Introduction
Reproducing the diverse heterogeneity of cancer in
preclinical models is important in mechanistic and functional
studies of tumor biology. Studying drug resistance mechanisms and
identifying biomarkers of therapeutic responses and biological
targets for treatment will be of increasing importance in the
future for individualized management. Among cancer types, lung
cancer has the highest global incidence and mortality. Non-small
cell lung cancer (NSCLC), which mainly comprises adenocarcinoma
(ADC) and squamous cell carcinoma (SCC), accounts for approximately
85% of all lung cancer cases and has a 5-year survival rate of less
than 20% (1–3). More recent studies have mainly focused
on lung ADC. However, SCC, a subgroup representing the second most
common type of lung cancer, accounts for over 30% of NSCLC
(4) but has received limited
attention. Traditionally, tumor biology studies have been conducted
mainly in cancer cell lines grown in vitro or implanted into
immunocompromised mice (e.g., nude, SCID, or NOD/SCID mice)
(5). Moreover, relatively few SCC
cell lines have been established (6).
The limitations of these models, including increased homogeneity
following long-term culture of established cell lines in
vitro, and cell line xenograft tumors rarely exhibiting the
tissue architecture of the original cancer, have become apparent
(7). Therefore, cell line xenograft
tumors frequently fail to adequately predict the efficacy of
anticancer agents in the clinic (8).
In theory, patient-derived tumor xenografts (PDTXs),
which are established by collecting fresh tissue specimens from
cancer patients and directly implanting them into immunocompromised
mice, may represent more realistic preclinical models as they
closely resemble the tissue architecture of primary tumors,
including interactions between other cell types such as the stroma
and endothelium (9). To date, several
PDTX models have been reported, including for NSCLC. These
model-related studies have demonstrated that PDTXs largely retain
the principal histological features and recapitulate the molecular
characterization of cancer biology (6,10–12). However, most have investigated overall
similarity between tumor and xenograft rather than differences,
which is the focus in the current study.
Long non-coding RNA (lncRNA) is a heterogeneous
class of transcripts with a minimum length of 200 bases and limited
protein-coding ability (13,14). However, increasing evidence indicates
that lncRNAs can affect multiple cellular functions and participate
in diverse physiological and pathological processes (15,16).
Moreover, emerging evidence supports the notion that aberrant
expression of lncRNAs plays a significant role in various human
malignant diseases, including NSCLC (17–22). For
this reason, we compared xenograft lncRNA profiles with those from
corresponding human tumors.
Materials and methods
Patients and tissue samples
Thirty-seven fresh tumor samples were obtained at
initial surgery from patients with pathologically confirmed SCC
between July 2013 and November 2014. No patient had received
chemotherapy or radiation therapy prior to surgery. Tumor samples
and clinical records were obtained from patients with their written
informed consent and the study was approved by the Anhui Provincial
Hospital Ethical Committee. The quality of the tumor samples was
assessed by histological evaluation, and tumor tissue accounted for
at least 50% of the sample. Sections of resected tumor samples for
the establishment of PDTX were immediately placed at 4°C in
RPMI-1640 solution (HyClone; GE Healthcare Life Sciences, Logan,
UT, USA) containing penicillin/streptomycin. Other tissue for
hematoxylin and eosin (H&E) staining and immunohistochemistry
(IHC) was processed by frozen sectioning and paraffin, and the
remaining tissue was immediately cryopreserved in liquid nitrogen
and stored at −80°C for future investigation.
Mouse use and care
Female 6–8-week-old Balb/c athymic nude mice (SLRC
Laboratory, Shanghai, China) were housed in a pathogen-free
environment and provided with sterile water, food, and litter.
Cages were replaced once per week, and temperature (24±2°C) and
humidity (55±5%) were controlled. All animal experiments were
approved by the Anhui Provincial Hospital Ethical Committee and
were carried out in accordance with the Guidelines for the Care and
Use of Laboratory Animals.
Establishment of PDTX models
The fresh tumor samples were placed into sterile
Petri dishes, washed three times with phosphate-buffered saline
(PBS), and cut into 3 mm3 fragments. Tumor fragments
were implanted subcutaneously into the left and right flanks of
mice (3–5 mice/patient specimen) and were monitored weekly using
Vernier calipers when the implanted tissue was palpable, with the
volume calculated as (length × width2)/2. When the volume of tumor
was 500 mm3 in any area of the mouse, the primary tumor
(P) was considered to have formed a xenograft tumor (X) and
designated ‘X-1,’ or not from xenograft tumors (no-X) if growth was
not detected by 4 months after implantation. Successfully engrafted
mice were euthanized and the tumor (X-1) was removed for serial
transplantation to the next generation (X-2, X-3). Following
removal of the tumor, necropsy was performed to determine the sites
of tumor metastasis (including lymph nodes, liver, and spleen). At
each xenograft passage, tumors were harvested, measured, fixed for
histopathologic analysis, cryopreserved (in 90% fetal bovine serum
and 10% DMSO), and snap-frozen in liquid nitrogen for future
investigation.
Histology and IHC
Paraffin-embedded tissue blocks from primary and
xenograft tumors were cut into 4 µm sections. The sections were
dewaxed in xylene, dehydrated in a graded alcohol series, and
stained with H&E for histological examination. Molecular marker
expression was assessed using IHC. Prior to application of the
primary antibody, sections were dewaxed and dehydrated, and antigen
retrieval was accomplished by heating in an autoclave according to
the antibody manufacturer's instructions. Endogenous peroxidase
activity was blocked with 3% H2O2. After
processing, sections were incubated overnight at 4°C with primary
antibodies against p53, p63, Ki-67, cytokeratin5/6, and E-cadherin
(ZsBio, Beijing, China). Sections were subsequently treated with
secondary antibody conjugated with horseradish peroxidase polymer
and DAB substrate according to the manufacturer's instructions.
Next, the slides were counterstained with hematoxylin and evaluated
by two pathologists using standard light microscopy. The proportion
of stained tumor cells was assessed by counting at least 1,000
cells in randomly selected ×400 magnification fields.
LncRNA microarray and computational
analysis
Samples
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. RNA quantity and quality
were measured with a NanoDrop ND-1,000 spectrophotometer (PeqLab,
Erlangen, Germany). RNA integrity was assessed by standard
denaturing agarose gel electrophoresis.
RNA microarray
The Arraystar Human LncRNA Microarray V3.0 is
designed for the global profiling of human LncRNAs and
protein-coding transcripts, and can detect 30,586 LncRNAs and
26,109 coding transcripts. The LncRNAs are carefully constructed
using established public transcriptome databases (e.g., Refseq,
UCSC knowngenes, Gencode) as well as landmark publications. Each
transcript is represented by a specific exon or splice junction
probe which can identify individual transcripts accurately.
Positive probes for housekeeping genes and negative probes are also
printed onto the array for hybridization quality control.
RNA labeling and array
hybridization
Sample labeling and array hybridization were
performed according to the Agilent One-Color Microarray-Based Gene
Expression Analysis protocol (Agilent Technologies, Inc., Santa
Clara, CA, USA) with minor modifications. Briefly, mRNA was
purified from total RNA after removal of rRNA (mRNA-ONLY™
Eukaryotic mRNA Isolation kit; Epicentre; Illumina, Inc., San
Diego, CA, USA). Then, each sample was amplified and transcribed
into fluorescent cRNA along the entire length of the transcripts
without 3′ bias utilizing a random priming method (Arraystar Flash
RNA Labeling kit; Arraystar, Rockville, MD, USA). The labeled cRNAs
were purified with an RNeasy Mini kit (Qiagen GmbH, Hilden,
Germany). The concentration and specific activity of the labeled
cRNAs (pmol Cy3/µg cRNA) were measured by NanoDrop ND-1,000. Next,
1 µg of each labeled cRNA was fragmented by adding 5 µl of 10X
blocking agent and 1 µl of 25X fragmentation buffer, heating the
mixture at 60°C for 30 min, and adding 25 µl 2X GE hybridization
buffer to dilute the labeled cRNA. Hybridization solution (50 µl)
was dispensed into the gasket slide and assembled with the LncRNA
expression microarray slide. The slides were incubated for 17 h at
65° in an Agilent hybridization oven. The hybridized arrays were
washed, fixed, and scanned using an Agilent DNA Microarray Scanner
(G2505C; Agilent Technologies, Inc.).
Data analysis
Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies, Inc.) was used to analyze the
acquired array images. Quantile normalization and subsequent data
processing were performed using the Gene Spring GX v12.1 software
package (Agilent Technologies, Inc.). After quantile normalization
of the raw data, LncRNAs and mRNAs from at least 6 out of 12
samples with flags in Present or Marginal (‘All Targets Value’)
were chosen for further data analysis. Differentially expressed
LncRNAs and mRNAs with statistical significance between the two
groups were identified through P-value/FDR filtering (P<0.05).
Differentially expressed LncRNAs and mRNAs between two samples were
identified through fold-change filtering (fold-change >2.0).
Hierarchical clustering and combined analysis were performed using
homemade scripts.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
To validate the selected mRNA expression levels in
primary tumors compared with third generation xenograft tumors,
RT-qPCR analysis was applied. β-actin was used as an internal
control. The primers used are listed in Table I. RT-qPCR was performed using the
SYBR-Green (Takara Biotechnology Co., Ltd., Dalian, China) dye
detection method on an ABI 7500 PCR instrument under the following
conditions: 95°C for 10 min, and 40 cycles of 95°C for 10 min and
60°C for 60 sec. Relative gene expression levels were analyzed by
the 2−ΔΔCq method as previously reported (23), where
ΔCq=Cqtarget-Cqβ-actin.
 | Table I.Primer sequences used in this
study. |
Table I.
