn2 are also
mitochondrial-dependent.
To conclude, the present study demonstrates that
Mnf2 functions as a suppressor of breast cancer and suggests
that elucidation of its complex mechanism may reveal a novel target
for breast cancer therapy.
Acknowledgements
The authors would like to thank Dr Hongmei Liu at
Tangshan People's Hospital (Tangshan, China) for providing the
5-aza-CdR.
Funding
The present study was partially supported by the
National Natural Science Foundation of China (grant no.
81301779).
Availability of data and materials
The raw data are available from the corresponding
author on reasonable request.
Authors' contributions
JZ and XZ designed the experiments and revised the
manuscript. YL analyzed the data and wrote the manuscript. WD, XS,
HH, XL, YanL and YankL performed the experiments. All authors
discussed the results and commented on the manuscript.
Ethics approval and consent to
participate
The protocol and use of the specimens in the present
study was approved by the Institutional Ethics Committee of
Tangshan People's Hospital (Tangshan, China), and written consent
was obtained from all participants.
Consent for publication
Written informed consent for the publication was
obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Mfn2
|
mitofusin 2
|
|
PKA
|
protein kinase A
|
|
HSG
|
hyperperplasia suppressor gene
|
References
|
1
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fojo T: Multiple paths to a drug
resistance phenotype: Mutations, translocations, deletions and
amplification of coding genes or promoter regions, epigenetic
changes and microRNAs. Drug Resist Updat. 10:59–67. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen KH, Guo X, Ma D, Guo Y, Li Q, Yang D,
Li P, Qiu X, Wen S, Xiao RP and Tang J: Dysregulation of HSG
triggers vascular proliferative disorders. Nat Cell Biol.
6:872–883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang D, Ma C, Li S, Ran Y, Chen J, Lu P,
Shi S and Zhu D: Effect of Mitofusin 2 on smooth muscle cells
proliferation in hypoxic pulmonary hypertension. Microvasc Res.
84:286–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang GE, Jin HL, Lin XK, Chen C, Liu XS,
Zhang Q and Yu JR: Anti-tumor effects of Mfn2 in gastric cancer.
Int J Mol Sci. 14:13005–13021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang W, Sun Q, Wu Z, Zhou D, Wei J, Xie H,
Zhou L and Zheng S: Mitochondrial dysfunction-related genes in
hepatocellular carcinoma. Front Biosci (Landmark Ed). 18:1141–1149.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin B, Fu G, Pan H, Cheng X, Zhou L, Lv J,
Chen G and Zheng S: Anti-tumour efficacy of mitofusin-2 in urinary
bladder carcinoma. Med Oncol. 28 Suppl 1:S373–S380. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma L, Liu Y, Geng C, Qi X and Jiang J:
Estrogen receptor β inhibits estradiol-induced proliferation and
migration of MCF-7 cells through regulation of mitofusin 2. Int J
Oncol. 42:1993–2000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia Y, Wu YQ, Zhang L, Li XL, Yuan HL, He
XJ, Tao DD, Gong JP and Qiu FZ: Effects of mitofusin-2 gene on
proliferation and chemosensitivity of human breast carcinoma cell
line MCF-7. Ai Zheng. 26:815–819. 2007.(In Chinese). PubMed/NCBI
|
|
10
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chik F and Szyf M: Effects of specific
DNMT gene depletion on cancer cell transformation and breast cancer
cell invasion; toward selective DNMT inhibitors. Carcinogenesis.
32:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Antequera F and Bird A: Number of CpG
islands and genes in human and mouse. Proc Natl Acad Sci USA.
90:11995–11999. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bergman Y and Cedar H: DNA methylation
dynamics in health and disease. Nat Struct Mol Biol. 20:274–281.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akhavan-Niaki H and Samadani AA: DNA
methylation and cancer development: Molecular mechanism. Cell
Biochem Biophys. 67:501–513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou W, Cao WJ, Chen LL, Xiaomei G and
Guanghui CH: Effect of mitofusin 2 gene with protein kinase A
phosphorylation site deletion on the proliferation of vascular
smooth muscle cells. J Clin Rehabilitative Tissue Engineering Res.
