Introduction
Breast cancer remains one of the most prevalent
types of cancer among women. It is estimated that there will be
255,180 new breast cancer cases and 41,070 deaths in the U.S. in
2017 (1). Most of the mortality is
caused by cancer invasion and metastasis. However, there are still
limited effective therapies for metastatic breast cancer patients.
In our previous study, we investigated the differentially expressed
proteins between parental MDA-MB-231 and highly metastatic
MDA-MB-231 (MDA-MB-231HM and MDA-MB-231BO) breast cancer cell lines
(2–4).
We found that forkhead box P2 (FOXP2), a transcription factor, had
a significantly reduced expression level in highly metastatic cell
lines. FOXP2 is a member of FOXP gene subfamily (FOXP1, FOXP2,
FOXP3, FOXP4) which all have a C-terminal winged helix forkhead DNA
binding domain. The structure is conserved in many species. It was
previously proved that the gene FOXP2, locating at 7q31, regulates
many neurogenesis signaling pathways critical in embryonic
development and cell cycle such as Hedgehog, WNT and Notch pathways
(5,6).
It is also involved in lung, heart and central nervous system (CNS)
development. Especially, it plays pivotal roles in human language
development. Depletion and mutation in FOXP2 can lead to speech and
linguistic impairment, aging and cancer (7,8).
Several researches have also reported the roles of
FOXP2 as a tumor suppressor in gastric cancer (9), osteosarcoma (10) and hepatocellular carcinoma (11) previously. It was revealed that the
expression of FOXP2 was usually regulated by a series of microRNAs
in cancer cells. However, there were not many studies investigating
the mechanisms of FOXP2 in breast cancer. In our current study, we
explored the effects and mechanisms of FOXP2 on breast cancer
metastasis and prognosis. We hope our study could provide deeper
insights into the diagnosis and treatment of metastatic breast
cancer.
Materials and methods
Tumor samples
Our study has already been approved by the Ethical
Committee and Institutional Review Board of Fudan University
Shanghai Cancer Centre (FDUSCC). The methods were performed in
accordance with the approved guidelines. All the participants
signed written informed consent forms. The tumour samples were
collected between May 2014 and May 2015. The breast cancer samples
and matched para-carcinoma samples were collected from 39 patients
pathologically diagnosed with breast cancer who undergone surgery.
The samples were frozen immediately in liquid nitrogen after
surgery.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
reverse transcribed with PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China). Subsequently,
RT-qPCR was performed with SYBR Premix Ex Taq (Takara
Biotechnology Co., Ltd.) using ABI Prism 7900 instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacture's protocol. The thermocycling condition was
pre-denaturation at 95°C for 30 sec, denaturation at 95°C for 5
sec, annealing at 60°C for 30 sec, with 40 cycles. The relative
gene expression was calculated using the 2−ΔΔCq method
(12). The primer sequences used in
this study are as follows: FOXP2: forward
5′-AACAACAGCAGGCTCTCCAG-3′, and reverse 5′-GGCACCTGCAGTGGTCTC-3′;
GAPDH: forward 5′-GGTGGTCTCCTCTGACTTCAACA-3′, and reverse
5′-GTTGCTGTAGCCAAATTCGTTGT-3′.
Cell culture
The cells in this study were obtained from the
Shanghai Cell Bank, Type Culture Collection Committee of Chinese
Academy of Science (Shanghai, China). The cells for experiments
were passaged for less than 6 months. MDA-MB-231, MDA-MB-231BO,
MDA-MB-231HM and MDA-MB-468 (all triple negative breast cancer)
cells were grown in Leibovitz L-15 medium (BasalMedia, Shanghai,
China). 293T, MCF7 (luminal positive breast cancer) and BT549
(triple-negative breast cancer) cells were cultured in high-glucose
Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare
Life Sciences, Logan, UT, USA). SKBR3 (HER2 positive breast
cancer), T47D, ZR7530 and ZR75-1 (all luminal positive breast
cancer) were maintained in RMPI-1640 medium (HyClone; GE Healthcare
Life Sciences). All the medium was supplemented with 10% FBS, 100
IU/ml penicillin, and 100 mg/ml streptomycin. All the cells were
maintained with 5% CO2 at 37°C. The MDA-MB-231HM was
developed from the parental MDA-MB-231 cell line via the tail vein
in mice for four cycles. We have patent application for the cell
line (patent no. 200910174455.4) which exhibited increased lung
metastasis compared to parental cells. The MDA-MB-231BO cells,
which have highly metastatic potential to bone, was obtained from
Dr Toshiyuki Yoneda (University of Texas, Houston, TX, USA). We
have done several studies with these cell lines (4,13).
Plasmids and lentivirus packaging
Human FOXP2 cDNA was purchased from fulenGen and
then subcloned into the pCDH-CMV-MCS-EF1-Puro lentiviral vector.