Primer sequences used in this
study.
| Target ID | Forward primer | Reverse primer |
|---|
| NRG1 |
CACTGGGACAAGCCATCTT |
AAGCACTCCCCTCCATTCA |
| CDC16 |
GGTCTTAGGCGAGATGATACA |
AATCCCACGGAGGTGAAATA |
| CCL20 |
GCGCAAATCCAAAACAGAC |
CCATTCCAGAAAAGCCACA |
| HLA-DPB1 |
CGGAGTAAGACATTGACGGG |
GGAGCCAGATGCTAACGAA |
| β-actin |
GTGGCCGAGGACTTTGATTG |
CCTGTAACAACGCATCTCATATT |
Chemosensitivity testing
For chemotherapeutic response assays,
second-generation xenograft tumor fragments were subcutaneously
transplanted in 6–8-week-old female mice. When tumor volume reached
50–200 mm3, 3–5 mice were randomly assigned to treatment
or control groups. Cisplatin (20 mg/ml; Jiangsu Hansoh
Pharmaceutical Co., Ltd., Jiangsu, China) preparations were diluted
to 0.5 mg/ml in saline and administered weekly (5 mg/kg/d, i.p.) to
treatment group xenograft tumors for 3 weeks. Control mice were
injected with an equivalent volume of saline. The injection volume
was 0.2 ml/20 g body weight. Tumor volume was monitored weekly
using Vernier calipers and tumor volume was calculated as for the
establishment of PDTXs. Relative tumor volume (RTV) and antitumor
activity were calculated as previously described (24). Briefly, RTV=(Vx/V1), where Vx is the
tumor volume on day × and V1 is the tumor volume upon initiation of
therapy (day 1). Antitumor activity was evaluated according to
tumor growth inhibition, percentage of growth
inhibition=100-(RTVt/RTVcx100), where RTVt is the mean RTV of
treatment groups and RTVc is the mean RTV of control groups. Tumor
growth inhibition of 50% was considered a meaningful biological
effect.
Statistical analysis
The unpaired two-tailed t-test, Wilcoxon rank sum
test or Chi-square test were used to determine the association of
individual tumor features with engraftment. The proportions of
Ki-67 stained tumor cells were presented as means ± standard
deviation and analyzed using Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were carried out using the statistical
analysis package SPSS version 11.0 (SPSS, Inc., Chicago, IL,
USA).
Results
Xenograft engraftment was associated
with tumor size, differentiation, and expression of Ki-67
To establish the PDTXs, surgically resected samples
from 37 SCC patients were each implanted subcutaneously into 3–5
athymic nude mice within 1 h of resection. Individual patient data
and tumor histopathology are presented in Table II. We successfully established 18
PDTXs (engraftment rate 48.6%, 18/37). The characteristics of
primary tumors that did or didn't form xenografts (X or no-X) were
compared retrospectively. Results showed that tumor engraftment was
not dependent on patient's age, smoking history, or tumor
pathological characteristics such as tumor TNM stage, grade, and
lymph node metastasis. However, tumors that formed xenografts had a
larger median tumor volume (87.5 vs. 15.8 cm3,
P<0.001; Table III). Poorly
differentiated tumors were notably easier to engraft than
moderately differentiated tumors (73.7 vs. 22.2%, P=0.002; Table III). In multivariate analysis, poor
differentiation and larger tumor volume were independently
associated with increased rates of engraftment. Therefore, we
speculate that the primary tumors which formed xenografts might
contain higher numbers of actively proliferating cancer cells.
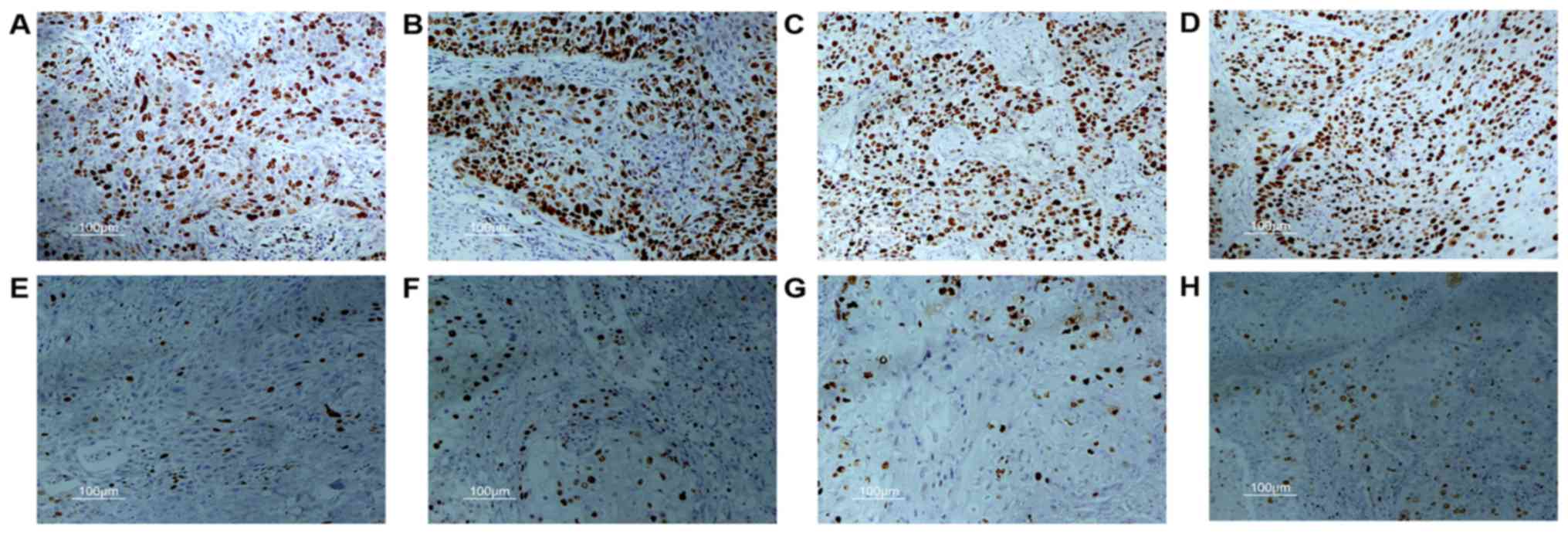
Ki-67 is a nuclear protein expressed by proliferating cells. The
labeling index of Ki-67 was determined in tumor samples by IHC. As
expected, the X group had a higher proportion of tumor cells
stained by Ki-67 than the no-X group (58.6±22.0 vs. 27.4±24.3%,
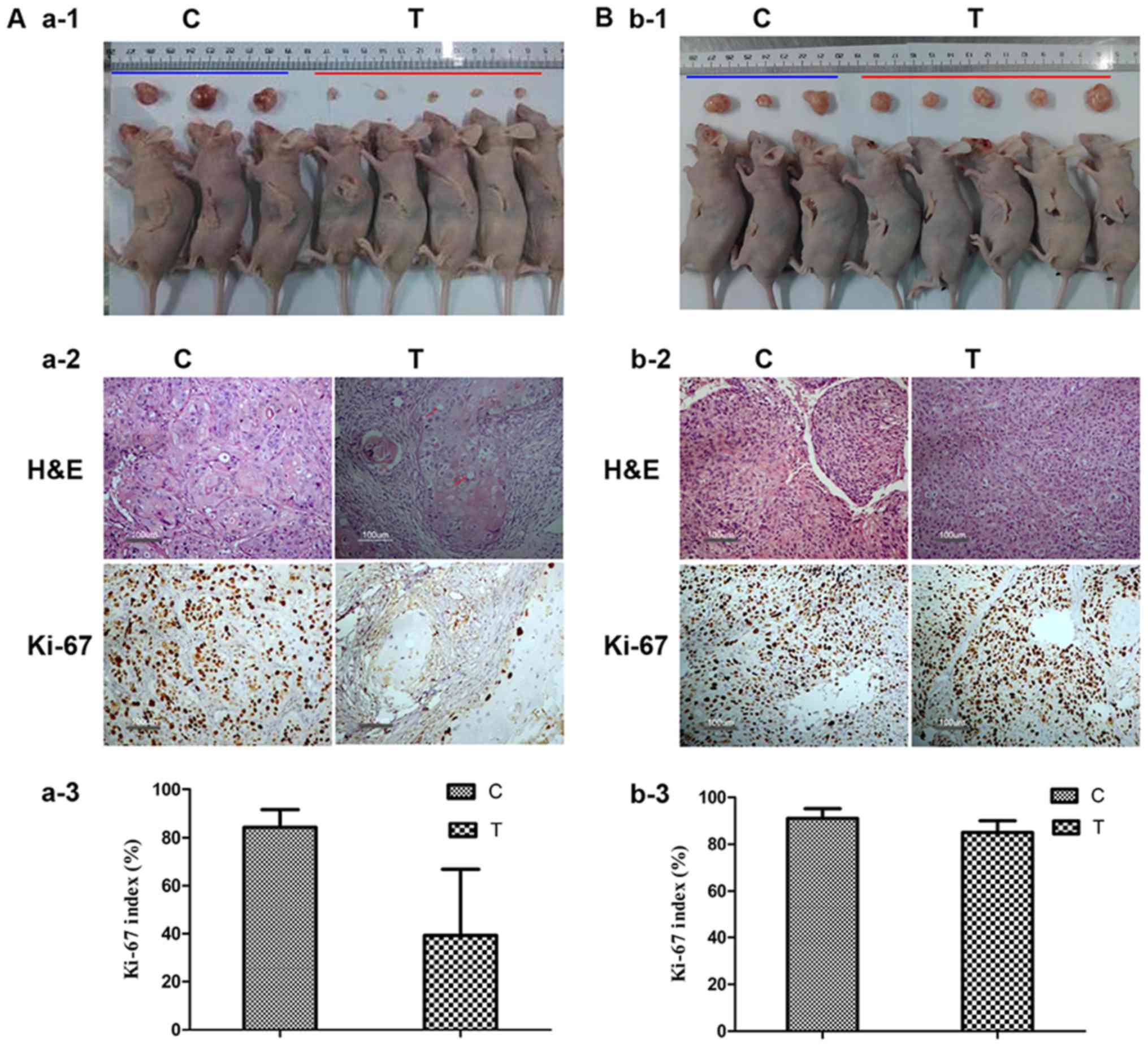
P<0.001; Fig. 1).
 | Table II.Clinical materials of 37 lung
squamous cell carcinoma patients and histopathology of their
tumors. |
Table II.
