Introduction
Breast cancer is highly heterogeneous, and its
biological behavior and response to therapy differ according to the
subtype of breast cancer (1,2). In 2000, according to the cancer gene
expression profiles, Sørlie et al (3) and Perou et al (4) divided breast cancer into luminal A,
luminal B+C, human epidermal growth factor receptor 2
(Her-2)-overexpressing, basal-like and normal-like subtypes. In
current clinical practice, immunohistochemical methods are used to
test for estrogen receptor (ER), progesterone receptor (PR) and
Her-2 expression due to the complexity, and high cost of performing
molecular profiling (5–8). Patients with TNBC or
Her-2-overexpressing subtypes exhibit the worst prognosis. In
addition to classic prognostic factors, including tumor size, lymph
nodes involved, and histological grade, some genetic and biological
factors have been investigated to determine their effects on
survival (9–11).
The Notch receptor family comprises four type I
membrane proteins. Of them, the role of Notch4 in epithelial tumors
was identified by insertional mutagenesis in mice infected with
mouse mammary tumor virus (12,13).
Notch4 has been identified to be expressed in stem cells of the
mammary gland terminal duct, and has been implicated in the
formation of branching structures that precede poorly
differentiated adenocarcinoma, the restraining of TAC-2 cells to
form duct branches, as well as growth factor β function, aggressive
tumor phenotype, and the enabling of the transition from normal
mouse mammary epithelial cells to heterotypic cells (12–15). These
results demonstrated that the Notch4 signaling pathway serves an
important role in the regulation of mammary gland growth and
development. Abnormal expression of Notch4 may inhibit the
differentiation of mammary stem cells, and mutations of the Notch4
gene may enhance mammary epithelial cell proliferation, thus
leading to the occurrence of breast cancer.
In the present study, different expression levels of
Notch4 were investigated in different subtypes of breast cancer. In
addition, the associations between Notch4 expression, and breast
cancer clinicopathological characteristics and the prognosis of
patients were analyzed. Furthermore, the present study aimed to
evaluate the potential of Notch4 as a prognostic marker for
patients with breast cancer.
Materials and methods
A total of 98 patients who were admitted to the
Cancer Hospital of Shantou University Medical College (Shantou,
China) between January 1996 and December 2008 were enrolled in the
current study. The study was approved by the Ethics Committee of
Shantou University Medical College and written informed consent was
obtained from all patients. All patients were female with a mean
age of 50.5 years old (range, 36–81 years old). All patients
received surgical treatment. Patients <50 years old accounted
for 41.8%; 64.3% of the patients were premenopausal; 38.8% of
patients had a tumor diameter reaching T1/T2, whereas 61.2% had
tumor diameters reaching T3/T4; 63.3% had an N0/N1 lymph node
grade, whereas 36.7% had N2/N3; 30.6% were at stage I/II, whereas
69.4% were at stage III/IV. Tumor stage was judged according to the
sixth edition of the breast cancer tumor node metastasis (TNM)
staging system of the American Cancer Federation (16), histological grade was judged according
to the Nottingham breast cancer grading system (17). Neoadjuvant chemotherapy and adjuvant
chemotherapy were all in accordance with guidelines.
ER, PR and Her-2 immunohistochemical detection was
performed on all specimens as previously reported (18). The criteria for ER- and PR-positive
staining was >10% of the cancer cell nuclei stained brown
(19), and Her-2 was considered
positive if >30% of the cancer cells presented with strong or
complete cell membrane brown coloring (20). Cases were then divided into three
groups according to ER, PR and Her-2 expression: i) Triple-negative
breast cancer: ER, PR and Her-2 were all negative (n=27); ii)
Her-2-overexpressing breast cancer: ER- and PR-negative,
Her-2-positive (n=24); iii) luminal breast cancer: ER- and/or
PR-positive, Her-2-negative or -positive (n=47).
