Introduction
Medulloblastoma is the most common childhood
intracranial malignant tumor. At present, therapies against
medulloblastoma comprise surgery, chemotherapy and radiotherapy.
Advances in technology have increased the long-term survival of
patients with this malignancy by up to ~40–70% (1,2). However,
the patients who survive often have long-term, treatment-associated
neurological complications (3).
Furthermore, the incidence rate of medulloblastoma has also
increased gradually over previous decades (3). One particular consensus is that
medulloblastoma has a high-metastatic nature (3). Based on a previous clinicopathological
study, ~40% of medulloblastoma was disseminated through the
cerebrospinal pathways (4). In a
previous report assessing metastasis of medulloblastoma, it was
revealed that medulloblastoma metastasized to multiple extraneural
locations, including bone, bone marrow, lung, liver and lymph nodes
(5). Therefore, there is an urgent
need to develop novel therapies for medulloblastoma.
The human tripartite motif (TRIM) family has >77
members, of which the majority of proteins are E3 ubiquitin
ligases, due to the highly conservative domain, namely, really
interesting new gene (RING). Proteins from this superfamily are
demonstrated to perform biological activities in a variety of
cellular processes, including transcriptional regulation, membrane
repair, cytoskeletal remodeling and oncogenesis (6,7). Of note,
specific members of this family, including TRIM13, TRIM19 and
TRIM25, are involved in human tumorigenesis, including leukemia,
prostate and breast cancers via mechanisms of transcriptional
regulation (8–11). Thereafter, updated evidence has
revealed that multiple TRIM family proteins are critical mediators
of human tumorigenesis, including TRIM59, which was initially
identified as a protein which interacts with Wnt signaling
(12) and was characterized by
oncogenic activity in a mouse model of prostate cancer (13).
TRIM59, a surface molecule, was remarkably increased
in gastric cancer and prominently associated with poor outcome of
patients (13). Ever since the
identification of these characteristics in 2011 (13), TRIM59 was identified to be a multiple
tumor biomarker in human tumorigenesis (14,15).
TRIM59 promotes tumor growth and migration in various cancer types,
including non-small cell lung cancer cells (16) and osteosarcoma (17), whereas the knockdown of TRIM59
inhibits cellular proliferation and migration in cervical cancer
cells (18). Knockdown of TRIM59
correlates with tumor growth inhibition in prostate cancer
(19). However, the exact role of
TRIM59 in human medulloblastoma has remained an enigma until
now.
The present study aimed to investigate the role of
TRIM59 in cell metastasis in medulloblastoma. The expression
profile of TRIM59 in clinical medulloblastoma tissues and in a
series of medulloblastoma cell lines was initially assessed.
Thereafter, the effects of TRIM59 modulation on cancerous cell
metastasis were systemically examined in vitro and in
vivo. The possible mechanisms that contribute to the biological
activity of TRIM59 in medulloblastoma were also investigated and
discussed.
Materials and methods
Human tissues and ethics
statements
In total, 14 patients with medulloblastoma, who were
admitted to Jining No. 1 People's Hospital (Shandong, China) were
included in the present study, patient specimens were collected
without any prior radiotherapy or chemotherapy between May 2012 and
October 2013. The population examined consisted of 14 patients with
9 males and 5 females and the median age at diagnosis was 12 years
(range, 8–17 years). For each case, cancerous tissues and the
matched adjacent non-cancerous tissues were obtained. All patients
gave their full consent to participate in the present study, and a
written consent form was obtained from each patient. All of the
experiments in the present study were in compliance with the
official policies and defined protocols. The research protocol was
approved by the Ethical Committee of Jining No. 1 People's
Hospital.
Cell lines and reagents
Human medulloblastoma cell lines Daoy, D283, D425,
D341 and D458 were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). The passage number of the
cell lines was 10. All cell lines were maintained in Dulbecco's
modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin. The culture medium was replaced every 2
days. Opti-MEM was purchased from Gibco (Gibco, Thermo Fisher
Scientific, Inc.). The primary antibodies were all commercially
purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), except for the antibodies used for
detection of phosphorylation, which were obtained from Cell
Signaling Technology Inc. (Danvers, MA, USA). For the knockdown of
TRIM59, two specific short hairpin (sh)RNAs were chemically
synthesized by GenePharma (Shanghai, China). The pcDNA3.1-TRIM59
expression plasmid was constructed using a pcDNA3.1 expression
vector (Invitrogen; Thermo Fisher Scientific, Inc.). TRIM59 gene
amplification was performed from cDNA of the human medulloblastoma
cell line D283 using the polymerase chain reaction (PCR). The
amplified fragments were digested using HindIII and
XhoI (Takara Biotechnology Co., Ltd., Dalian, China).
Cell transfection
A day before transfection, 1×105 cells
were plated per well in 2 ml complete growth medium (DMEM with 10%
FBS and 100 U/ml penicillin/streptomycin). Cells were 50–60%
confluent on the day of transfection. For each well of cells to be
transfected, 2 µg DNA expression plasmid or shRNA plasmid was
diluted in 250 µl Opti-MEM without serum. For each well of cells, 5
µl Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was added to the media. After gentle mixing the
cells were incubated for 5 min at room temperature, prior to the
addition 500 µl transfection media and another 5–15 min incubation
at room temperature. The cells were transferred to a 37°C, 5%
CO2 incubator for 6 h, before replacing the media with
complete media. A total of 48 h post-transfection, subsequent
experimentation took place.
Total RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of each sample was isolated using TRIzol
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The quality and concentration of extracted
RNAs were identified by measuring the absorbance at 260 nm.
First-strand cDNAs were generated with the PrimeScript RT Master
Mix Perfect Real Time kit (Takara Biotechnology Co., Ltd.) by
mixing the components and incubate at 42°C for 1 h. All qPCRs were
performed with SYBR Premix Ex Taq kit (Takara Biotechnology Co.,
Ltd.) on an ABI PRISM 7500 Real-Time system (Thermo Fisher
Scientific, Inc.). Initial denaturation took place at 95°C for 5
min, followed by annealing at 60°C for 30 sec, and a final
extension at 72°C for 5 min. These conditions were cycled 40 times.