Clinical materials of 37 lung
squamous cell carcinoma patients and histopathology of their
tumors.
| Cases | Age/sex | TNM stage |
Differentiation | Smoking | LN metastasis | Xenograft
formationa |
|---|
| 1 | 58/M | II | Poor | No | No | Yes |
| 2 | 70/M | II | Poor | No | No | Yes |
| 3 | 58/M | III | Poor | No | Yes | Yes |
| 4 | 65/M | II | Poor | Yes | No | Yes |
| 5 | 57/M | III | Poor | No | No | Yes |
| 6 | 72/M | I | Poor | Unknown | No | Yes |
| 7 | 59/M | III | Poor | No | Yes | Yes |
| 8 | 60/M | I | Poor | No | No | Yes |
| 9 | 73/M | II | Moderate | Yes | No | Yes |
| 10 | 70/M | III | Poor | Yes | No | Yes |
| 11 | 61/M | III | Poor | No | Yes | Yes |
| 12 | 67/M | III | Poor | Yes | Yes | Yes |
| 13 | 73/M | III | Moderate | Yes | Yes | Yes |
| 14 | 64/M | I | Moderate | No | No | Yes |
| 15 | 51/M | I | Poor | Yes | No | Yes |
| 16 | 77/M | II | Poor | No | No | Yes |
| 17 | 65/M | I | Moderate | Yes | No | Yes |
| 18 | 55/M | III | Poor | No | Yes | Yes |
| 19 | 56/M | III | Moderate | Yes | Yes | No |
| 20 | 75/M | II | Moderate | Yes | No | No |
| 21 | 72/M | III | Poor | Unknown | Yes | No |
| 22 | 74/M | I | Poor | Yes | No | No |
| 23 | 69/M | I | Moderate | No | No | No |
| 24 | 71/M | II | Moderate | Yes | No | No |
| 25 | 71/M | II | Moderate | No | No | No |
| 26 | 59/M | III | Moderate | Yes | Yes | No |
| 27 | 69/M | II | Moderate | Yes | Yes | No |
| 28 | 72/M | I | Moderate | Yes | No | No |
| 29 | 73/M | IV | Poor | Yes | No | No |
| 30 | 64/F | I | Moderate | No | No | No |
| 31 | 63/F | I | Moderate | No | No | No |
| 32 | 69/M | III | Moderate | Yes | Yes | No |
| 33 | 59/M | I | Poor | No | No | No |
| 34 | 70/M | II | Poor | Yes | No | No |
| 35 | 49/M | III | Moderate | Yes | Yes | No |
| 36 | 60/M | I | Moderate | Unknown | No | No |
| 37 | 66/M | II | Moderate | Unknown | No | No |
 | Table III.Association between tumorigenicity of
SCC in nude mice and the clinicopathological parameters of
patients. |
Table III.
Association between tumorigenicity of
SCC in nude mice and the clinicopathological parameters of
patients.
| Clinicopathological
parameter | X (n=18) | No-X (n=19) | P-value |
|---|
| Age, years | 64.2±7.3 | 66.4±7.0 | 0.136a |
| Median tumor
volume, cm3 (25%, 75%) | 87.5 (44.8,
130.5) | 15.8 (6.0,
48.0) |
<0.001b |
| TNM stage |
|
| 0.484c |
| I | 5
(27.8) | 8 (42.1) |
|
| II | 5
(27.8) | 6
(31.6) |
|
|
III/IV | 8
(44.4) | 5
(26.3) |
|
|
Differentiation |
|
| 0.002c |
|
Moderate | 4
(22.2) | 14 (73.7) |
|
|
Poor | 14 (77.8) | 5
(26.3) |
|
| Smoking |
|
| 0.286c |
|
Yes | 7
(38.9) | 10 (52.6) |
|
| No | 10 (55.6) | 6
(31.6) |
|
|
Unknown | 1 (5.6) | 3
(15.8) |
|
| Lymph node
metastasis |
|
| 0.641c |
|
Positive | 6
(33.3) | 5
(26.3) |
|
|
Negative | 12 (66.7) | 14 (73.7) |
|
Passaged xenografts proliferated
faster than first-generation xenografts
Among the 18 successfully established xenografts, 16
were passaged to second generation (engraftment rate of 89%) and
all 16 sec-generation xenografts were serially passaged to third
generation (engraftment rate of 100%). A comparison between primary
tumors and the corresponding xenograft tumors revealed similar
histological architecture within three passages, particularly in
terms of cell type and grade of nuclear atypia (Fig. 2, H&E). However, second-generation
xenograft tumors required significantly less time to reach a size
of 500 mm3 compared with first-generation tumors
(average time of 6.8 and 9.2 weeks, respectively, P<0.001), but
the difference between the second and third generation was not
significant (average time of 6.3 and 6.8 weeks, respectively,
P>0.05), which suggested that passaged xenografts were more
proliferative than the first-generation tumor and that PDTXs had
increased proliferative activity of cancer cells. Moreover, for
primary tumors obtained from six patients with lymph node
metastasis, as a result of short time and relatively small numbers
of mice, all xenograft tumors showed only local growth, without
distant metastasis at autopsy.
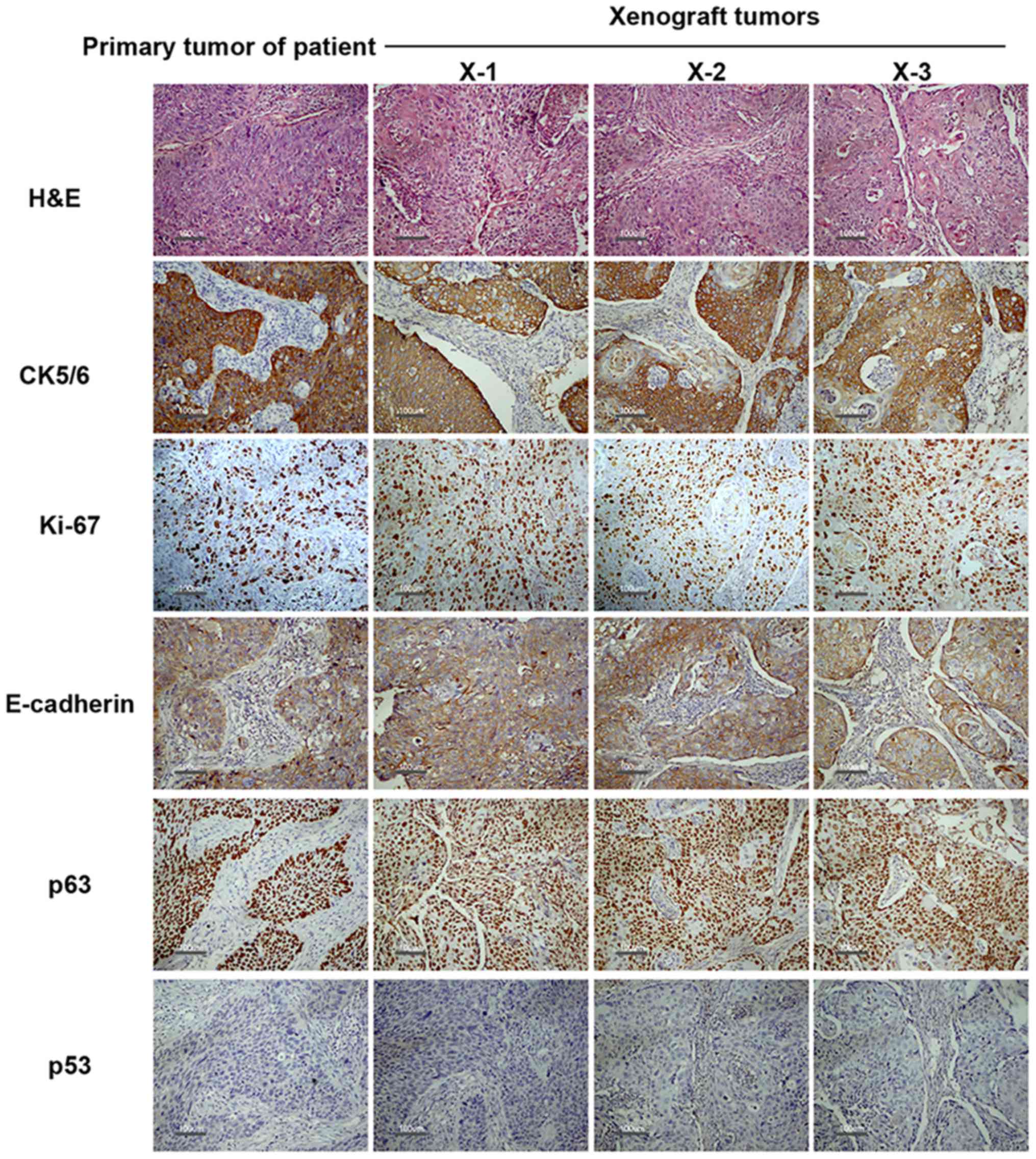 | Figure 2.Representative findings of each
histology/immunophenotypic in each passage. Histology and tumor
molecular markers (p53, p63, Ki-67, cytokeratin5/6 and E-cadherin)
expression of primary tumor (P) and corresponding xenograft tumors
(X-1, X-2 and X-3) were detected by H&E and IHC. No major
differences are seen in the tumor structure and cancer molecular
markers expression between the primary tumor and the xenograft
tumors except Ki-67. In the case of Ki-67 expression, compared with
P, an increase was observed in X-1 and also in serially passages
(X-2, X-3), suggesting a more proliferative phenotype characterized
by over expression of Ki-67. Scale bars, 100 µm. X-1, X-2 and X-3
represents the first, second and third generation xenograft tumor,
respectively. |
PDTXs largely retained histological
and key immunophenotypic features apart from increased expression
of Ki-67 in primary tumors with relatively low expression
Whether PDXTs suitable for the preclinical studies,
12 representative xenograft tumors and matched primary tumors were
evaluated by IHC for markers of proliferation (p53 and Ki-67),
aggressiveness (E-cadherin), and differentiation (p63 and
cytokeratin5/6). A representative sample is shown in Fig. 2, and the overall expression of
E-cadherin, p53, p63, and cytokeratin5/6 in primary tumors was
highly similar to that in matched xenograft tumors. Furthermore, in
general, the primary and xenograft tumors shared a strong
expression of Ki-67 (proportion of the Ki-67-stained tumor cells
>75%). However, some primary tumors (cases 2, 9, 10, 11, 14 and
15) with lower levels of Ki-67 expression (average proportion of
Ki-67 stained tumor cells 41.7±8.8%) were observed, but expression
was significantly elevated in xenograft tumors (75.0±14.0,
74.2±15.6 and 83.3±7.5% for first, second, and third generation,
respectively, P<0.05). This tendency was particularly pronounced
in the third-generation xenograft tumors. Altogether, these data
suggest that xenograft tumors maintain the essential
immunophenotypic features of the primary tumor, but that in the
process of establishing PDTXs, the selection of a more
proliferative phenotype characterized by overexpression of Ki-67
may be accomplished.