The cloned primer sequence is as follows: FOXP2: forward
5′-CCGGAATTCGCCACCATGATGCAGGAATCTGCGAC-3′, and reverse
5′-CGCGGATCCTCATTCCAGATCTTCAGATA-3′.
FOXP2 shRNAs and the negative control were purchased
from GeneChem and expressed in the GV248 backbone. The target
sequences are as follows: shRNA NC 5′-TTCTCCGAACGTGTCACGT-3′;
shRNA1: 5′-TTAACAATGAACACGCATT-3′; shRNA2:
5′-AGCAAACAAGTGGATTGAA-3′.
293T cells were cotransfected with lentiviral
vectors and the packaging vectors pCDH (GV248), psPAX2 and pMD2G.
Virus-containing medium was collected 48 h after the transfection
and added to the cancer cells.
Kinetic wound-healing assay
MDA-MB-231 and MDA-MB-231BO cells
(3.6×104) were plated on 96-well plates (Essen
ImageLock; Essen Biosciences, Ann Arbor, MI, USA), and a wound was
scratched with wound scratcher (Essen Instruments). Wound
confluence was monitored with Live-Cell Imaging System and software
(Essen Biosciences). The wound closure was observed after 28 h by
comparing the mean relative wound density of at least three
biological replicates in each experiment.
Transwell assay
Cell migration and invasion was examined with
Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA). The
cells were seeded in the upper chamber in serum-free medium. The
cell density was 5×105 for migration assay and
1×106 for Matrigel-coated invasion assay. Medium
supplemented with 20% serum was added in the lower chamber. The
cells were incubated for 15 to 20 h. After that, the cells remained
on the upper chamber were removed with cotton swabs. The cells on
the lower surface of the membrane were stained with 0.4% methanol
and 0.25% crystal violet.
Western blot analysis
Whole cell extracts were isolated with Pierce T-PER
(Tissue Protein Extraction Reagent; Thermo Fisher Scientific, Inc.)
containing protease inhibitor cocktail tablets (Roche Diagnostics,
Indianapolis, IN, USA) and phosphatase inhibitors (Roche
Diagnostics). Proteins (30 µg) were resolved by sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene fluoride membrane (Pall
Corporation, Port Washington, NY, USA). The membranes were blocked
in 5% BSA and incubated in various primary antibodies followed by
the corresponding horseradish peroxidase-conjugated secondary
antibodies (Proteintech Group, Inc., Chicago, IL, USA). The primary
antibody against FOXP2 (21608) and vimentin (40477) was obtained
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), fibronectin
(66042-1-Ig) and GAPDH (60004-1-Ig) from Proteintech Group, Inc.,
N-cadherin (610920) from BD Biosciences, p-SMAD3 (ab52903) from
Abcam (Cambridge, UK), SMAD3 (1735-s) from Epitomics (Burlingame,
CA, USA), SMAD4 (9515p) from Cell Signaling Technology, Inc.
(Danvers, MA, USA) and TGFβR1 (sc-398) from Santa Cruz
Biotechnology, Inc. Antibody binding was detected using
chemiluminescence (Amersham Imager 600; GE Healthcare Life
Sciences), according to the manufacturer's instructions.
Statistical analysis
SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was
used to conduct the statistical analysis. Data were expressed as
means ± standard deviation. The student's t-test was used to
evaluate the differences between two groups and one-way analysis of
variance (ANOVA) was used among multiple groups. LSD test was used
after ANOVA to compare the differences between two groups.
P<0.05 was considered to indicate a statistically significant
difference. Graphs were created with GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA).