Immunohistochemical staining
Formalin-fixed and paraffin-embedded breast cancer
tissues were cut into 4-µm thick sections. Immunohistochemical
staining was performed using Envision's two-step method to assess
Notch4 expression, as previously reported (21). Briefly, tissue sections were
deparaffinized with xylene and rehydrated via incubation with
gradient dilutions of ethanol. Antigen retrieval was achieved
through microwaving in 0.01 mol/l citrate buffer (pH=6.0) for 15
min and then allowing cooling to room temperature (RT). Endogenous
peroxidase activity was subsequently blocked by incubation with 3%
H2O2 for 10 min at RT, and then blocked with
10% normal goat serum (OriGene Technologies, Inc., Rockville, MD,
USA) in PBS (pH=7.4) for 30 min at RT. Following blocking, sections
were incubated with anti-Notch4 polyclonal antibody (catalog no.
SC-5594; H-225; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) overnight at 4°C. Then, sections were incubated with
Supervision™ Universal goat anti-rabbit horseradish
peroxidase-conjugated Detection reagent (catalog no. SC-2004; Santa
Cruz Biotechnology, Inc.) for 30 min at room temperature. Three
washes with PBS were performed between each step of the procedure.
Staining was developed with 3,3′-diaminobenzidine at room
temperature and counterstained with hematoxylin at room
temperature. Negative controls were evaluated by replacing the
primary antibody with PBS.
Evaluation of immunohistochemistry and
statistical analysis
Expression of Notch4 was primarily detected in the
cytoplasm and nucleus of tumor cells, and was evaluated using a
semi-quantitative scoring system as previously reported (22). Firstly, the extent of
positively-labeled cells was ranked into four semi-quantitative
grades: <5%, 0; 5–35%, 1; 36–70%, 2; 71–100%, 4. Secondly, the
intensity of staining was categorized into four classes as follows:
No staining, 0; weak staining, 1; intermediate staining, 2; and
strong staining, 3. The four groups were categorized according to
the multiplied score of the two classifications: Negative (−), ≤1;
+, 2–3; ++, 4–5; and +++, ≥6. Based on the final score, tumor
tissues that were negative (−) and weakly-positive (+) were defined
as low Notch4-expressing, while tissues with moderately-(++) and
strongly-positive (+++) Notch4 expression were defined as high
Notch4-expressing.