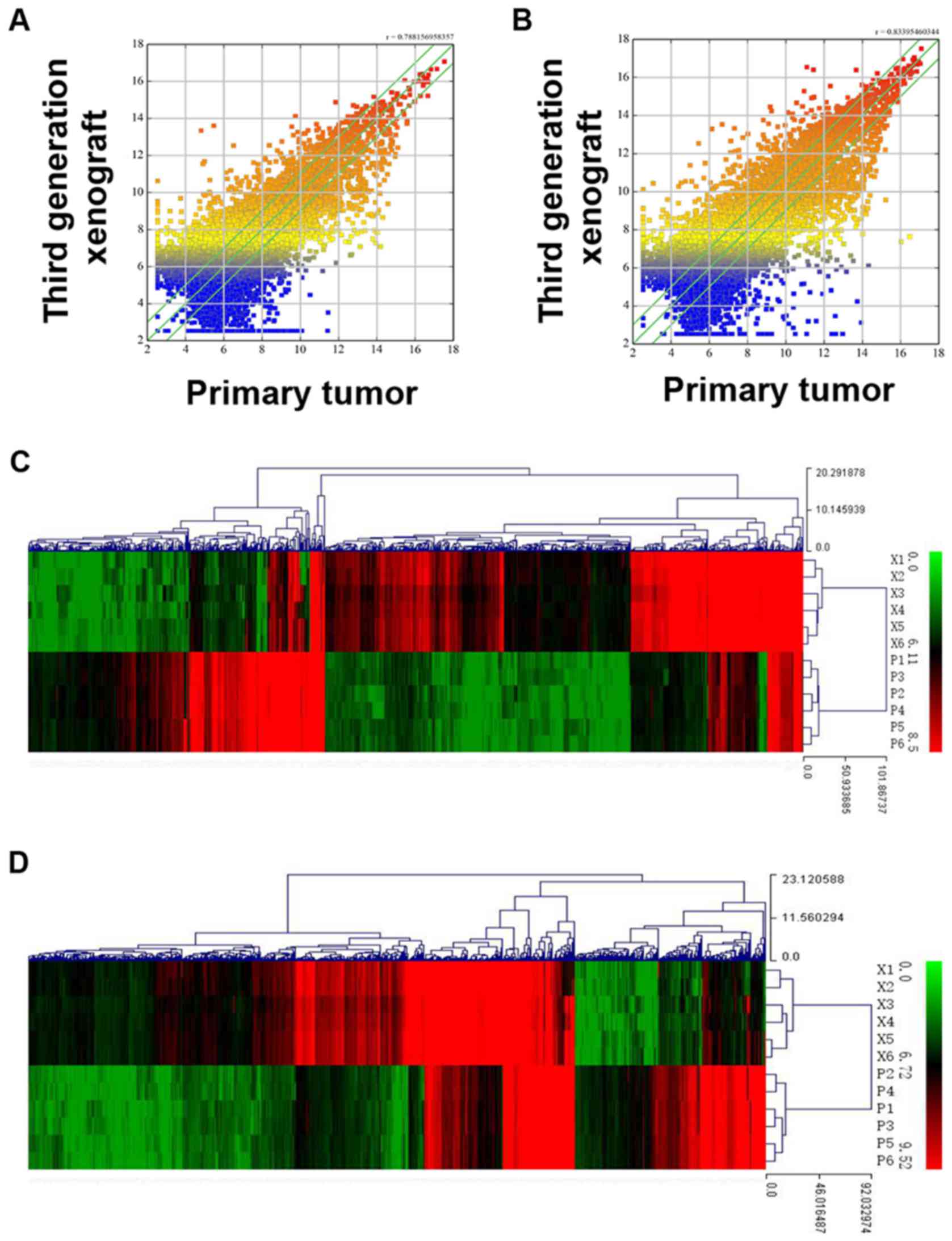
LncRNA and mRNA expression in
third-generation xenograft tumors differed from primary tumors
To determine whether the genetic features of
xenograft tumors changed during the serial passage of PDTX models,
a gene chip study was performed in 6 primary tumors (P) and their
corresponding third-generation xenograft tumors (X-3) using an
Arraystar probe dataset of 30,586 lncRNAs and 26,109 mRNAs. Linear
regression analysis was performed to analyze the lncRNA and mRNA
expression patterns in xenograft tumors and primary tumors. Gene
expression (including lncRNAs and mRNAs) scatter plots between
primary tumors and third-generation xenograft tumors indicated that
the majority of probes were not differentially expressed. An
example of the correlation between lncRNA and mRNA expression
patterns is shown in Fig. 3A and B.
The correlation coefficient of lncRNAs between P and X-3 ranged
from 0.788 to 0.851. Similarly, mRNA expression levels were also
well correlated between P and X-3 tumors, with a correlation
coefficient ranging from 0.834 to 0.895 (listed in Table IV). However, lncRNA or mRNA
expression profiling in the xenograft tumor and primary tumor was
separated into two distinct clusters by unsupervised hierarchical
clustering, with all xenograft tumors within the same branch and
primary tumors within the other branch. Differentially expressed
lncRNAs or mRNAs with statistical significance were identified by
Volcano Plot filtering between third-generation xenograft tumors
(X-3) and primary tumors (fold-change >2.0, P<0.05). Compared
with primary tumors, 2,109 lncRNAs were consistently upregulated
and 2,129 lncRNAs were consistently downregulated in the X-3
groups. In addition, the expression of 2,582 mRNAs increased and
1,122 decreased in the X-3 groups. Fold-change analysis between the
primary and xenograft tumors revealed 941 differentially expressed
lncRNAs (585 upregulated, 356 downregulated) and 695 differentially
expressed mRNAs probe sets (474 upregulated, 221 downregulated)
with fold-changes >5.0. Clustering based on these probe sets
showed a clear distinction between primary tumors and xenografts
(Fig. 3C and D). The expression
levels of the 20 top-ranked lncRNAs and mRNAs (xenograft tumors vs.
primary tumors) are listed in Tables
V and VI.
 | Table IV.Correlation coefficient of lncRNAs
and mRNAs between primary tumors and corresponding third generation
xenografts (X-3). |
Table IV.
Correlation coefficient of lncRNAs
and mRNAs between primary tumors and corresponding third generation
xenografts (X-3).
|
| Correlation
coefficient |
|---|
|
|
|
|---|
| Cases | lncRNA | mRNA |
|---|
| 3 | 0.85 | 0.89 |
| 6 | 0.84 | 0.88 |
| 9 | 0.79 | 0.83 |
| 10 | 0.81 | 0.87 |
| 12 | 0.81 | 0.87 |
| 15 | 0.85 | 0.89 |
 | Table V.Deregulated mRNAs detected using
microarray in 6 primary tumors and corresponding third generation
xenografts (X-3). |
Table V.
Deregulated mRNAs detected using
microarray in 6 primary tumors and corresponding third generation
xenografts (X-3).
| A, Upregulated in
X-3 group |
|---|
|
|---|
| Seqname | GeneSymbol | Fold-change |
|---|
| NM_018661 | DEFB103B | 152 |
| NM_006158 | NEFL | 142 |
| NM_001017920 | DAPL1 | 105 |
| NM_001062 | TCN1 | 104 |
| NM_022097 | CHP2 | 76 |
| NM_001024372 | BAALC | 73 |
| NM_001144940 | VMO1 | 72 |
| NM_001014291 | SPRR2G | 65 |
| NM_001144941 | VMO1 | 56 |
| NM_173178 | IL36B | 52 |
| NM_020958 | PPP4R4 | 51 |
| NM_005557 | KRT16 | 48 |
| NM_001146055 | SNCA | 48 |
| NM_018159 | NUDT11 | 47 |
|
ENST00000263182 | BBOX1 | 45 |
|
ENST00000315238 | CALML3 | 43 |
| NM_182566 | VMO1 | 40 |
| NM_001144939 | VMO1 | 39 |
| NM_013964 | NRG1 | 39 |
|
| B, Downregulated
in X-3 group |
|
| Seqname |
GeneSymbol |
Fold-change |
|
| NM_002196 | INSM1 | 847 |
| NM_153488 | MAGEA2B | 135 |
| NM_006183 | NTS | 123 |
| NM_172313 | CSF3R | 122 |
| NM_001025199 | CHI3L2 | 117 |
| NM_001127592 | FCGR3A | 68 |
| NM_021048 | MAGEA10 | 61 |
| NM_000569 | FCGR3A | 58 |
| NM_006172 | NPPA | 52 |
| NM_018643 | TREM1 | 49 |
| NM_002364 | MAGEB2 | 49 |
| NM_001161728 | PLA2G2A | 44 |
| NM_002338 | LSAMP | 43 |
| NM_001130046 | CCL20 | 41 |
| NM_002121 | HLA-DPB1 | 39 |
| NM_005306 | FFAR2 | 38 |
| NM_015424 | CHRDL2 | 34 |
| NM_003353 | UCN | 34 |
| NM_152997 | FDCSP | 31 |
 | Table VI.Deregulated lncRNAs detected using
microarray in 6 primary tumors and corresponding third generation
xenografts (X-3). |
Table VI.