Results
Low expression of FOXP2 indicated
poorer prognosis in breast cancer
The information of FOXP2 expression and patients'
survival was acquired from an online Kaplan-Meier plotter
(http://kmplot.com/analysis/). This
online tool was based on data of 1,809 patients downloaded from GEO
(Affymetrix HGU133A and HGU133+2 microarrays) (14). The low and high expression was divided
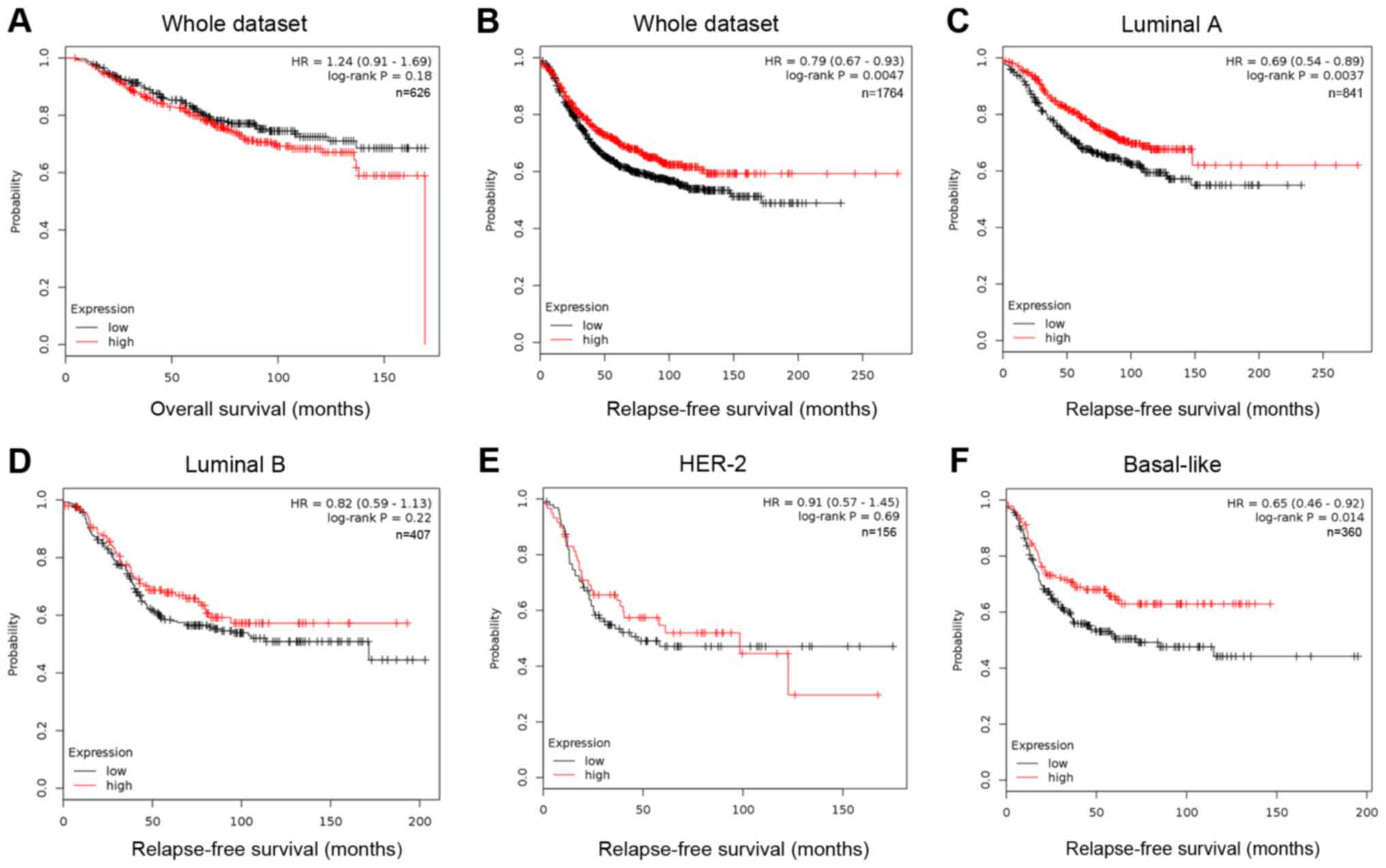
by median expression. In Kaplan Meier analysis (Fig. 1), the expression level of FOXP2 was
significantly correlated to the patients' relapse-free survival
(RFS, P=0.0047, HR=0.79; Fig. 1B),
but not to overall survival (OS, P=0.18, Fig. 1A). Among patients with different
molecular subtypes, low FOXP2 expression only predicted higher risk
of relapse among luminal A (P=0.0037) and basal-like breast cancer
patients (P=0.014). The results suggested that low expression of
FOXP2 may be associated with recurrence and metastasis in some
breast cancer patients.