The primers used are listed below, and GAPDH was included as the
internal control. Each experiment was performed in triplicate at
least three times. The primer sequences used are as follows: TRIM59
forward, 5′-TACGAGAGCAGCAGCTTGAA-3′; and reverse,
5′-ACGGGTTGAACCTCAGGAAG-3′; GAPDH forward,
5′-GTGGACATCCGCAAAGAC-3′; and reverse, 5′-AAAGGGTGTAACGCAACTA-3′.
The sequence of the short hairpin RNAs against Trim59 were as
follows: shTrim59#1, 5′-ACATTACAGGCAACCATTAAA-3′; shTrim59#2,
5′-TCCTCGTGTACTGCCATGCTCTCAT-3′; sh (negative control) NC:
5′-GGGTGAACTCACGTCAGAA-3′.
Western blot analysis
Total protein was extracted using a lysis buffer (50
mM Tris-HCl, 150 mM NaCl, 1% NP-40 and 1 mM EDTA, pH 7.5)
containing a Complete protein inhibitor cocktail (Roche
Diagnostics, Indianapolis, IN, USA) to generate the whole protein
lysate following centrifugation at 14,000 × g at 4°C for 15 min. An
equal amount of 50 µg protein/lane was loaded into each lane in a
12% SDS-PAGE gel. The proteins were then transferred onto a
polyvinylidene fluoride (PVDF) membrane by electrophoresis.
Following blocking using 5% milk in Tris-buffered saline with 0.1%
Tween for 1 h at 25°C, the membranes were incubated with primary
antibodies against Trim59 (1:1,000; catalog no. ab166793; Abcam,
Cambridge, UK) overnight at 4°C. A secondary antibody conjugated
with horseradish peroxidase (goat-anti rabbit; 1:5,000; catalog no.
SC-2030; Santa Cruz Biotechnology Inc.) that recognizes the primary
antibody was then added at room temperature for 1 h, and the
immunoreactivity was determined with enhanced chemiluminescent
autoradiography (Thermo Fisher Scientific, Inc.). β-actin was used
as a loading control. Each experiment was repeated at least three
times.
Immunofluorescent assay
Briefly, 1×105 D283 cells treated with 2
µg Trim59 shRNA or shNC for 48 h were seeded on the sterile
coverslips in a 24-well plate in DMEM with 10% FBS. After 24 h, the
cells were rinsed with PBS and fixed for 20 min in 4%
paraformaldehyde at 25°C. The cells were washed using PBS. Membrane
penetration was accomplished with 2% Triton X-100 and the membrane
was incubated for 20 min in 0.3% (v/v) H2O2
at 25°C. Following washing with PBS, the cells were blocked in 5%
FBS for 1 h and then incubated with a primary antibody against
F-actin (1:500; Santa Cruz Biotechnology Inc.) at 4°C overnight.
Following washing with PBS, the cells were incubated with the
corresponding secondary antibody conjugated with rhodamine
phalloidin (goat-anti rabbit; 1:5,000; Santa Cruz Biotechnology
Inc.) in the dark for 1 h and counterstained at 25°C for 30 min
with DAPI (1:1,000). Finally, the samples were mounted and
observed, and images were captured using an inverted microscope
(IX71; Olympus Corporation, Tokyo, Japan).
Wound-healing and Borden chamber
assays
The wound-healing assay and Boyden chamber assay
were performed according to previous literature (20). Briefly, 5×105 D283 cells
were plated on 6-well plates to form a confluent monolayer. Wounds
made with sterile pipette tips were observed after 6 h. The rate of
wound recovery was then calculated at each time point (0 and 24 h).
A migration assay was carried out using Boyden chambers (tissue
culture-treated; diameter, 6.5 mm; pore size, 8 µm; Transwell,
Corning Incorporated, Corning, NY, USA) containing polycarbonate
membrane. For the invasion assay, the upper surface of the insert
was pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA) to
mimic basement membrane. Thereafter, 100 µl 1×106 cells
in serum-free DMEM was added to the upper chamber, and 600 µl DMEM
with 10% FBS was added to the lower chamber. The cells were
incubated for 12 h at 37°C. The migrated cells on the under-surface
of the membrane were fixed and stained with crystal violet for 10
min at room temperature. Images of five random regions were
captured, and the number of cells was counted to calculate the mean
number of migrated cells per plate.
Statistical analysis
The data are expressed as the mean ± standard
deviation (SD). Statistical analysis was performed using SPSS
software package (version 16.0; SPSS, Inc., Chicago, IL, USA).
Trim59 mRNA statistical comparisons were made using unpaired
Student's t-test. Comparisons between two groups were analyzed
using Mann-Whitney U test. Multiple group comparisons were analyzed
using one-way analysis of variance test with post hoc contrasts by
Student-Newman-Keuls test. All the experiments were repeated at
least three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
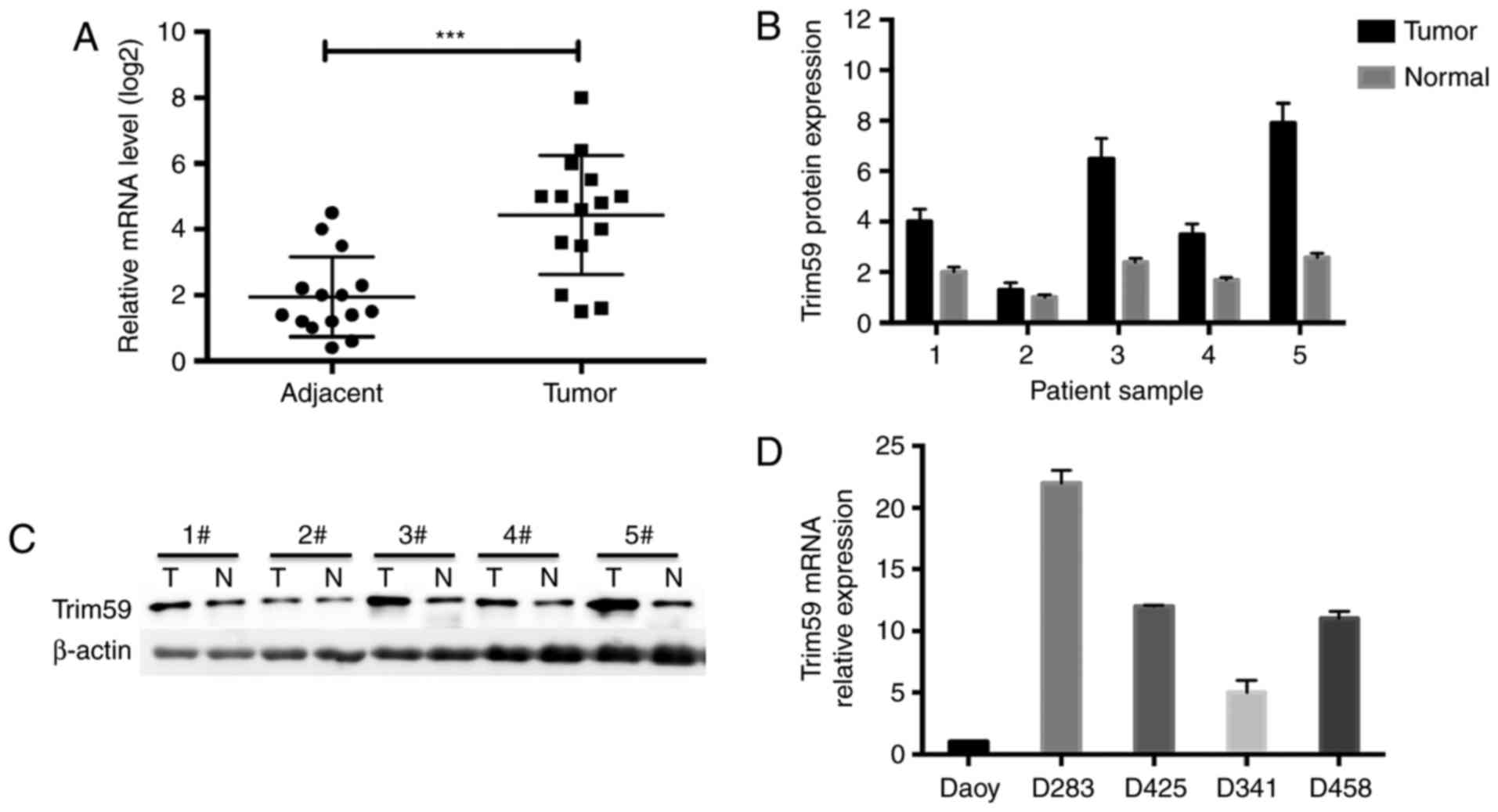
TRIM59 is upregulated in
medulloblastoma
The expression profile of TRIM59 was initially
determined. In the 15 cases of clinical medulloblastoma tissues, it
was detected that the relative mRNA of TRIM59 was ~2-fold in
cancerous tissues compared with adjacent non-cancerous tissues
(Fig. 1A). qPCR also validated the
high expression of TRIM59 in medulloblastoma tissues compared with
adjacent noncancerous tissues (Fig.
1B). Western blot analysis further indicated that TRIM59
protein levels were consistently upregulated in cancerous tissues
compared with adjacent non-cancerous tissues (Fig. 1C). In a series of medulloblastoma cell
lines, it was notable that TRIM59 was differentially expressed in
multiple cell lines, with the highest expression observed in D283
cells and the lowest level in Daoy cells (Fig. 1D). These data suggested that TRIM59
was upregulated in medulloblastoma.
Knockdown of TRIM59 in D283 cells
inhibits epithelial-to-mesenchymal transition (EMT)
Two specific shTrim59s were synthesized and used to
infect D283 cells. As compared with the negative control shRNA
(shNC), the two different specific shTrim59s were effective in
depleting TRIM59 gene expression, with the second shTrim59
exhibiting higher efficiency (Fig.
2A). Western blot analysis also validated that shTrim59#2 was
more effective in depleting TRIM59 expression (Fig. 2B). Therefore, the second shTrim59 was
used for subsequent analyses. Notably, it was observed that
knockdown of TRIM59 inhibited D283 cell pseudopodia formation and
led to oval morphological shapes (Fig.
2C), therefore indicating epithelial characteristics following
the depletion of TRIM59. Furthermore, it was demonstrated that the
expression of the epithelial marker E-cadherin was increased, while
mesenchymal markers vimentin, SMA and N-cadherin were decreased
following the knockdown of TRIM59 in D283 cells.
Epithelial-to-mesenchymal transition (EMT) signaling molecules
ZEB1, Slug and Twist were consistently depleted by shTrim59
(Fig. 2D). Immunofluorescent assay
also revealed that knockdown of TRIM59 inhibited pseudopodia
formation of D283 cells, and the expression of cytoskeleton protein
F-actin was markedly decreased, compared with shNC (Fig. 2E). These data strongly indicate that
TRIM59 is a mediator of EMT in D283 cells.