All data were analyzed using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA). The Chi-squared and two-sided
Fisher's exact tests were used to assess the clinicopathological
characteristics categorized by breast cancer subgroups and levels
of Notch4 expression. Disease-free survival (DFS) duration was
defined as the time between the date of first diagnosis and the
date of the last follow-up or the date of cancer relapse. Overall
survival (OS) duration was defined as the time between the date of
first diagnosis and the date of the last follow-up or the date of
cancer-relative death. Survival analysis was performed in
sub-groups using the Kaplan-Meier survival analysis and log-rank
test. Univariate and multivariate analyses were applied to quantify
the effect of variables on patient survival. P<0.05 was
considered to indicate a statistically significant difference.
Results
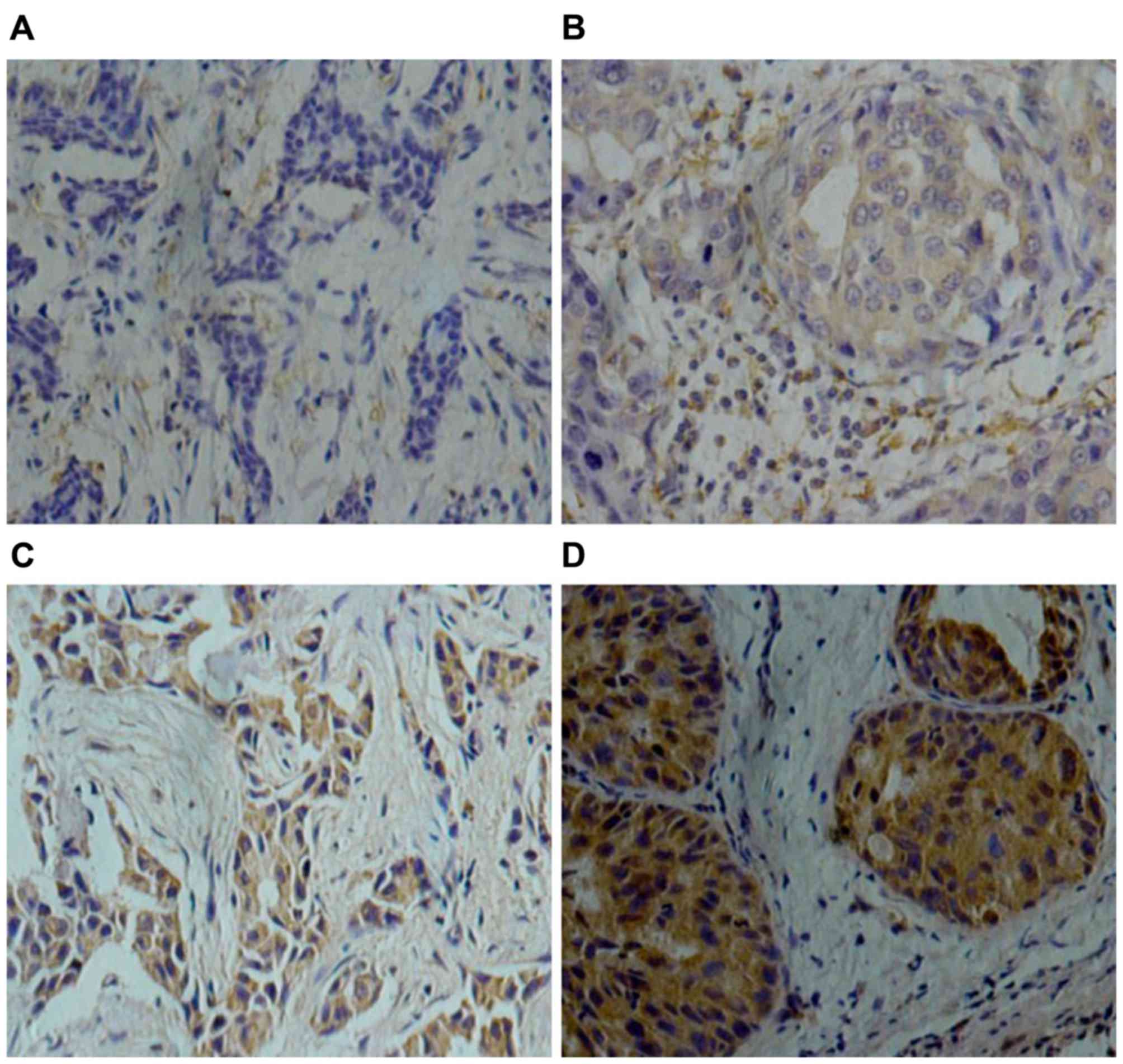
Notch4 protein is expressed in the
cytoplasm and nucleus of tumor cells
According to the evaluation system of
Notch4-staining, 60/98 patients (61%) exhibited low Notch4
expression and 38/98 patients (39%) had high Notch4 expression
(Fig. 1). Notably, high Notch4
high-expression was detected in 55.6% (15/27) of TNBC cases, in
45.8% (11/24) of Her-2-overexpression cases and in 25.2% (12/47) of
luminal breast cancer cases. Patients with triple-negative or
Her-2-overexpressing breast cancer exhibited significantly higher
Notch4 high-expression compared with patients with the luminal type
(P=0.028), but there was no significant difference between
triple-negative breast cancer and Her-2-overexpressing groups
(P=0.808; data not shown) (Table
I).
 | Table I.Expression of Notch4 in different
subtypes of breast cancer. |
Table I.
Expression of Notch4 in different
subtypes of breast cancer.
|
| Notch4 (%) |
|
|
|---|
|
|
|
|
|
|---|
| Breast cancer
subtype | Low expression | High expression |
χ2 | P-value |
|---|
| Triple-negative | 12 (44.4) | 15 (55.6) |
|
|
| Her-2
overexpression | 13 (54.2) | 11 (45.8) | 7.178 | 0.028 |
| Luminal | 35 (74.5) | 12 (25.5) |
|
|
Association between Notch4 expression
and clinicopathological characteristics of breast cancer
High Notch4 expression was associated with lower ER
(52.8 vs. 22.2%; P=0.002) and PR (47.6 vs. 22.9%; P=0.016), larger
tumor size (46.7 vs. 26.3%; P=0.044), greater lymph node metastasis
(51.9 vs. 22.7%; P=0.003) and advanced TNM stage (III/IV) (47.1 vs.