FOXP2 expression was downregulated in
breast cancer compared to normal breast tissue
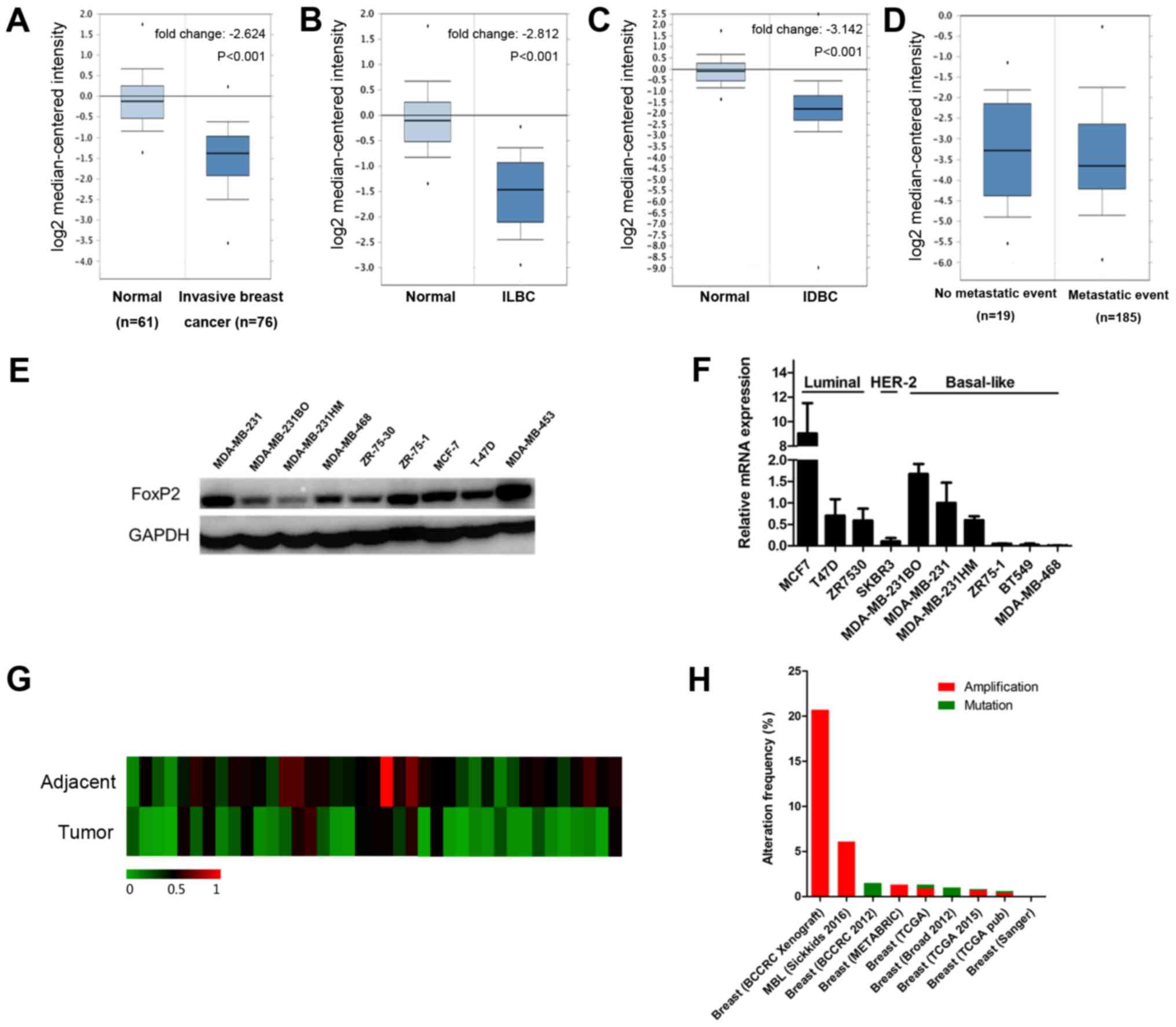
According to the data from oncomine (www.oncomine.org), the expression of FOXP2 was
significantly lower in breast cancer than normal breast tissue
(P<0.001; Fig. 2A). The results
remained same both in invasive ductal carcinoma and invasive
lobular carcinoma (Fig. 2B and C).
However, there was no significant difference among patients with
and without metastasis (Fig. 2D). We
also evaluate the expression level of FOXP2 in different breast
cancer cell lines (Fig. 2E and F) as
well as 39 patients' samples (Fig.
2G). Interestingly, FOXP2 was significantly less expressed in
breast cancer tissue compared to adjacent control tissue
(P=0.0005). The results were consistent with the data from the
public database. Table I summarized
the clinicopathological characteristics of the patients. In this
cohort, the FOXP2 expression was not significantly associated with
these clinical features except age.
 | Table I.Correlation between FOXP2 expression
in patients with breast cancer and their clinicopathologic
characteristics. |
Table I.
Correlation between FOXP2 expression
in patients with breast cancer and their clinicopathologic
characteristics.