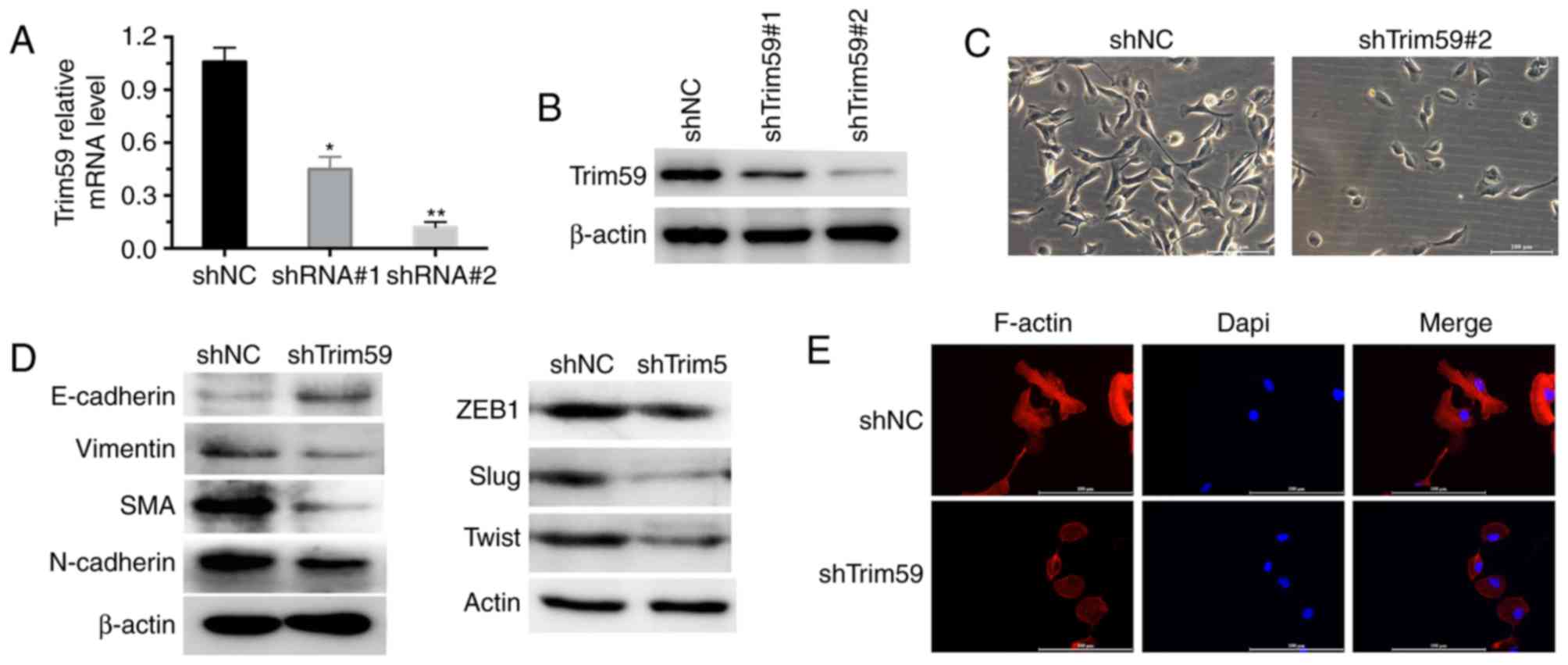 | Figure 2.Knockdown of TRIM59 inhibits
epithelial-to-mesenchymal transition in D283 cells. (A) Two
specific shRNAs against TRIM59 were synthesized. RT-qPCR revealed
that the two shTrim59 worked efficiently to deplete TRIM59, with
the second shRNA being more effective. (B) Western blot analysis
confirmed that shTrim59#2 was effective in depleting TRIM59 protein
compared with shTrim59#1. (C) Cell morphology was analyzed in
negative control shRNA or shTrim59-infected D283 cells
(magnification, ×100). (D) In D283 cells, with or without TRIM59
knockdown, the expression of epithelial and mesenchymal markers was
analyzed. (E) The cytoskeleton protein F-actin was analyzed using
immunofluorescent assay in D283 cells (magnification, ×200).
*P<0.05, **P<0.01 vs. shNC. shNC, negative control shRNA;
shRNA, short hairpin RNA; shTrim59, shRNA against TRIM59; TRIM59,
tripartite motif containing 59; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
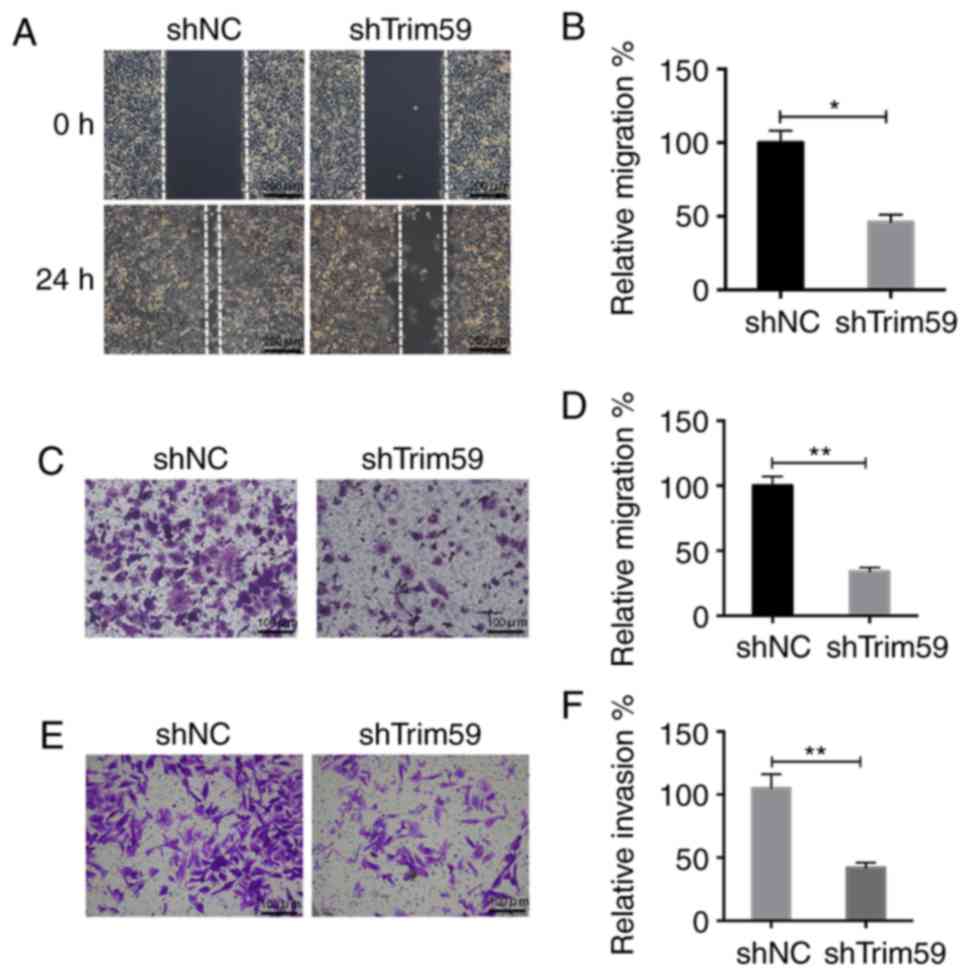
Knockdown of TRIM59 inhibits motility
of D283 cells
In the wound-healing assay, it was observed that
after 24 h, the wound was barely observable in the shNC-treated
D283 cells, whereas a significant wound remained in
shTrim59-infected D283 cells (Fig.
3A). Quantification of wound-recovery area further elucidated
that only half of the wound was recovered in shTrim59 group,
whereas the wound was almost fully recovered area in the control
group. Knockdown of TRIM59 markedly suppressed the cell mobility
ability by 50% in D283 cells (Fig.