Deregulated lncRNAs detected using
microarray in 6 primary tumors and corresponding third generation
xenografts (X-3).
| A, Upregulated in
X-3 group |
|---|
|
|---|
| Seqname | GeneSymbol | Fold-change |
|---|
| NR_038340 | LOC100505817 | 184 |
|
ENST00000583942 | CTD-2354A18.1 | 118 |
|
ENST00000448991 | RP1-214M20.2 | 114 |
|
ENST00000435813 | RP11-346D6.6 | 96 |
| uc022cje.1 | CYorf16 | 81 |
|
ENST00000584612 | KRT16P2 | 77 |
|
ENST00000498616 | RP11-85M11.2 | 74 |
|
ENST00000583748 | AC022596.6 | 69 |
| uc001uzl.3 | BC025370 | 66 |
|
ENST00000425820 | RP4-694A7.2 | 65 |
|
ENST00000425820 | RP4-694A7.2 | 65 |
| TCONS_00010362 | XLOC_004859 | 60 |
|
ENST00000576842 | CTD-2034I21.2 | 46 |
|
ENST00000579062 | KRT16P2 | 39 |
| NR_024475 | LOC100216001 | 35 |
|
ENST00000504916 | RP11-78C3.1 | 35 |
|
ENST00000433377 | RP5-866L20.1 | 34 |
|
ENST00000526487 | RP11-839D17.3 | 33 |
|
ENST00000434541 | AC147651.1 | 30 |
|
ENST00000509399 | RP11-297P16.4 | 30 |
|
| B, Downregulated
in X-3 group |
|
| Seqname |
GeneSymbol |
Fold-change |
|
| NR_034129 | LOC100128098 | 399 |
|
ENST00000451190 | RP11-414K1.3 | 128 |
|
ENST00000420058 | RP11-645N11.2 | 94 |
|
ENST00000420058 | RP11-645N11.2 | 94 |
| TCONS_00026385 | XLOC_012735 | 68 |
|
ENST00000440221 | AL773572.7 | 68 |
|
ENST00000440221 | AL773572.7 | 68 |
|
ENST00000440221 | AL773572.7 | 68 |
|
ENST00000440221 | AL773572.7 | 68 |
|
ENST00000451225 | RP11-414K1.3 | 64 |
| uc002ywn.1 | AL109792 | 62 |
|
ENST00000429730 | AC079767.4 | 61 |
| uc001gzl.3 | BC034684 | 58 |
| NR_033863 | FLJ41200 | 49 |
|
ENST00000470997 | HLA-DPB2 | 43 |
|
ENST00000579923 | RP11-106E15.1 | 39 |
|
ENST00000538294 | RP11-230G5.2 | 37 |
|
ENST00000446557 | RP13-16H11.2 | 36 |
|
ENST00000523523 | CTD-2024D23.1 | 26 |
| TCONS_00010211 | XLOC_004680 | 24 |
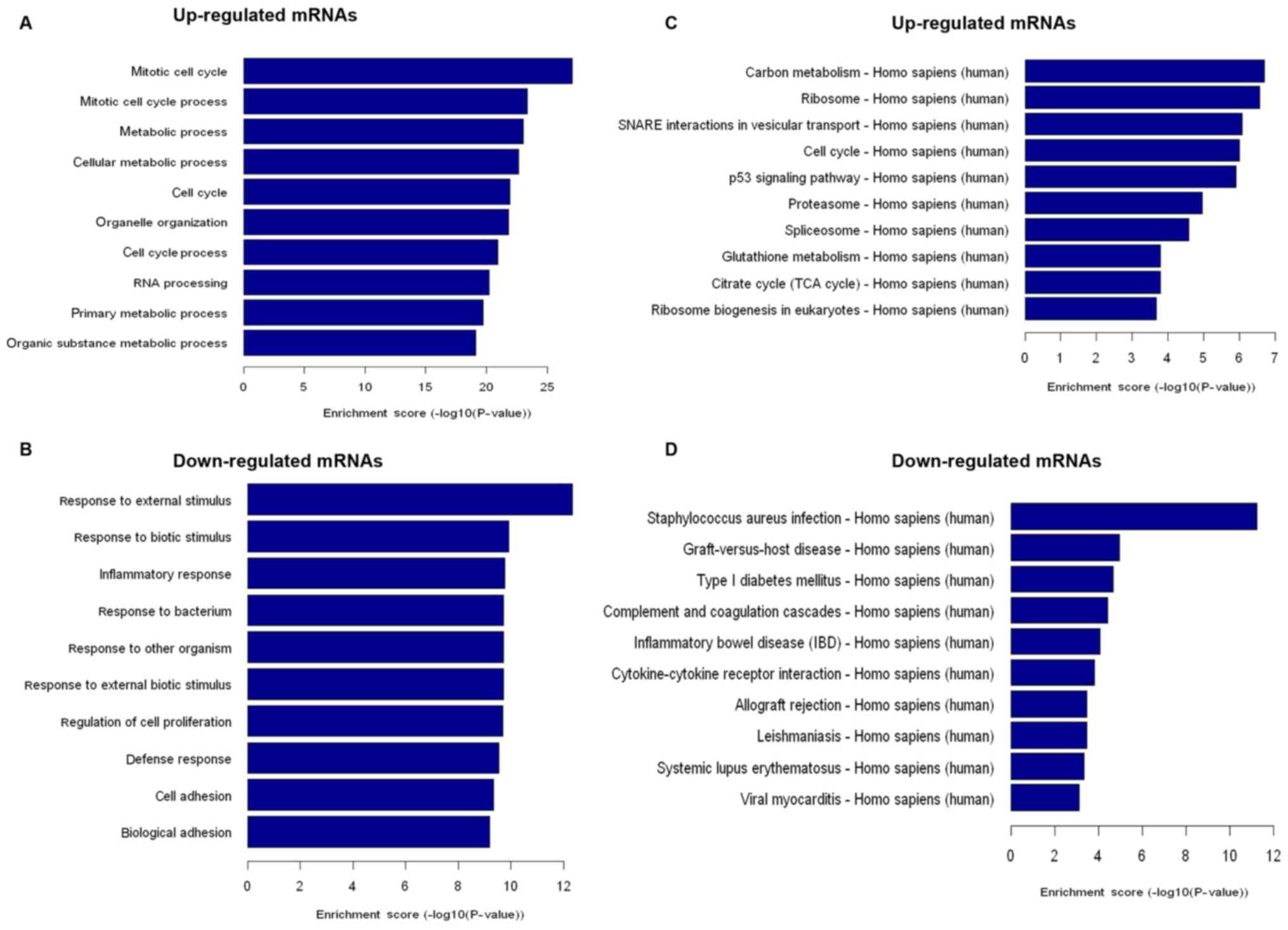
Upregulated mRNAs in PDTXs were mainly
associated with cell proliferation while downregulated mRNAs
appeared to be responsible for immune response
GO analysis showed that upregulated mRNAs in PDTXs
were mainly associated with cell cycle and metabolic processes
(Fig. 4A), while downregulated mRNAs
were mainly involved in the immune response and cell adhesion
(Fig. 4B). Similarly, pathway
analysis indicated that 45 enriched pathways corresponded to
upregulated mRNAs in xenograft groups. Among these, we found that
several enriched networks including ‘Cell cycle’, ‘p53 signaling
pathway’, ‘Carbon metabolism’, and ‘Glutathione metabolism’ were
associated with cell proliferation (Fig.
4C). In addition, 32 enriched pathways corresponding to
downregulated mRNAs in xenograft groups were responsible for
‘Graft-versus-host’, ‘Allograft rejection’, and ‘Cytokine-cytokine
receptor interaction’ networks, which are associated with
immune-mediated processes (Fig.
4D).