| Characteristic | Group | Low expression | High expression | Total | P-value |
|---|
| Age | ≤50 | 12 | 4 | 16 | 0.006a |
|
| >50 | 7 | 16 | 23 |
|
| Tumor size (cm) | ≤2 | 5 | 11 | 16 | 0.097 |
|
| 2<n≤5 | 12 | 9 | 21 |
|
|
| >5 | 2 | 0 | 2 |
|
| Lymph nodes | Negative | 2 | 1 | 3 | 0.347 |
|
| Positive | 14 | 22 | 36 |
|
| Menopausal
status | Premenopausal | 10 | 8 | 18 | 0.497 |
|
| Postmenopausal | 9 | 11 | 20 |
|
|
| NA | 0 | 1 | 1 |
|
| Histological
grade | 1 | 1 | 0 | 1 | 0.260 |
|
| 2 | 10 | 7 | 17 |
|
|
| 3 | 8 | 13 | 21 |
|
| ER | Negative | 4 | 6 | 10 | 0.522 |
|
| Positive | 15 | 14 | 29 |
|
| PR | Negative | 6 | 11 | 17 | 0.140 |
|
| Positive | 13 | 9 | 22 |
|
| HER-2 | Negative | 8 | 9 | 17 | 0.855 |
|
| Positive | 11 | 11 | 22 |
|
In addition, we also investigated the frequency of
FOXP2 alterations, including amplification and mutations, in breast
cancer (Fig. 2H) from cbioprotal
(www.cbioportal.org) (15). We found that among 4,786 invasive
breast cancer cases from 9 studies, 144 patients had amplification
(3%) and 17 (0.4%) had mutation of this gene.
FOXP2 inhibited breast cancer cells
migration and invasion
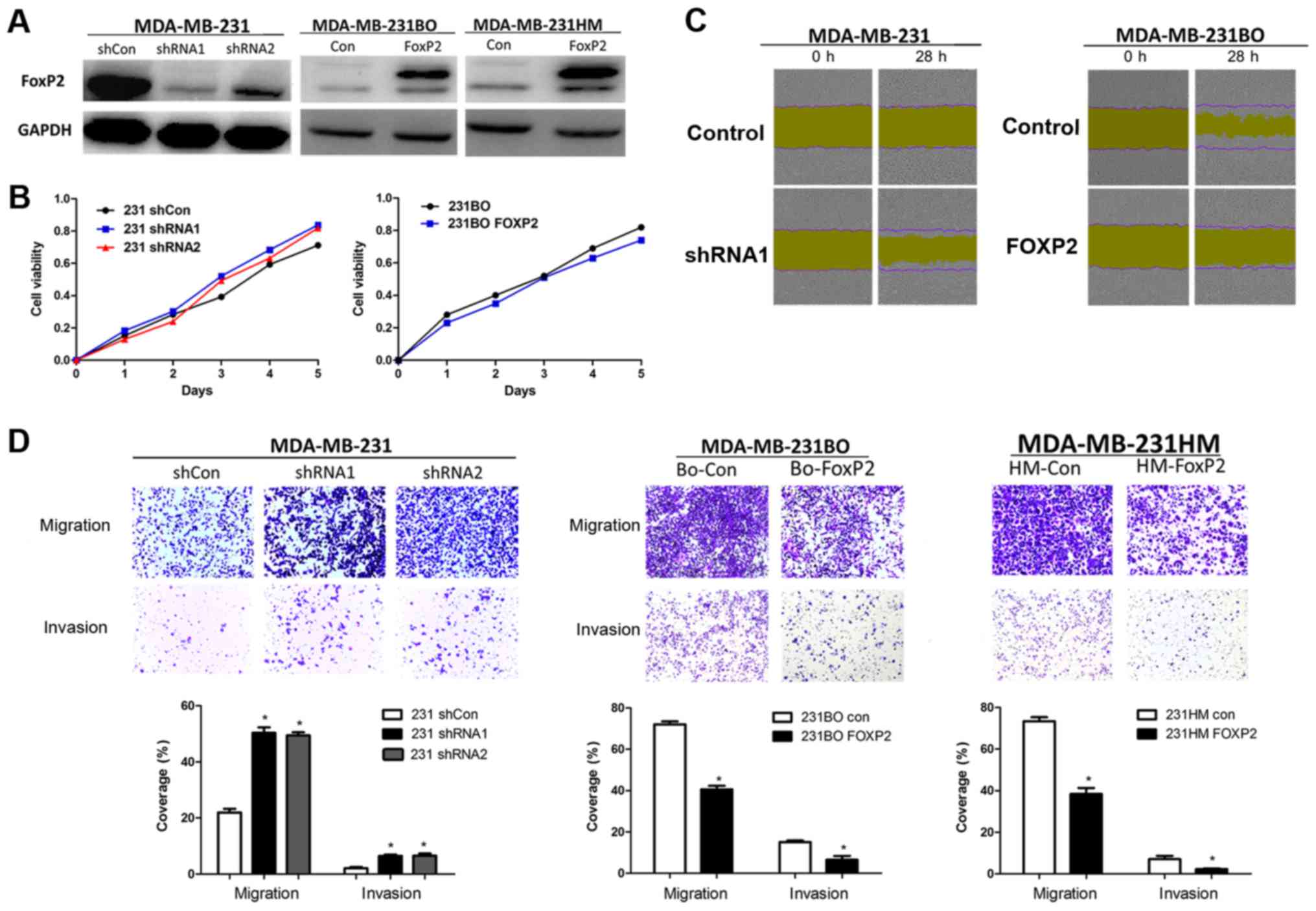
To validate the function of FOXP2 in breast cancer
metastasis and proliferation, we knocked down FOXP2 in MDA-MB-231
and overexpressed it in MDA-MB-231BO and MDA-MB-231HM breast cancer
cells via lentivirus infection. The efficiency of infection was
confirmed by western blot analysis (Fig.