3B). Similarly, cell migration was significantly inhibited by
shTrim59 as indicated by the Transwell migration assay, compared
with shNC (Fig. 3C). An average of
~40 cells in the TRIM59-knocked down group migrated to the lower
surface, whereas ~100 control D283 cells transmigrated. Knockdown
of TRIM59 inhibited the cell migratory ability by 60% in D283 cells
(Fig. 3D). Furthermore, as indicated
by the Transwell invasion assay, there was a significant decrease
in the number of cells that invaded following the knockdown of
TRIM59 (Fig. 3E and F). Therefore,
the depletion of TRIM59 is attributable for the inhibition of
migration and invasion of D283 cells.
Upregulation of TRIM59 induces
epithelial-to-mesenchymal transition in Daoy cells
The pcDNA3.1-TRIM59 expression plasmid was utilized
to upregulate the protein level of TRIM59 in Daoy cells (Fig. 4A). Overexpression of TRIM59 led to
EMT-mediated morphological changes, for example
TRIM59-overexpressing Daoy cells exhibited a shuttle shape or
multiple angle shape (Fig. 4B). This
suggests that EMT formation was mediated by the overexpression of
TRIM59. Western blot analysis also revealed that following the
overexpression of TRIM59, the expression of epithelial marker
E-cadherin was decreased, whereas the expression of mesenchymal
markers, vimentin, SMA and N-cadherin was increased, compared with
the expression in the cells that were transfected with the control
vector (Fig. 4C). The transcription
factors ZEB1, Slug and Twist, which suppress epithelial genes and
activate mesenchymal genes, were consistently upregulated by TRIM59
(Fig. 4D). Together with the data
acquired from TRIM59 knockdown, it may be concluded that TRIM59 may
a mediator of EMT in medulloblastoma cells.
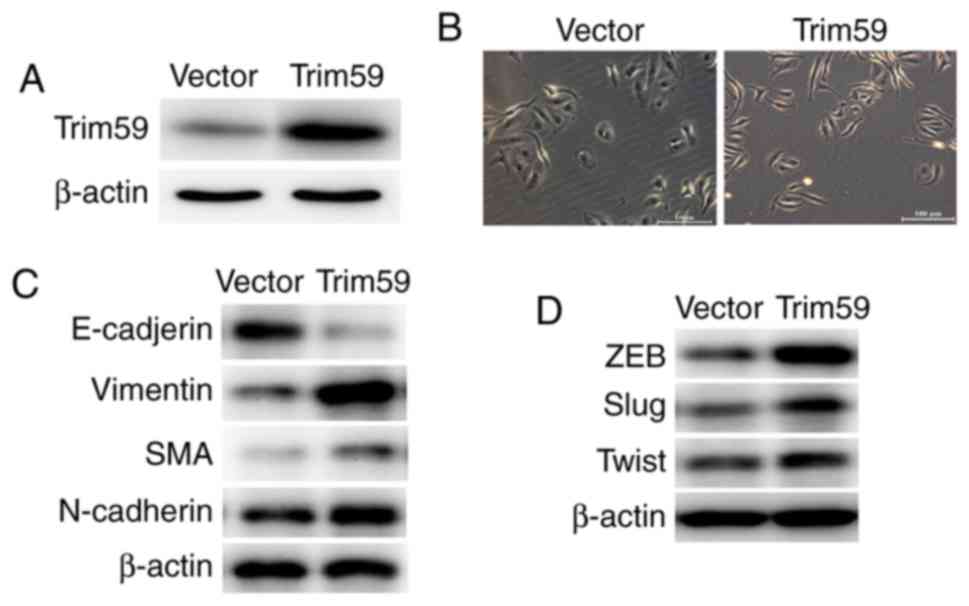 | Figure 4.Upregulation of TRIM59 induces
epithelial-to-mesenchymal transition in Daoy cells. (A) An
expression plasmid was constructed to upregulate TRIM59 in Daoy
cells. (B) Following the overexpression of TRIM59, Daoy cells were
observed to be morphologically changed. Control cells exhibited an
oval shape, with more epithelial characteristics, whereas
TRIM59-upregulated Daoy cells were shuttle-shaped or multiple
angle-shaped, with more mesenchymal characteristics (magnification,
×100). Western blot analysis of the epithelial markers (C)
E-cadherin, mesenchymal markers vimentin, SMA and N-cadherin, and
(D) EMT-associated transcription factors ZEB1, Slug and Twist in
control and TRIM59-overexpressed Daoy cells. SMA, smooth muscle
actin; TRIM59, tripartite motif containing 59; ZEB1, zinc finger
E-box-binding homeobox 1. |
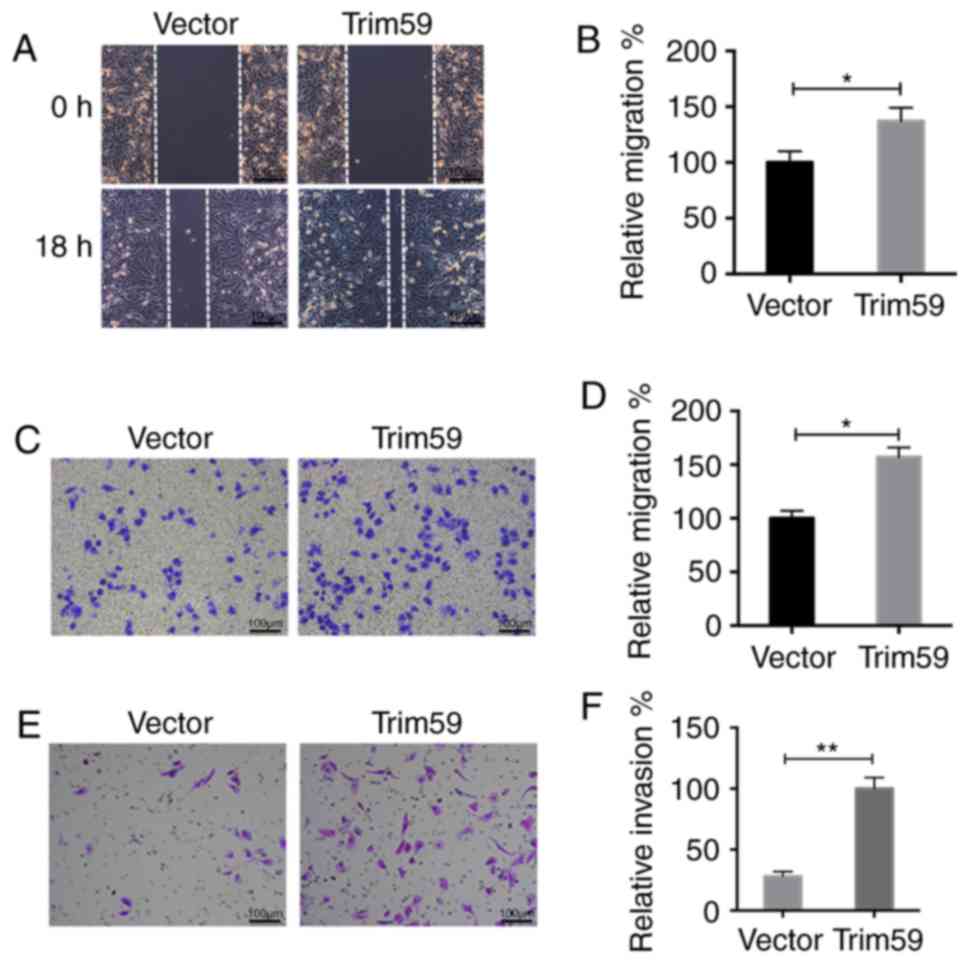
Upregulation of TRIM59 promotes
motility in Daoy cells
Furthermore, in TRIM59-overexpressing Daoy cells,
the wound recovery process was accelerated, as indicated by the
closure of the wound after 18 h (Fig. 5A
and B). In the Transwell migration assay, the cells that had
migrated to the lower surface were increased following
overexpression of TRIM59 (Fig. 5C).