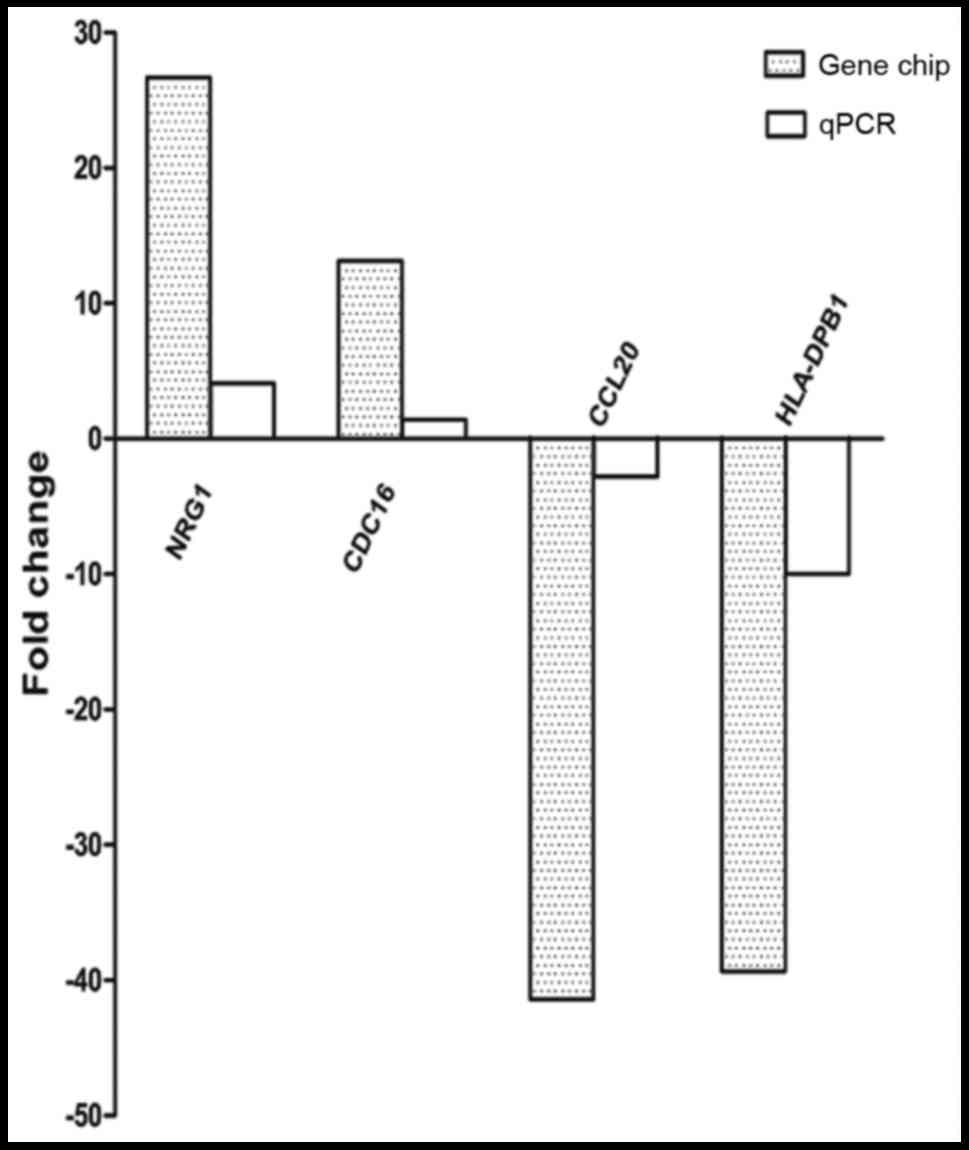
To validate the microarray and pathway analysis
results, the expression level of 4 mRNAs (NRG1, CDC16, HLA-DPB1,
and CCL-20) thought to play important roles in cell proliferation
or immune response were analyzed by RT-qPCR. The expression levels
of NRG1 and CDC16 were clearly increased (all P<0.05), while
HLA-DPB1 and CCL-20 were dramatically decreased in xenograft tumors
(all P<0.05). These RT-PCR results were consistent with those
observed in the microarray analysis (Fig.
5).
Association between cisplatin response
and p53 expression
Platinum-based chemotherapy is currently the
standard first-line treatment for SCC, and most patients in the
study had received cisplatin. To further characterize the
established xenograft models, the third-generation xenograft tumors
from 12 representative patients were treated with cisplatin.
Fig. 6 presents the growth curves of
the 12 xenograft tumors treated with cisplatin or saline, and the
results show that 7 of 12 xenograft tumors were responsive to
cisplatin chemotherapy (response rate 58%) while the remaining 5
were resistant. Furthermore, the expression of p53 was positive in
4 of 5 nonresponding tumors and negative in 6 of 7 responding
tumors (Table VII), indicating the
benefit of administrating cisplatin-based adjuvant chemotherapy
according to the expression status of p53.
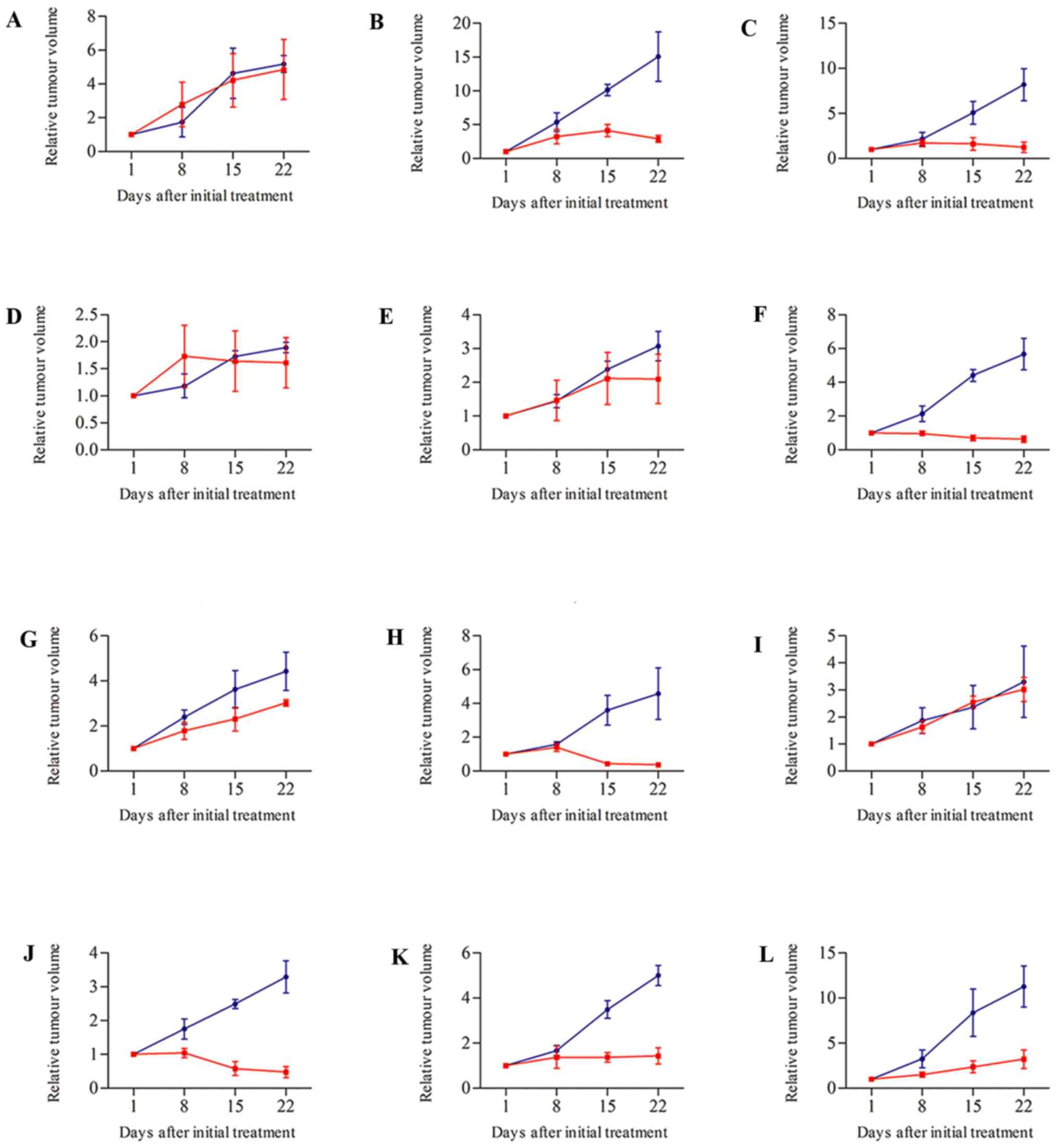 | Figure 6.Therapeutic response of PDTXs treated
with cisplatin or saline. Cispaltin (red) was administered at a
dose of 5 mg/kg every week. Mice in the control group (blue) were
injected with the same volumes of saline. Mice were treated weekly
for 3 wk (i.p.). Treatment started when subcutaneous growing tumor
volumes were 50–200 mm3; tumor volume and RTV were
calculated as described in materials and methods; growth curves
were obtained by plotting mean RTV against time. Compared with
control groups, 7 of 12 xenograft tumors were responsive to
cisplatin chemotherapy (B, C, F, H, J, K and L), the others (A, D,
E, G and I) were resistant. Error bars represent calculated
standard deviation. PDTX, patient-derived tumor xenograft; RTV,
relative tumor volume. |
 | Table VII.Comparison of clinical outcome
(post-operatively treated with platinum-based regime) with
responses of xenografts to cisplatin. |
Table VII.