3A). We then found that the depletion of FOXP2 did not have
significant effects on cell proliferation (Fig. 3B). But the wound-healing assay and
Transwell assay showed that the knockdown of FOXP2 could promote
cell migration and invasion in vitro (P=0.001; Fig. 3C and D). Meanwhile, the cell migration
and invasion was inhibited after the overexpression of FOXP2
(P<0.001). Taken together, the results suggested that FOXP2 may
play a certain role in breast cancer as a tumor suppressor
gene.
Suppression of FOXP2 induced EMT and
activated TGFβ/SMAD signaling pathway
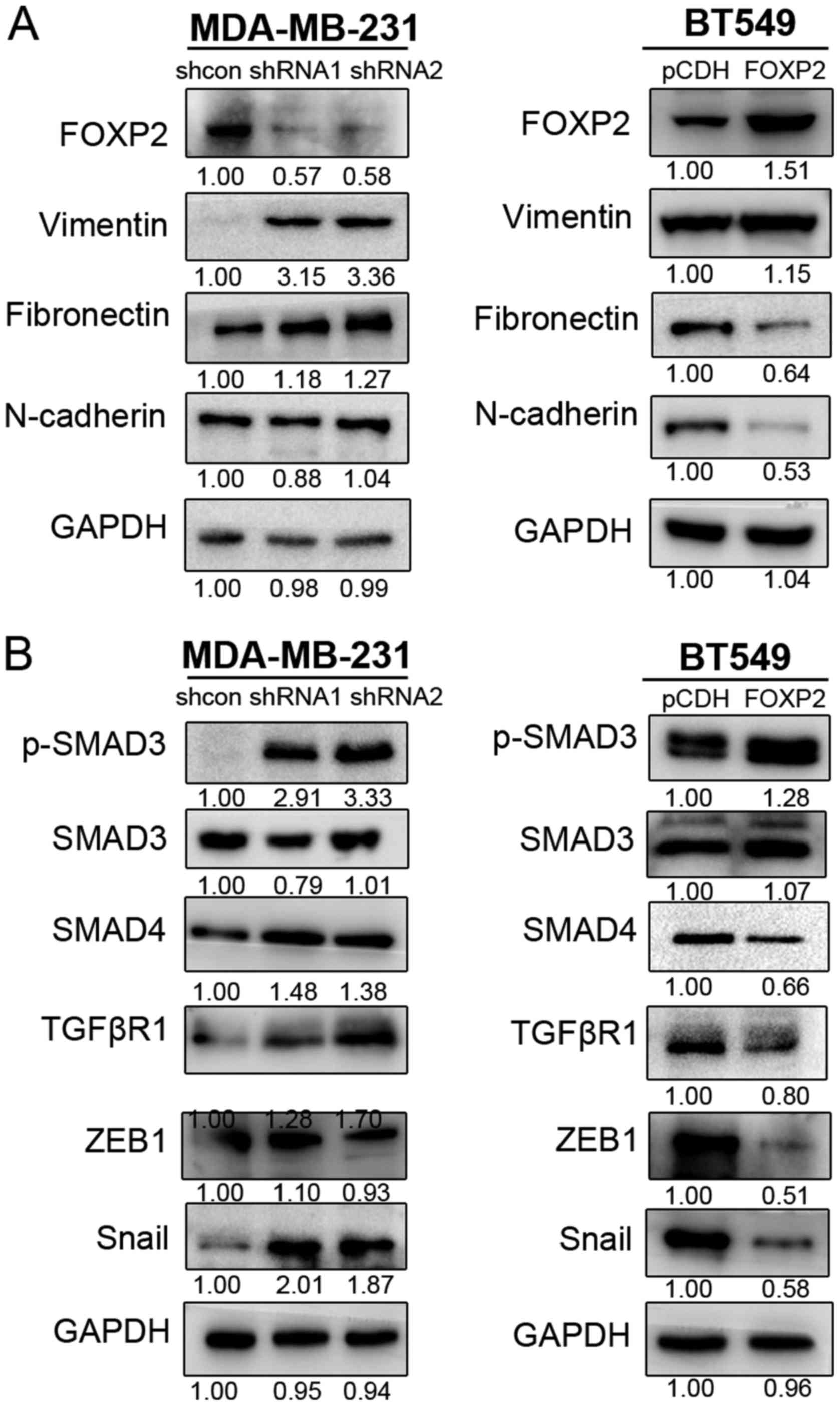
Epithelial-mesenchymal transition (EMT) was known as
one of crucial mechanisms in cancer progression and metastasis
(16). In order to further
investigate the mechanisms of FOXP2 inhibiting breast cancer
metastasis, we detected the expression level of several
EMT-associated biomarkers in MDA-MB-231 and BT549 (Fig. 4A). The results revealed that
suppression of FOXP2 promoted the expression of vimentin and
fibronectin in MDA-MB-231. In BT549 cell line, fibronectin and
N-cadherin was inhibited after overexpression of FOXP2. The results
suggested that FOXP2 could inhibit EMT in breast cancer in
vitro.