14:1322–1325. 2010.
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gnyszka A, Jastrzebski Z and Flis S: DNA
methyltransferase inhibitors and their emerging role in epigenetic
therapy of cancer. Anticancer Res. 33:2989–2996. 2013.PubMed/NCBI
|
|
19
|
Yoo CB and Jones PA: Epigenetic therapy of
cancer: Past, present and future. Nat Rev Drug Discov. 5:37–50.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee S, Kim HS, Roh KH, Lee BC, Shin TH,
Yoo JM, Kim YL, Yu KR, Kang KS and Seo KW: DNA methyltransferase
inhibition accelerates the immunomodulation and migration of human
mesenchymal stem cells. Sci Rep. 5:80202015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu J, Huo D, Chen Y, Nwachukwu C, Collins
C, Rowell J, Slamon DJ and Olopade OI: CpG island methylation
affects accessibility of the proximal BRCA1 promoter to
transcription factors. Breast Cancer Res Treat. 120:593–601. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo Y, Stacey DW and Hitomi M:
Post-transcriptional regulation of cyclin D1 expression during G2
phase. Oncogene. 21:7545–7556. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sorianello E, Soriano FX,
Fernandez-Pascual S, Sancho A, Naon D, Vila-Caballer M,
Gonzalez-Navarro H, Portugal J, Andres V, Palacin M and Zorzano A:
The promoter activity of human Mfn2 depends on Sp1 in vascular
smooth muscle cells. Cardiovasc Res. 94:38–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vucic EA, Brown CJ and Lam WL: Epigenetics
of cancer progression. Pharmacogenomics. 9:215–234. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tada M, Kanai F, Tanaka Y, Tateishi K,
Ohta M, Asaoka Y, Seto M, Muroyama R, Fukai K, Imazeki F, et al:
Down-regulation of hedgehog-interacting protein through genetic and
epigenetic alterations in human hepatocellular carcinoma. Clin
Cancer Res. 14:3768–3776. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duffy MJ, Napieralski R, Martens JW, Span
PN, Spyratos F, Sweep FC, Brunner N, Foekens JA and Schmitt M:
EORTC PathoBiology Group: Methylated genes as new cancer
biomarkers. Eur J Cancer. 45:335–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tost J, Hamzaoui H, Busato F, Neyret A,
Mourah S, Dupont JM and Bouizar Z: Methylation of specific CpG
sites in the P2 promoter of parathyroid hormone-related protein
determines the invasive potential of breast cancer cell lines.
Epigenetics. 6:1035–1046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caffarelli E and Filetici P: Epigenetic
regulation in cancer development. Front Biosci (Landmark Ed).
16:2682–2694. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Coppedè F: The role of epigenetics in
colorectal cancer. Expert Rev Gastroenterol Hepatol. 8:935–948.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Milavetz BI and Balakrishnan L: Viral
epigenetics. Methods Mol Biol. 1238:569–596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu Y, Sarkissyan M and Vadgama JV:
Epigenetics in breast and prostate cancer. Methods Mol Biol.
1238:425–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou X, Zhang L, Zheng B, Yan Y, Zhang Y,
Xie H, Zhou L, Zheng S and Wang W: MicroRNA-761 is upregulated in
hepatocellular carcinoma and regulates tumorigenesis by targeting
Mitofusin-2. Cancer Sci. 107:424–432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Santel A and Fuller MT: Control of
mitochondrial morphology by a human mitofusin. J Cell Sci.