20.0%; P=0.011), compared with low Notch4 expression. However, no
significant difference was identified between the <50 and ≥50
years old age groups, pre- and post-menopausal groups, low and high
Her-2 groups, and with or without distant metastasis (Table II).
 | Table II.Association between Notch4 expression
and clinicopathological characteristics in 98 cases of breast
cancer. |
Table II.
Association between Notch4 expression
and clinicopathological characteristics in 98 cases of breast
cancer.
|
| Notch4 (%) |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low expression | High expression |
χ2 | P-value |
|---|
| Age, years |
|
|
|
|
| ≤50 | 22 (53.7) | 19 (46.3) | 1.7 | 0.192 |
| ≥50 | 38 (66.7) | 19 (33.3) |
|
|
| Menstrual status |
|
|
|
|
|
Premenopausal | 42 (66.7) | 21 (33.3) | 2.201 | 0.138 |
|
Postmenopausal | 18 (51.4) | 17 (48.6) |
|
|
| ER status |
|
|
|
|
|
Negative | 25 (47.2) | 28 (52.8) | 9.604 | 0.002 |
|
Positive | 35 (77.8) | 10 (22.2) |
|
|
| PR status |
|
|
|
|
|
Negative | 33 (52.4) | 30 (47.6) | 5.811 | 0.016 |
|
Positive | 27 (77.1) | 8 (22.9) |
|
|
| Her-2 status |
|
|
|
|
|
Negative | 37 (67.3) | 18 (32.7) | 1.932 | 0.165 |
|
Positive | 23 (53.5) | 20 (46.5) |
|
|
| Tumor size |
|
|
|
|
|
T1/T2 | 28 (73.7) | 10 (26.3) | 4.059 | 0.044 |
|
T3/T4 | 32 (53.3) | 28 (46.7) |
|
|
| Lymph node
involvement |
|
|
|
|
|
N0/N1 | 34 (77.3) | 10 (22.7) | 8.663 | 0.003 |
|
N2/N3 | 26 (48.1 | 28 (51.9) |
|
|
| Distant
metastasis |
|
|
|
|
| No | 49 (61.3) | 31 (38.7) | 0.009 | 0.925 |
|
Yes | 10 (62.5) | 6 (37.5) |
|
|
| Tumor stage |
|
|
|
|
|
I/II | 24 (80.0) | 6 (20.0) | 6.42 | 0.011 |
|
III/IV | 36 (52.9) | 32 (47.1) |
|
|
Prognostic significance of
clinicopathological factors for breast cancer
Factors, including patient age, tumor size, axillary
lymph node metastasis, distant metastasis, clinical stage, ER, PR,
Her-2 and Notch4 expression, were used for univariate, and
multivariate analysis of OS. Univariate analysis demonstrated that
patients with high Notch4 expression possessed a 3.8-fold increase
in relative risk of cancer-associated mortality (95% confidence
interval, 0.892–16.204; P=0.071, data not shown) compared with
patients with low Notch4 expression. These results demonstrated
that four variable groups: Large tumor sizes, axillary lymph node
metastasis, distant metastasis present, and advanced clinical TNM
stage were associated with worse prognosis (Table III).
 | Table III.Univariate and multivariate overall
survival analyses in 98 patients with breast cancer. |
Table III.