Comparison of clinical outcome
(post-operatively treated with platinum-based regime) with
responses of xenografts to cisplatin.
|
| Patients | Xenografts |
|---|
|
|
|
|
|---|
| Cases | TNM stage | Treatment
regimes | Clinical
outcome | Disease-free time
(mo) | p53 protein | Response to
cisplatin (n) |
|---|
| 1 | T3N0M0 | Cisplatin 75 mg/m2,
day 1+vinorelbine 30 mg/m2, day 1, 8, every 21 days, 4
cycle | Recurrence | 11.4 | Positive | NR (6) |
| 2 | T3N0M0 | Cisplatin 75 mg/m2,
day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21 days,
3 cycle | No recurrence | >41.0 | Negative | R (81) |
| 3 | T4N2M0 | – | – | Lost in
follow-up | Negative | R (85) |
| 4 | T3N0M0 | Cisplatin 75 mg/m2,
day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21 days,
3 cycle | Recurrence | 4.8 | Positive | NR (41) |
| 6 | T2N0M0 | – | – | Lost in
follow-up | Negative | NR (32) |
| 9 | T3N0M0 | Carboplatin 300
mg/m2, day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21
days, 2 cycle | Recurrence | 2.4 | Positive | R (89) |
| 10 | T4N0M0 | Cisplatin 75 mg/m2,
day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21 days,
3 cycle | Recurrence | 8.6 | Positive | NR (32) |
| 11 | T3N2M0 | Cisplatin 75 mg/m2,
day 1+docetaxel 75 mg/m2, day 1 every 21 days, 3
cycle | No recurrence | >35.0 | Negative | R (92) |
| 12 | T3N1M0 | Cisplatin 75 mg/m2,
day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21 days,
1 cycle | Recurrence | 4.5 | Positive | NR (9) |
| 13 | T2N2M0 | Carboplatin 300
mg/m2, day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21
days, 3 cycle | No recurrence | >32.0 | Negative | R (86) |
| 14 | T2N0M0 | – | – | Lost in
follow-up | Negative | R (72) |
| 15 | T2N0M0 | Cisplatin 75 mg/m2,
day 1+gemcitabine 1,000 mg/m2, day 1, 8, every 21 days,
3 cycle | No recurrence | >28.0 | Negative | R (71) |
Altered tissue structure and
downregulated Ki-67 expression in tumors responsive to
cisplatin
Histologic changes after chemotherapy were analyzed
in xenograft tumors, and showed that the segmental tumor tissue in
xenograft tumors responsive to cisplatin chemotherapy was replaced
by scar tissue or necrosis, with the remaining cancer cells
generally characterized by foamy cytoplasm. Moreover, IHC staining
with anti-Ki-67 antibody showed that the proportion of Ki-67
stained tumor cells was dramatically decreased compared with
controls (39.3±27.5 vs. 84.3±7.3, P<0.01), indicating reduced
cell proliferation. In contrast, cancer cells in nonresponding
tumors had few histologic changes compared with controls and the
expression of Ki-67 was not significantly affected by cisplatin
chemotherapy (85.0±5.0 vs. 91.0±4.2) (Fig. 7).
Comparison of tumor responses in mice
with clinical outcome of patients
Given that none of the SCC patients had received
chemotherapy prior to surgery, individual comparison of clinical
and xenograft outcomes could not be performed. Nevertheless,
following surgical resection, the survival times of donor patients
were followed up for a minimum of 2 years (Table VII). Among the 12 patients, 9
received adjuvant platinum-based (cisplatin/carboplatin +
gemcitabine/docetaxel/vinorelbine) chemotherapy and 3 were lost to
follow-up. Xenografts from 4 of 5 patients who developed recurrence
during the 2-year follow-up were nonresponsive in mice. Four
patients who had survived at the 2-year follow-up were responsive
to chemotherapy in mice.
Discussion
In this study, a total of 37 fresh SCC tissues were
directly transplanted into athymic nude mice, and 18 PDTX models
were successfully established. However, our success rate (48.6%)
was lower than that reported previously (64%, 29/45) (6) This difference may be attributable to
sample size and the use of different mice strains, as this previous
study used more severely immunocompromised mice (NOD/SCID, lacking
functional T-, B-, and NK cells). However, studies with
neuroblastoma patient-derived xenografts have shown that lymphoid
cells and lymphatic vessels were present in PDTXs grown in athymic
nude mice but not in NOD/SCID mice. Therefore, the choice of mouse
strain dictated tumor microenvironmental components, which are
implicated as crucial mediators of tumor growth, metastasis, and
therapeutic responses and resistance (25).
The relationship between SCC engraftment in nude
mice and clinicopathological parameters of patients was analyzed
retrospectively, and we found that tumor volume and differentiation
were closely correlated with the take rate of PDTXs, which was
similar to that of a prior study in NSCLC (6) In addition, our results showed that the
expression of Ki67 in original SCC patient tumors was a predictive
factor for implanted tumor growth and the success of serial
passages in PDTX mice, as reported previously for prostate cancer
and hepatic metastasis uveal melanoma (26,27).
Moreover we observed that the average time for xenograft tumors to
reach a volume of 500 mm3 in the first generation was
longer than that of the other two passages (9.2, 6.8 and 6.3 weeks,
respectively), which suggested that passaging xenograft tumors were
more proliferative than first-generation tumors and that PDTXs
increased cancer cell proliferative activity, as reported
previously in human malignant pleural mesothelioma (28).
To determine the histological and immunophenotypic
features of the primary and xenograft tumors, 12 primary tumors and
their corresponding xenograft tumors were analyzed by H&E and
IHC. The majority of xenograft tumors maintained the histological
and key immunophenotypic features of the primary tumor, with the
exception of case 15, which showed strong expression of p63 and
cytokeratin5/6 by IHC in the primary tumor which decreased in
corresponding passaging xenograft tumors, possibly because of the
high heterogeneity of SCC. Additionally, variations in the
expression of Ki-67 were observed. In some primary tumors (cases 2,
9, 10, 11, 14 and 15) the average proportion of Ki-67 stained tumor
cells was 41.7±8.8%, but was significantly elevated in
first-generation xenograft tumors and maintained during serial
passaging (75.0±14.0, 74.2±15.6 and 83.3±7.5%, respectively).
Third-generation xenograft tumors had a further increased
expression of Ki-67 (83.3±7.5%), indicating a tendency to select
for more rapidly-growing cells during the engraftment process, as
reported previously in PDTX models for urothelial cancer (29). This result may partially explain why
the growth of xenograft tumors in mice was faster in serial
passages.
The global profiling of human lncRNA and mRNA
expression patterns in the SCC specimens and the corresponding
third-generation xenograft tumors were evaluated by microarray
analysis. The calculated correlation coefficients showed a high
concordance of gene expression between the third-generation
xenograft tumors and the corresponding primary tumors. However, all
6 third-generation xenograft tumors clustered together within the
primary tumors by unsupervised hierarchical clustering,
demonstrating that the xenograft tumors were more similar to each
other than to the primary tumors. These results indicated that,
despite similarities, there were also changes to a certain extent
in the RNA expression of xenograft tumors (including lncRNAs and
mRNAs) compared with primary tumors. Because the stroma in the
xenograft tumor was largely replaced by murine stroma, as expected,
GO and pathway analysis showed that decreased mRNAs in third
xenograft tumors might be mainly involved in immune-mediated
processes, consistent with previous reports (30,31). In
addition, increased expression of mRNAs mainly involved in the
regulation of cell cycle and metabolic processes in the
third-generation xenograft tumors was consistent with the study of
Wang et al (32). Their
results showed that genomic alterations in cell cycle pathways were
the most frequently found changes in PDTXs of NSCLC. In a similar
study with proteomic characterization of head and neck cancer
(33), proteins associated with
proliferative signaling appeared to be preferentially selected
during the process of creating PDTXs. Taken together, changes in
cell proliferation and immune-mediated processes may represent an
important reason for the faster growth of xenograft tumors in
serial passages. Moreover, we found that lncRNA expression was
altered in xenograft tumors. It seems likely that some of the
separation seen in the lncRNA clustering analysis reflects
variations in tumor stroma. Furthermore, the functions of many of
the differentially expressed lncRNAs observed in our study have not
yet been established, and it is possible that they do not
significantly impact carcinoma phenotype. However, whether they
influence cell proliferation and immune-mediated processes needs to
be explored. Similarly, the mRNA profile in lung cancer xenograft
tumors has previously been shown to differ from that of original
tumors (34).
Given the alterations in cell cycle and metabolic
processes of xenograft tumors, and the faster growth of xenograft
tumors in serial passages, proliferation and DNA synthesis in
xenografts may be faster than that of primary tumors.
Platinum-based chemotherapy is the first-line treatment for SCC.
Platinum-based drugs bind to nuclear DNA, forming a variety of
structural adducts and triggering cellular apoptosis (35,36). We
therefore hypothesize that changes in cell proliferation may affect
the response to platinum-based chemotherapy. To test this
hypothesis, we investigated the responses of 12 established
third-generation xenograft tumors to single cisplatin treatments,
and found that 60% of the tumors were responsive and that
chemotherapy-induced histologic changes in third-generation
xenograft tumors were consistent with changes documented previously
in clinical studies in NSCLC specimens following preoperative
chemotherapy (37). Secondly, the
International Adjuvant Lung Cancer Trial (IALT) demonstrated that
cell cycle regulators (p16INK4A, cyclin D1, cyclin D3, cyclin E)
and Ki-67 did not predict the benefit of adjuvant cisplatin-based
chemotherapy in NSCLC patients, except for p27Kip1 (38). However, in our study, when compared
with primary tumors, the mRNA expression of p27Kip1 was not changed
in the 6 third-generation xenograft tumors. In addition, we found
that the response rates to cisplatin-based chemotherapy in
xenograft tumors related to the expression status of p53, which was
positive in 4 of 5 non-responding tumors and negative in 6 of 7
responding tumors. Similarly, p53 was assessed using IHC in a large
subset of NSCLC patients (n=769) from the IALT trial who had
received cisplatin-based chemotherapy. The results suggested that
p53 was predictive of a cisplatin-based therapeutic benefit in
patients with SCC but not ADC (39).
As none of the SCC donors in our study had received chemotherapy
prior to surgery, individual comparison of clinical and xenograft
outcomes could not be performed. However, the differential
responses of the 12 observed xenograft tumors to the individual
cisplatin drug treatments would strongly justify an individual
prediction of responsiveness. Moreover, we compared tumor responses
in xenograft tumors with clinical outcomes of patients who received
platinum-based chemotherapy after surgery, and the results
suggested that four patients had survived to the 2-year follow-up
and that the corresponding third-generation xenograft tumors were
responsive to cisplatin chemotherapy. Xenograft tumors from four of
five patients who developed recurrence were nonresponsive in mice.
Nevertheless, in view of the small number of SCC patients included,
further studies are required to verify this observation.