The analysis of the migrated cells further demonstrated that
>150 TRIM59-overexpressing Daoy cells migrated, which was by
contrast with only 100 control cells that exhibited migratory
abilities. Overexpression of TRIM59 promoted the migratory ability
by 45% in Daoy cells (Fig. 5D).
Similarly, invasion was significantly increased in
TRIM59-overxpressing Daoy cells compared with control Daoy cells
(Fig. 5E). A total of ~100
TRIM59-overexpressing Daoy cells were stained, whereas only 40
control cells were observed under the lower surface of the
membrane. Overexpression of TRIM59 promoted the invasive ability by
70% in Daoy cells (Fig. 5F). These
data strongly suggest that the overexpression of TRIM59
significantly promotes cell motility in Daoy cells.
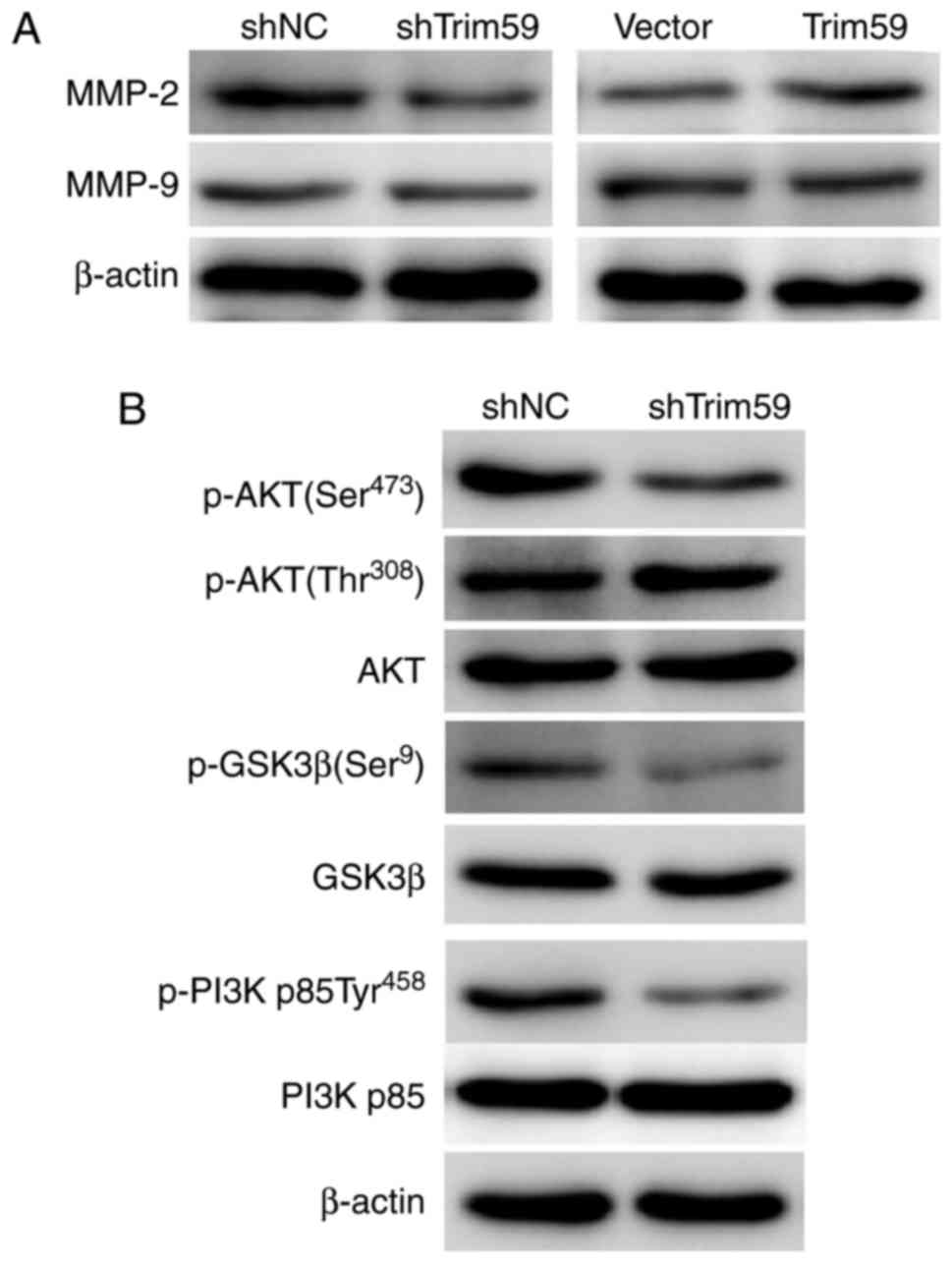
TRIM59 positively regulates MMP-2 and
phosphoinositide 3-kinase (PI3K)/AKT signaling
MMPs are upregulated in the process of tumor
invasion-metastasis cascade (21).