The activation of TGFβ/SMAD signaling was known as
one of the critical pathways in EMT process and cancer metastasis
(17,18). Therefore, we also detected the
expression of some important proteins in TGFβ signaling (Fig. 4B). We found that upregulation of FOXP2
in BT549 could markedly decrease the phosphorylation of SMAD3,
enhanced the expression of SMAD4, TGFβR1, ZEB1 and Snail.
Meanwhile, suppression of the gene in MDA-MB-231 could cause the
reverse effects except ZEB1. The expression of Twist and p-SMAD2
did not seem to show significant differences in the cell lines in
this research. The phenomenon indicated that inhibition of FOXP2
induce EMT process in breast cancer probably via TGFβ/SMAD
signaling pathway.
Discussion
The mechanisms of breast cancer invasion and
metastasis are still not fully understood. In our study, we
demonstrated FOXP2 might be a potential target for the prevention
and treatment of breast cancer metastasis. The function of FOXP2
was initially identified in neurodevelopmental disorders,
especially inherited speech-and-language disorders (19,20). Here,
we first reported the role of FOXP2 in inhibiting EMT in breast
cancer via TGFβ/SMAD signaling pathway. Upon FOXP2 depletion, the
cell mesenchymal biomarkers decreased which indicated higher
potential of cell motility and migration. Interestingly, we found
that the TGFβ pathway related proteins (TGFβR1, p-SMAD3, SMAD4,
Snail) were obviously elevated after downregulation of FOXP2.
However, the detailed mechanisms of FOXP2 regulating TGFβ pathway
are still unclear. TGFβ/SMAD signaling is a well-known pathway
activated during cancer metastasis and transmits signals from cell
surface receptors to nuclear transcription (18). Tumor cells usually produce abundant
amount of TGFβ which activates downstream SMAD2/3/4. Then the
activated SMADs translocate to nucleus and activated EMT-related
transcription factors including Snail, ZEB1/2, Twist, (21). But effective drugs targeting this
pathway are still limited clinically, partially because of the side
effects (22).
There have been only a few studies investigating the
role of FOXP2 in cancer so far. Some studies suggested FOXP2
mediated the crosstalk between breast cancer cells and
tumor-associated mesenchymal stem cells (MSCs). For example, Cuiffo
et al (23,24) reported that the expression of FOXP2
was regulated by a series of miRNA including miR-199a. They also
found that FOXP2 could inhibited breast cancer initiation and
colonization through inhibiting a series of cancer stem cell
associated factors including c-Myc, Oct-4, CD44, etc. FOXP2 could
also be regulated by miR-190 in gastric cancer (9). Upregulation of miR-190 can cause the
downregulation of FOXP2 protein expression and then lead to gastric
cancer cells migration and invasion. Another study (10) observed transient overexpression of
FOXP2 in pre-osteoblast mesenchymal cells influenced a
p21-dependent growth arrest checkpoint, which played important
roles in the growth of osteosarcoma. These results all confirmed
the function of FOXP2 in tumor suppression.
There were also several limitations in our study.
For example, the targeted genes regulated by FOXP2 in breast cancer
was still unclear. As a transcription factor, FOXP2 need to combine
the promoters of the targeted genes to regulate the downstream
pathway. But we failed to find the promoters directly targeted by
FOXP2. Moreover, we only involved a limited number of patients,
most of which had lymph nodes invasion. We did not establish ideal
tumour metastatic models in this study. Therefore, the results
still need further validation in larger number of cohorts and in
vivo.
In conclusion, we identified FOXP2 as a novel tumor
suppressor gene in human breast cancer using clinical correlation
analysis and in vitro functional metastasis assays. Drugs
might be developed to mimic the functions of FOXP2 as a tumor
suppressor to prevent or treat breast cancer metastasis in the
future. We hope our findings could contribute to the diagnosis and
treatment of breast cancer to some extent. Further studies still
need to be done in order to investigate the underlying mechanisms
of the functions of FOXP2 in the future.
Acknowledgements
Not applicable.
Funding
This study was supported by the grant from National
Natural Science Foundation of China (no. 81472669) and the Hundred
Talents Program of the Health System in Shanghai (no.