114:867–874. 2001.PubMed/NCBI
|
|
34
|
Rojo M, Legros F, Chateau D and Lombès A:
Membrane topology and mitochondrial targeting of mitofusins,
ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J
Cell Sci. 115:1663–1674. 2002.PubMed/NCBI
|
|
35
|
Karbowski M, Lee YJ, Gaume B, Jeong SY,
Frank S, Nechushtan A, Santel A, Fuller M, Smith CL and Youle RJ:
Spatial and temporal association of Bax with mitochondrial fission
sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 159:931–938.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen H, Detmer SA, Ewald AJ, Griffin EE,
Fraser SE and Chan DC: Mitofusins Mfn1 and Mfn2 coordinately
regulate mitochondrial fusion and are essential for embryonic
development. J Cell Biol. 160:189–200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stefansson OA, Jonasson JG, Olafsdottir K,
Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT and
Eyfjord JE: CpG island hypermethylation of BRCA1 and loss of pRb as
co-occurring events in basal/triple-negative breast cancer.
Epigenetics. 6:638–649. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang W, Zhu F, Wang S, Wei J, Jia C, Zhang
Y, Zhou L, Xie H and Zheng S: HSG provides antitumor efficacy on
hepatocellular carcinoma both in vitro and in vivo. Oncol Rep.
24:183–188. 2010.PubMed/NCBI
|
|
39
|
Xia Y, Wu Y, He X, Gong J and Qiu F:
Effects of mitofusin-2 gene on cell proliferation and chemotherapy
sensitivity of MCF-7. J Huazhong Univ Sci Technolog Med Sci.
28:185–189. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang W, Zhou D, Wei J, Wu Z, Cheng X, Sun
Q, Xie H, Zhou L and Zheng S: Hepatitis B virus X protein inhibits
p53-mediated upregulation of mitofusin-2 in hepatocellular
carcinoma cells. Biochem Biophys Res Commun. 421:355–360. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rehman J, Zhang HJ, Toth PT, Zhang Y,
Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C and Archer SL:
Inhibition of mitochondrial fission prevents cell cycle progression
in lung cancer. FASEB J. 26:2175–2186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lou Y, Li R, Liu J, Zhang Y, Zhang X, Jin
B, Liu Y, Wang Z, Zhong H, Wen S and Han B: Mitofusin-2
over-expresses and leads to dysregulation of cell cycle and cell
invasion in lung adenocarcinoma. Med Oncol. 32:1322015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lou Y, Zhang Y, Li R, Gu P, Xiong L, Zhong
H, Zhang W and Han B: Transcriptional profiling revealed the
anti-proliferative effect of MFN2 deficiency and identified risk
factors in lung adenocarcinoma. Tumour Biol. 37:8643–8655. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ansurudeen I, Willenberg HS, Kopprasch S,
Krug AW, Ehrhart-Bornstein M and Bornstein SR: Endothelial factors
mediate aldosterone release via PKA-independent pathways. Mol Cell
Endocrinol. 300:66–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Martini CN, Plaza MV and Vila Mdel C:
PKA-dependent and independent cAMP signaling in 3T3-L1 fibroblasts
differentiation. Mol Cell Endocrinol. 298:42–47. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Omar B, Zmuda-Trzebiatowska E, Manganiello
V, Göransson O and Degerman E: Regulation of AMP-activated protein
kinase by cAMP in adipocytes: Roles for phosphodiesterases, protein
kinase B protein kinase A, Epac and lipolysis. Cell Signal.
21:760–766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
McConnachie G, Langeberg LK and Scott JD:
AKAP signaling complexes: Getting to the heart of the matter.
Trends Mol Med. 12:317–323. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lorenowicz MJ, Fernandez-Borja M, Kooistra
MR, Bos JL and Hordijk PL: PKA and Epac1 regulate endothelial
integrity and migration through parallel and independent pathways.
Eur J Cell Biol. 87:779–792. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kannan A, Wells RB, Sivakumar S, Komatsu
S, Singh KP, Samten B, Philley JV, Sauter ER, Ikebe M, Idell S, et
al: Mitochondrial reprogramming regulates breast cancer
progression. Clin Cancer Res. 22:3348–3360. 2016. View Article : Google Scholar : PubMed/NCBI
|

