Western blot analysis revealed that the major MMP, MMP-2 but not
MMP-9, was downregulated following the knockdown of TRIM59 in D283
cells, and upregulated by the overexpression of TRIM59 in Daoy
cells compared with shNC cells (Fig.
6A), which supported the findings of the aforementioned
invasion assays. Notably, it was observed that following the
knockdown of TRIM59, the levels of p-AKT (Ser473), p-GSK3β (Ser9)
and p-PI3K p85 (Tyr458) were consistently decreased, while the
total protein levels remained largely unaltered. The
phosphorylation level of AKT at the site Thr308 also remained
unchanged (Fig. 6B). These data
suggest that the PI3K/AKT signaling cascade is activated by TRIM59
in medulloblastoma.
Discussion
Medulloblastoma is the most common malignant
pediatric tumor in the brain, and it is often resistant to
traditional therapy. Medulloblastoma is also characterized by
frequent extraneural metastasis (5).
Therefore, novel therapeutic strategies for intercepting critical
regulatory pathways in cancer development and progression are
warranted. Here, we present in vitro and in vivo data
that TRIM59 displays potent metastatic activity via the PI3K/AKT
signaling pathway in medulloblastoma and may be a potential
therapeutic target for the treatment of medulloblastoma.
In the present study, TRIM59 was initially observed
to be upregulated in medulloblastoma tissues and differentially
expressed in a series of medulloblastoma cell lines. Notably, it
was observed that the knockdown of TRIM59 in D283 cells resulted in
prominent epithelial features, which was accompanied with decreased
cell migratory and invasive capacity. By contrast, the
overexpression of TRIM59 in Daoy cells induced mesenchymal
features, which was accompanied with increased cell migratory and
invasive abilities. The corresponding regulation of MMP-2 by TRIM59
further supported the metastatic potency of TRIM59 in
medulloblastoma cells. The present data suggested that TRIM59 was
able to induce the EMT process in medulloblastoma.
EMT is a critical manifestation of epithelial cell
plasticity, where multiple regulatory molecules are involved,
including the Zeb family and the Snail family (22). The EMT process is induced by various
cellular procedures, including increased expression of mesenchymal
markers (N-cadherin and vimentin), decreased protein levels of
epithelial markers (E-cadherin) and the overexpression of ECM
molecules (fibronectin) (23). EMT
formation is also associated with changes in cell morphology (the
cells exhibited a shuttle shape or multiple angle shape) (24). The EMT process has been associated
with human tumorigenesis in a wide range of literature. For
instance, it was shown that in ovarian cancer, the activation of
EMT is associated with chemoresistance, the latter of which can
cause cancer recurrence and metastasis following conventional
treatment against ovarian cancer (25,26).
Additionally, benzophenone-3 has been demonstrated to increase
metastasis potential in lung cancer via the induction of EMT
(27). Following full review of the
EMT data in the present study, together with the cell migration and
invasion assays, it is concluded that TRIM59 may promote metastasis
in medulloblastoma.
The PI3K/AKT signaling pathway has been identified
as a key driver of proliferation, migration and angiogenesis in
human tumorigenesis, including medulloblastoma, where the
activation of PI3K/AKT signaling has been associated with enhanced
tumor growth, metastasis and chemoresistance (28–30). In
addition, the present study also identified that PI3K/AKT signaling
was involved in TRIM59-mediated cell metastasis. Following the
knockdown of TRIM59, the levels of p-AKT, p-GSK3β and p-PI3K p85
were decreased, suggesting the positive regulation of PI3K/AKT
signaling by TRIM59. A previous study indicated that the PI3K
inhibitor GDC-0941 displayed promising in vitro and in
vivo efficacy for targeted medulloblastoma therapy (31). In addition, PI3K/AKT signaling serves
as an integration node in a network of tumor-promoting signaling
pathways (32). It is therefore
likely that any compound or reagent targeting TRIM59 and
consequently inhibiting the PI3K/AKT signaling pathway may serve as
a promising therapeutic strategy for the treatment of
medulloblastoma.
In conclusion, the present study identified TRIM59
as a critical mediator of cell metastasis in medulloblastoma. The
loss-of-function and gain-of-function assays highlighted the strong
metastatic potential of TRIM59 in medulloblastoma. TRIM59 may be
able to induce the EMT process and promote cell migration and
invasion via the PI3K/AKT signaling pathway. The present study
provided data to indicate that TRIM59 may serve as a molecular
target that is useful for targeted therapeutic strategies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RG and LJC conceived the study; RG, GQL, XW and CZ
collected the cancer tissues; RG and GQL performed the experiment;
RG and LC calculated the data and wrote the paper.
Ethics approval and consent to
participate
All patients gave their full consent to participate
in the present study, and a written consent form was obtained from
each patient. All of the experiments in the present study were in
compliance with the official policies and defined protocols. The
research protocol was approved by the Ethical Committee of Jining
No. 1 People's Hospital.
Consent for publication
A written consent form was obtained from each
patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gajjar A, Chintagumpala M, Ashey D, Kellie
S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin MJ, et
al: Risk-adapted craniospinal radiotherapy followed by high-dose
chemotherapy and stem-cell rescue in children with newly diagnosed
medulloblastoma (St Jude Medulloblastoma-96): Ong-term results from
a prospective, multicentre trial. Lancet Oncol. 7:813–820. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Packer RJ: Risk-adapted craniospinal
radiotherapy followed by high-dose chemotherapy and stem-cell
rescue in children with newly diagnosed medulloblastoma. Curr
Neurol Neurosci Rep. 7:130–132. 2007.
|
|
3
|
Aref D and Croul S: Medulloblastoma:
Recurrence and metastasis. CNS Oncol. 2:377–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horten BC and Rubinstein LJ: Primary
cerebral neuroblastoma. A clinicopathological study of 35 cases.
Brain. 99:735–756. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mazloom A, Zangeneh AH, Teh BS and Paulino
AC: Extraneural metastasis of medulloblastoma. J Clin Oncol.