2017BR028).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WJ, HFS and MTC conceived and designed the study.
MTC, LDL and SPG conducted the in vitro experiments. LDL and
YZ constructed the plasmids. LPY helped with the tissue preparation
and statistical analysis. MTC analyzed the results and wrote the
main manuscript. All of the authors reviewed the manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethical Committee and
Institutional Review Board of Fudan University Shanghai Cancer
Centre and all participants provided written informed consent.
Consent for publication
Written informed consent was obtained from all
participants for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gao SP, Sun HF, Jiang HL, Li LD, Hu X, Xu
XE and Jin W: Loss of COX5B inhibits proliferation and promotes
senescence via mitochondrial dysfunction in breast cancer.
Oncotarget. 6:43363–43374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li LD, Sun HF, Liu XX, Gao SP, Jiang HL,
Hu X and Jin W: Down-regulation of NDUFB9 promotes breast cancer
cell proliferation, metastasis by mediating mitochondrial
metabolism. PLoS One. 10:e01444412015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang HL, Sun HF, Gao SP, Li LD, Huang S,
Hu X, Liu S, Wu J, Shao ZM and Jin W: SSBP1 suppresses TGFβ-driven
epithelial-to-mesenchymal transition and metastasis in
triple-negative breast cancer by regulating mitochondrial
retrograde signaling. Cancer Res. 76:952–964. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chiu YC, Li MY, Liu YH, Ding JY, Yu JY and
Wang TW: Foxp2 regulates neuronal differentiation and neuronal
subtype specification. Dev Neurobiol. 74:723–738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsui D, Vessey JP, Tomita H, Kaplan DR and
Miller FD: FoxP2 regulates neurogenesis during embryonic cortical
development. J Neurosci. 33:244–258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fisher SE and Scharff C: FOXP2 as a
molecular window into speech and language. Trends Genet.
25:166–177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ostrow AZ, Kalhor R, Gan Y, Villwock SK,
Linke C, Barberis M, Chen L and Aparicio OM: Conserved forkhead
dimerization motif controls DNA replication timing and spatial
organization of chromosomes in S. cerevisiae. Proc Natl Acad Sci
USA. 114:E2411–E2419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X
and Xiao G: MicroRNA-190 regulates FOXP2 genes in human gastric
cancer. Onco Targets Ther. 9:3643–3651. 2016.PubMed/NCBI
|
|
10
|
Gascoyne DM, Spearman H, Lyne L, Puliyadi
R, Perez-Alcantara M, Coulton L, Fisher SE, Croucher PI and Banham
AH: The forkhead transcription factor FOXP2 is required for
regulation of p21WAF1/CIP1 in 143B osteosarcoma cell growth arrest.
PLoS One. 10:e01285132015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang HL, Sun HF, Gao SP, Li LD, Hu X, Wu
J and Jin W: Loss of RAB1B promotes triple-negative breast cancer
metastasis by activating TGF-β/SMAD signaling. Oncotarget.
6:16352–16365. 2015.PubMed/NCBI
|
|
14
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye X, Brabletz T, Kang Y, Longmore GD,
Nieto MA, Stanger BZ, Yang J and Weinberg RA: Upholding a role for
EMT in breast cancer metastasis. Nature. 547:E1–E3. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Strzyz P: Cancer biology: TGFβ and EMT as
double agents. Nat Rev Mol Cell Biol. 17:202–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gupta S and Maitra A: EMT: Matter of life
or death? Cell. 164:840–842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Konopka G, Friedrich T, Davis-Turak J,
Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM and
Geschwind DH: Human-specific transcriptional networks in the brain.
Neuron. 75:601–617. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Newbury DF and Monaco AP: Genetic advances
in the study of speech and language disorders. Neuron. 68:309–320.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moustakas A and Heldin CH: Mechanisms of
TGFβ-induced epithelial-mesenchymal transition. J Clin Med. 5:pii:
E63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cohn A, Lahn MM, Williams KE, Cleverly AL,
Pitou C, Kadam SK, Farmen MW, Desaiah D, Raju R, Conkling P and
Richards D: A phase I dose-escalation study to a predefined dose of
a transforming growth factor-β1 monoclonal antibody (TβM1) in
patients with metastatic cancer. Int J Oncol. 45:2221–2231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cuiffo BG, Campagne A, Bell GW, Lembo A,
Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al:
MSC-regulated microRNAs converge on the transcription factor FOXP2
and promote breast cancer metastasis. Cell Stem Cell. 15:762–774.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cuiffo BG and Karnoub AE: Silencing FOXP2
in breast cancer cells promotes cancer stem cell traits and
metastasis. Mol Cell Oncol. 3:e10190222015. View Article : Google Scholar : PubMed/NCBI
|