Univariate and multivariate overall
survival analyses in 98 patients with breast cancer.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Prognostic
factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Agea | 1.039 | 0.371–2.911 | 0.942 |
|
|
|
| Tumor
sizeb | 3.194 | 1.103–9.253 | 0.032 |
|
|
|
| Lymph node
stagec | 3.944 | 1.429–10.884 | 0.008 |
|
|
|
| Distant
metastasisd | 6.82 | 2.369–19.632 | <0.001 | 4.421 | 1.502–13.012 | 0.007 |
| Clinical
stagee | 6.305 | 1.784–22.283 | 0.004 | 3.92 | 1.027–14.963 | 0.046 |
| ERf | 1.131 | 0.420–3.043 | 0.808 |
|
|
|
| PRg | 0.891 | 0.323–2.454 | 0.823 |
|
|
|
| Her-2h | 0.723 | 0.249–2.094 | 0.550 |
|
|
|
| Notch4i | 0.499 | 0.160–1.554 | 0.231 |
|
|
|
Association between Notch4 expression
and survival
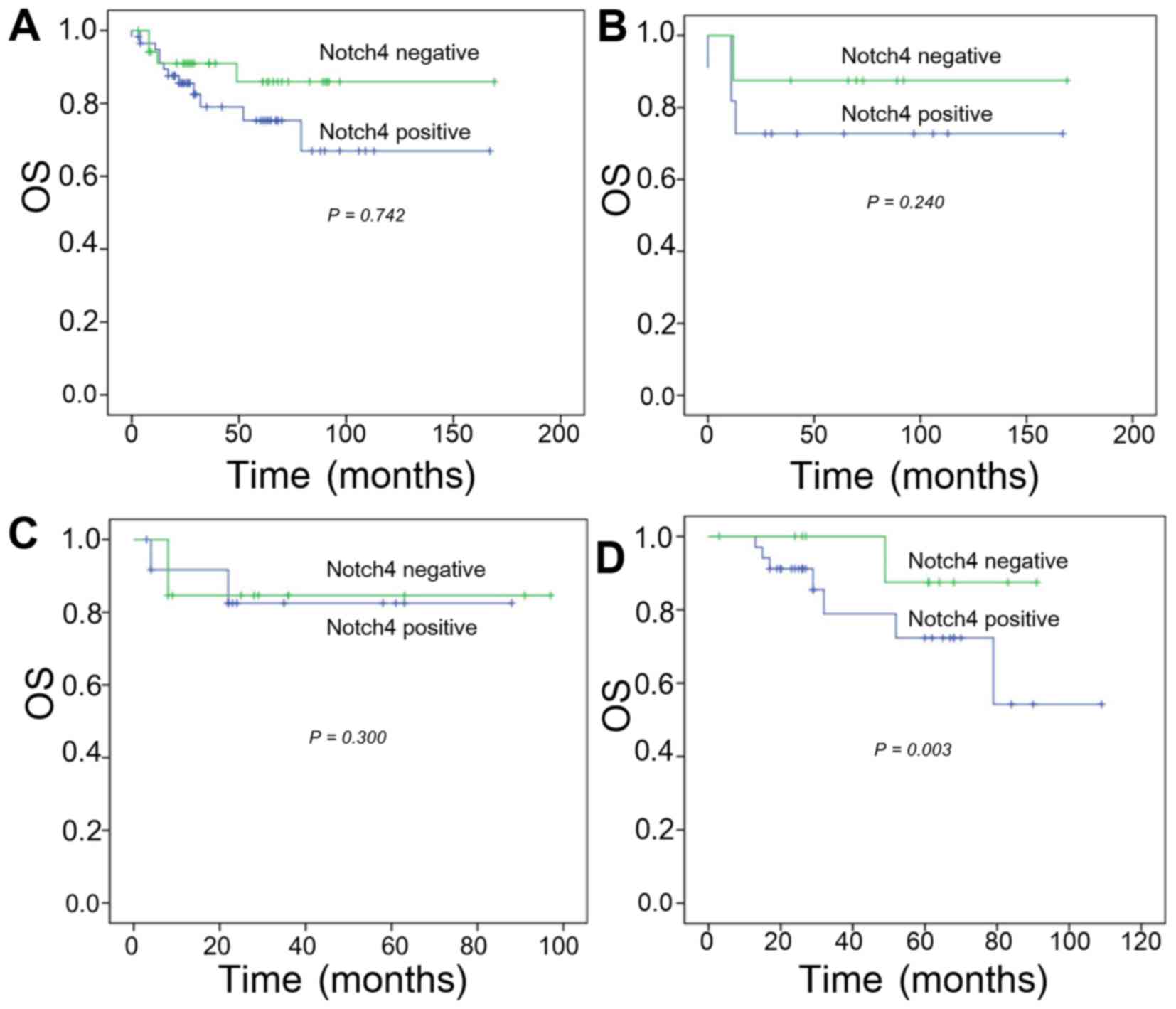
The OS rates at 2, 3, and 5 years in the high
Notch4-expressing group were 63.2, 36.8, and 31.6%, respectively.