27:20652009.
|
|
6
|
Elabd S, Meroni G and Blattner C: TRIMming
p53's anticancer activity. Oncogene. 35:5577–5584. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lazzari E and Meroni G: TRIM32 ubiquitin
E3 ligase, one enzyme for several pathologies: From muscular
dystrophy to tumours. Int J Biochem Cell Biol. 79:469–477. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Thé H, Lavau C, Marchio A, Chomienne C,
Degos L and Dejean A: The PML-RAR alpha fusion mRNA generated by
the t(15;17) translocation in acute promyelocytic leukemia encodes
a functionally altered RAR. Cell. 66:675–684. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cambiaghi V, Giuliani V, Lombardi S,
Marinelli C, Toffalorio F and Pelicci PG: TRIM proteins in cancer.
Adv Exp Med Biol. 770:77–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Le Douarin B, Zechel C, Garnier JM, Lutz
Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P and Losson
R: The N-terminal part of TIF1, a putative mediator of the
ligand-dependent activation function (AF-2) of nuclear receptors,
is fused to B-raf in the oncogenic protein T18. EMBO J.
14:2020–2033. 1995.PubMed/NCBI
|
|
12
|
Licchesi JD, Van Neste L, Tiwari VK, Cope
L, Lin X, Baylin SB and Herman JG: Transcriptional regulation of
Wnt inhibitory factor-1 by Miz-1/c-Myc. Oncogene. 29:5923–5934.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valiyeva F, Jiang F, Elmaadawi A, Moussa
M, Yee SP, Raptis L, Izawa JI, Yang BB, Greenberg NM, Wang F and
Xuan JW: Characterization of the oncogenic activity of the novel
TRIM59 gene in mouse cancer models. Mol Cancer Ther. 10:1229–1240.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khatamianfar V, Valiyeva F, Rennie PS, Lu
WY, Yang BB, Bauman GS, Moussa M and Xuan JW: TRIM59, a novel
multiple cancer biomarker for immunohistochemical detection of
tumorigenesis. BMJ Open. 2:pii: e001410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhan W, Han T, Zhang C, Xie C, Gan M, Deng
K, Fu M and Wang JB: TRIM59 promotes the proliferation and
migration of non-small cell lung cancer cells by upregulating cell
cycle related proteins. PLoS One. 10:e1425962015. View Article : Google Scholar
|
|
17
|
Liang J, Xing D, Li Z, Shen J, Zhao H and
Li S: TRIM59 is upregulated and promotes cell proliferation and
migration in human osteosarcoma. Mol Med Rep. 13:5200–5206. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aierken G, Seyiti A, Alifu M and Kuerban
G: Knockdown of tripartrtite-59 (TRIM59) inhibits cellular
proliferation and migration in human cervical cancer cells. Oncol
Res. 25:381–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin WY, Wang H, Song X, Zhang SX, Zhou PS,
Sun JM and Li JS: Knockdown of tripartite motif 59 (TRIM59)
inhibits tumor growth in prostate cancer. Eur Rev Med Pharmacol
Sci. 20:4864–4873. 2016.PubMed/NCBI
|
|
20
|
Li LL, Xue AM, Li BX, Shen YW, Li YH, Luo
CL, Zhang MC, Jiang JQ, Xu ZD, Xie JH and Zhao ZQ: JMJD2A
contributes to breast cancer progression through transcriptional
repression of the tumor suppressor ARHI. Breast Cancer Res.
16:R562014. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strauss R, Li ZY, Liu Y, Beyer I, Persson
J, Sova P, Möller T, Pesonen S, Hemminki A, Hamerlik P, et al:
Analysis of epithelial and mesenchymal markers in ovarian cancer
reveals phenotypic heterogeneity and plasticity. PLoS One.
6:e161862011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shenoy AK, Jin Y, Luo H, Tang M, Pampo C,
Shao R, Siemann DW, Wu L, Heldermon CD, Law BK, et al:
Epithelial-to-mesenchymal transition confers pericyte properties on
cancer cells. J Clin Invest. 126:4174–4186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abdullah LN and Chow EK: Mechanisms of
chemoresistance in cancer stem cells. Clin Transl Med. 2:32013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iwatsuki M, Mimori K, Yokobori T, Ishi H,
Beppu T, Nakamori S, Baba H and Mori M: Epithelial-mesenchymal
transition in cancer development and its clinical significance.
Cancer Sci. 101:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Phiboonchaiyanan PP, Busaranon K,
Ninsontia C and Chanvorachote P: Benzophenone-3 increases
metastasis potential in lung cancer cells via epithelial to
mesenchymal transition. Cell Biol Toxicol. 33:251–261. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hartmann W, Digon-Söntgerath B, Koch A,
Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sörensen N,
Wiestler OD and Pietsch T: Phosphatidylinositol 3′-kinase/AKT
signaling is activated in medulloblastoma cell proliferation and is
associated with reduced expression of PTEN. Clin Cancer Res.
12:3019–3027. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baryawno N, Sveinbjörnsson B, Eksborg S,
Chen CS, Kogner P and Johnsen JI: Small-molecule inhibitors of
phosphatidylinositol 3-kinase/Akt signaling inhibit
Wnt/beta-catenin pathway cross-talk and suppress medulloblastoma
growth. Cancer Res. 70:266–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guerreiro AS, Fattet S, Fischer B, Shalaby
T, Jackson SP, Schoenwaelder SM, Grotzer MA, Delattre O and Arcaro
A: Targeting the PI3K p110alpha isoform inhibits medulloblastoma
proliferation, chemoresistance, and migration. Clin Cancer Res.
14:6761–6769. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ehrhardt M, Craveiro RB, Holst MI, Pietsch
T and Dilloo D: The PI3K inhibitor GDC-0941 displays promising in
vitro and in vivo efficacy for targeted medulloblastoma therapy.
Oncotarget. 6:802–813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang F, Li M, Wu X, Hu Y, Cao Y, Wang X,
Xiang S, Li H, Jiang L, Tan Z, et al: 20(S)-ginsenoside Rg3
promotes senescence and apoptosis in gallbladder cancer cells via
the p53 pathway. Drug Des Devel Ther. 9:3969–3987. 2015.PubMed/NCBI
|