In conclusion, we evaluated the characteristics of
SCC xenograft tumors in this study, and our findings suggest that
these tumors closely resembled the original tumor and largely
retained key immunophenotypic features. Moreover, we showed that
LSCC xenografting altered tumor cell proliferation but maintained
the responsiveness to platinum-based chemotherapy. In expression
profiling of human lncRNAs and mRNAs, however, xenografts clustered
separately from the original tumors. This result suggests that
lncRNA and mRNA expression may be altered in xenografts, a
possibility which warrants further investigation.
Acknowledgments
Not applicable.
Funding
This study was supported by Province science and
technology in the Anhui offends pass item (no. 1604a0802072, 2016),
Natural Science Foundation of Anhui province, China (no.
1708085QH220, 2017), National Natural Science Foundation of China
(no. 81172172, 2011).
Availability of data and materials
The datasets generated or analyzed during the
current study are available from the corresponding author upon
written request.
Authors' contributions
BW designed the study and participated in writing
the manuscript and approved the final version of the manuscript to
be published. JZ, YY, QW, ML, HZ and MX performed the laboratory
work. DL and PL contributed to the acquisition, analysis and
interpretation of data for the study, and participated in writing
the manuscript. JZ, YY and QW participated in designing the
statistical analysis and preparing the manuscript. ML, HZ and MX
participated in designing the study and provided clinical data. All
authors contributed to the manuscript and approved the final
version of it.
Ethics approval and consent to
participate
Tumor samples and clinical records were obtained
from patients with their written informed consent and the study was
approved by Anhui Provincial Hospital Ethical Committee. Consent
was obtained to publish from the participant to report individual
patient data. All animal experiments were approved by Anhui
Provincial Hospital Ethical Committee and were carried out in
accordance with Guidelines for the Care and Use of Laboratory
Animals.
Consent for publication
Consent for publication was obtained through the
Institutional Review Board approved protocol with an institutional
consent form.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pikor LA, Ramnarine VR, Lam S and Lam WL:
Genetic alterations defining NSCLC subtypes and their therapeutic
implications. Lung Cancer. 82:179–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kerbel RS: Human tumor xenografts as
predictive preclinical models for anticancer drug activity in
humans: Better than commonly perceived-but they can be improved.
Cancer Biol Ther. 2 4 Suppl 1:S134–S139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
John T, Kohler D, Pintilie M, Yanagawa N,
Pham NA, Li M, Panchal D, Hui F, Meng F, Shepherd FA and Tsao MS:
The ability to form primary tumor xenografts is predictive of
increased risk of disease recurrence in early-stage non-small cell
lung cancer. Clin Cancer Res. 17:134–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin D, Wyatt AW, Xue H, Wang Y, Dong X,
Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, et al: High fidelity
patient-derived xenografts for accelerating prostate cancer
discovery and drug development. Cancer Res. 74:1272–1283. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johnson JI, Decker S, Zaharevitz D,
Rubinstein LV, Venditti JM, Schepartz S, Kalyandrug S, Christian M,
Arbuck S, Hollingshead M and Sausville EA: Relationships between
drug activity in NCI preclinical in vitro and in vivo models and
early clinical trials. Br J Cancer. 84:1424–1431. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garber K: From human to mouse and back:
‘tumorgraft’ models surge in popularity. J Natl Cancer Inst.
101:6–8. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong X, Guan J, English JC, Flint J, Yee
J, Evans K, Murray N, Macaulay C, Ng RT, Gout PW, et al:
Patient-derived first generation xenografts of non-small cell lung
cancers: promising tools for predicting drug responses for
personalized chemotherapy. Clin Cancer Res. 16:1442–1451. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perez-Soler R, Kemp B, Wu QP, Mao L, Gomez
J, Zeleniuch-Jacquotte A, Yee H, Lee JS, Jagirdar J and Ling YH:
Response and determinants of sensitivity to paclitaxel in human
non-small cell lung cancer tumors heterotransplanted in nude mice.
Clin Cancer Res. 6:4932–4938. 2000.PubMed/NCBI
|
|
12
|
Hao C, Wang L, Peng S, Cao M, Li H, Hu J,
Huang X, Liu W, Zhang H, Wu S, et al: Gene mutations in primary
tumors and corresponding patient-derived xenografts derived from
non-small cell lung cancer. Cancer Lett. 357:179–185. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guttman M, Russell P, Ingolia NT, Weissman
JS and Lander ES: Ribosome profiling provides evidence that large
noncoding RNAs do not encode proteins. Cell. 154:240–251. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Bio. 14:699–712. 2013. View Article : Google Scholar
|
|
16
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun M, Nie FQ, Wang YF, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan JH, Liu XN, Wang TT, Pan W, Tao QF,
Zhou WP, Wang F and Sun SH: The MBNL3 splicing factor promotes
hepatocellular carcinoma by increasing PXN expression through the
alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 19:820–832.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Y, Lyu H, Liu H, Shi X, Song Y and Liu
B: Downregulation of the long noncoding RNA GAS5-AS1 contributes to
tumor metastasis in non-small cell lung cancer. Sci Rep.
6:310932016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao S, Lin Z, Li C, Wang Y, Yang L, Zou B,
Chen J, Li J, Song Z and Liu G: TFPI2AS1, a novel lncRNA that
inhibits cell proliferation and migration in lung cancer. Cell
Cycle. 16:2249–2258. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Julien S, Merino-Trigo A, Lacroix L,
Pocard M, Goéré D, Mariani P, Landron S, Bigot L, Nemati F,
Dartigues P, et al: Characterization of a large panel of
patient-derived tumor xenografts representing the clinical
heterogeneity of human colorectal cancer. Clin Cancer Res.
18:5314–5328. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Braekeveldt N, Wigerup C, Tadeo I, Beckman
S, Sandén C, Jönsson J, Erjefält JS, Berbegall AP, Börjesson A,
Backman T, et al: Neuroblastoma patient-derived orthotopic
xenografts reflect the microenvironmental hallmarks of aggressive
patient tumours. Cancer Lett. 375:384–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lawrence MG, Pook DW, Wang H, Porter LH,
Frydenberg M, Kourambas J, Appu S, Poole C, Beardsley EK, Ryan A,
et al: Establishment of primary patient-derived xenografts of
palliative TURP specimens to study castrate-resistant prostate
cancer. Prostate. 75:1475–1483. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kageyama K, Ohara M, Saito K, Ozaki S,
Terai M, Mastrangelo MJ, Fortina P, Aplin AE and Sato T:
Establishment of an orthotopic patient-derived xenograft mouse
model using uveal melanoma hepatic metastasis. J Transl Med.
15:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu LC, Allo G, John T, Li M, Tagawa T,
Opitz I, Anraku M, Yun Z, Pintilie M, Pitcher B, et al:
Patient-derived xenograft establishment from human malignant
pleural mesothelioma. Clin Cancer Res. 23:1060–1067. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abe T, Tada M, Shinohara N, Okada F, Itoh
T, Hamada J, Harabayashi T, Chen Q, Moriuchi T and Nonomura K:
Establishment and characterization of human urothelial cancer
xenografts in severe combined immunodeficient mice. Int J Urol.
13:47–57. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fichtner I, Rolff J, Soong R, Hoffmann J,
Hammer S, Sommer A, Becker M and Merk J: Establishment of
patient-derived non-small cell lung cancer xenografts as models for
the identification of predictive biomarkers. Clin Cancer Res.
14:6456–6468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reyal F, Guyader C, Decraene C, Lucchesi
C, Auger N, Assayag F, De Plater L, Gentien D, Poupon MF, Cottu P,
et al: Molecular profiling of patient-derived breast cancer
xenografts. Breast Cancer Res. 14:R112012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang D, Pham NA, Tong JF, Sakashita S,
Allo G, Kim L, Yanagawa N, Raghavan V, Wei Y, To C, et al:
Molecular heterogeneity of non-small cell lung carcinoma
patient-derived xenografts closely reflect their primary tumors.
Int J Cancer. 140:662–673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li H, Wheeler S, Park Y, Ju Z, Thomas SM,
Fichera M, Egloff AM, Lui VW, Duvvuri U, Bauman JE, et al:
Proteomic characterization of head and neck cancer patient-derived
xenografts. Mol Cancer Res. 14:278–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bogner PN, Patnaik SK, Pitoniak R,
Kannisto E, Repasky E, Hylander B, Yendamuri S and Ramnath N: Lung
cancer xenografting alters microRNA profile but not
immunophenotype. Biochem Biophys Res Commun. 386:305–310. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Crino L, Weder W, van Meerbeeck J and
Felip E: ESMO Guidelines Working Group: Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO clinical
practice guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21 Suppl 5:v103–v115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Azzoli CG, Temin S, Aliff T, Baker S Jr,
Brahmer J, Johnson DH, Laskin JL, Masters G, Milton D, Nordquist L,
et al: 2011 focused update of 2009 American society of clinical
oncology clinical practice guideline update on chemotherapy for
stage IV non-small-cell lung cancer. J Clin Oncol. 29:3825–3831.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Junker K: Therapy-induced tumor regression
and regression grading in lung cancer. Pathologe. 35:574–577.
2014.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Filipits M, Pirker R, Dunant A, Lantuejoul
S, Schmid K, Huynh A, Haddad V, André F, Stahel R, Pignon JP, et
al: Cell cycle regulators and outcome of adjuvant cisplatin-based
chemotherapy in completely resected non-small-cell lung cancer: The
international adjuvant lung cancer trial biologic program. J Clin
Oncol. 25:2735–2740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pierceall WE, Olaussen KA, Rousseau V,
Brambilla E, Sprott KM, Andre F, Pignon JP, Le Chevalier T, Pirker
R, Jiang C, et al: Cisplatin benefit is predicted by
immunohistochemical analysis of DNA repair proteins in squamous
cell carcinoma but not adenocarcinoma: Theranostic modeling by
NSCLC constituent histological subclasses. Ann Oncol. 23:2245–2252.
2012. View Article : Google Scholar : PubMed/NCBI
|