In addition, the 2-, 3-, and 5-year survival rates for the low
Notch4 expression group were 81.8, 57.6, and 48.5%, respectively.
No significant difference was identified in the OS rates between
the high- and low-Notch4 expressing groups (P=0.742; Fig. 2A).
Categorization by tumor type also did not reveal a
significant effect of Notch4 on OS in the TNBC or
Her-2-overexpressing groups. In patients with TNBC, the 2-, 3-, and
5-year OS rates for the high Notch4 expression group were 70.0,
50.0, and 40.0%, compared with 85.7, 85.7, and 71.4% in the low
Notch4 expression group (P=0.240; Fig.
2B). In patients with Her-2-overexpressing breast cancer, the
2-, 3-, and 5-year OS rates were 45.5, 27.3 and 18.2% in the high
Notch4 expression group vs. 66.7, 25.0 and 16.7% in the low Notch4
expression group (χ2, 2.408; P=0.300; Fig. 2C).
In contrast, Notch4 expression was significantly
associated with a reduced survival rate in patients with luminal
breast cancer. The 2-, 3-, and 5-year survival rates for the high
Notch expression group were 69.7, 33.3 and 30.3%, compared with
100, 100 and 85.7% in the low Notch4 expression group. Survival was
significantly lower in the high Notch4 expression group compared
with in the low Notch4 expression group (P=0.003; Fig. 2D).
Discussion
In the present study, the expression level of Notch4
among different subtypes of breast cancer was explored, and the
association between Notch4 expression and clinicopathological
characteristics in patients with breast cancer was analyzed. The
Notch4 immunohistochemical staining results reveals that Notch4 was
located in the cytoplasm. Notably, cytoplasmic (perinuclear) Notch
staining primarily represents newly synthesized receptors; whereas
nuclear Notch staining may represent an activated receptor.
Following release into nuclei, the Notch intracellular domain is
rapidly phosphorylated, ubiquitinated and degraded, and seldom
accumulates in the nucleus (23).
Thus, the cytoplasmic expression of Notch4 detected in the present
study may represent the functional protein newly synthesized.
Numerous studies have confirmed that the expression
level of Notch receptor and its ligands in breast cancer tissue is
increased compared with in normal breast tissue (24–26). For
example, Rizzo et al (26)
demonstrated that Notch1 and Notch4 expression is low in normal
breast tissue, while invasive ductal carcinoma and invasive lobular
carcinoma exhibited 81, and 93% Notch4-positivity, respectively. In
the present study on 98 cases of breast cancer tissue,
Her-2-overexpressing breast cancer and TNBC exhibited higher Notch4
high-expression compared with luminal breast cancer, which is
consistent with a previous study (27). It was further demonstrated that Notch4
expression was inversely associated with ER and/or PR. Yao et
al (27) and Rizzo et al
(26) revealed that estrogen causes
the accumulation of uncleaved Notch4 at the cell membrane while
preventing Notch activation. Estrogen-treated ERα-positive breast
cancer cells exhibit high levels of membrane-bound Notch, and
relatively lower levels of nuclear and cytoplasmic Notch.
Furthermore, Magnifico et al (28) confirmed that Notch4 in
Her-2-overexpressing breast cancer cells is highly active. These
results supported the findings in the present study regarding the
association between Notch4 expression, and ER and Her2.
In the current series of patients, Notch4 expression
was identified to be inversely associated with ER and/or PR, and
positively associated with tumor size, lymph node involvement and
clinical TNM stage. Shawber et al (22) identified that Notch4 signaling is able
to upregulate vascular endothelial growth factor-3 and promote
cancer lymph node metastases. Yao et al (27) also demonstrated that cytoplasmic
Notch4 expression is associated with Ki67 expression, suggesting
that tumor tissues with high Notch4 expression have higher
proliferation rates.
In the survival analysis, Notch4 expression does not
exhibit prognostic significance in the Her-2 overexpression group.
In luminal breast cancers, patients with high Notch4 expression
demonstrated significantly lower OS rates compared with the low
Notch4 expression group. Rizzo et al (26) revealed that the simultaneous use of
tamoxifen and a Notch inhibitor to treat ERα-positive breast cancer
cells, inhibited cell proliferation and triggered apoptosis more
effectively in Notch4-expressing and ER-positive breast cancer
cells. They further indicated that combinations of antiestrogens
and Notch inhibitors may be effective in ERα (+) breast cancers and
that Notch signaling is also a potential therapeutic target in ERα
(−) breast cancers. The finding that Notch4 is able to predict the
prognosis of luminal type of breast cancer suggests Notch4 may
cause hormone therapy resistance, and may serve as a therapeutic
target. The limitation of the present study was the relatively
small number of cases included, and in certain instances, a shorter
follow-up period.
In conclusion, the results of the present study
demonstrated that Notch4 protein was primarily expressed in the
cytoplasm in triple-negative and Her-2-overexpressing breast
cancer. Notch4 expression was also identified to be inversely
associated with better prognostic factors, such as small tumor
size, less lymph nodes involved and positive p53 expression. In
patients with luminal breast cancer, high Notch4 expression may be
an important indicator and predict poor prognosis, but Notch4 is
not an independent prognostic factor in patients with breast
cancer. With the further understanding of its functions, the Notch4
maybe a predictor for aggressive behavior in breast cancer, and
inhibition of Notch4 signaling using Notch4 antagonists may be a
novel strategy to develop targeted therapy.
Acknowledgments
The present study was partially supported by the
Major State Basic Research Development Program (grant no.
2011CB707705); Natural Science Foundation Committee (grant nos.
31271068 and 81302331), Major International Collaborative Research
Project (grant no. 81320108015) and Guangdong Provincial Key
Laboratory on Breast Cancer Diagnosis and Treatment Research.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brenton JD, Carey LA, Ahmed AA and Caldas
C: Molecular classification and molecular forecasting of breast
cancer: Ready for clinical application? J Clin Oncol. 23:7350–7360.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Ruijter TC, Veeck J, de Hoon JP, van
Engeland M and Tjan-Heijnen VC: Characteristics of triple-negative
breast cancer. J Cancer Res Clin Oncol. 137:183–192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hernandez-Aya LF, Chavez-MacGregor M, Lei
X, Meric-Bernstam F, Buchholz TA, Hsu L, Sahin AA, Do KA, Valero V,
Hortobagyi GN, et al: Nodal status and clinical outcomes in a large
cohort of patients with triple-negative breast cancer. J Clin
Oncol. 29:2628–2634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Comen EA, Norton L and Massagué J: Breast
cancer tumor size, nodal status, and prognosis: Biology trumps
anatomy. J Clin Oncol. 29:2610–2612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cianfrocca M and Goldstein LJ: Prognostic
and predictive factors in early-stage breast cancer. Oncologist.
9:606–616. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gallahan D and Callahan R: Mammary
tumorigenesis in feral mice: Identification of a new int locus in
mouse mammary tumor virus (Czech II)-induced mammary tumors. J
Virol. 61:66–74. 1987.PubMed/NCBI
|
|
13
|
Smith GH, Gallahan D, Diella F, Jhappan C,
Merlino G and Callahan R: Constitutive expression of a truncated
INT3 gene in mouse mammary epithelium impairs differentiation and
functional development. Cell Growth Differ. 6:563–577.
1995.PubMed/NCBI
|
|
14
|
Uyttendaele H, Soriano JV, Montesano R and
Kitajewski J: Notch4 and Wnt-1 proteins function to regulate
branching morphogenesis of mammary epithelial cells in an opposing
fashion. Dev Biol. 196:204–217. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weijzen S, Rizzo P, Braid M, Vaishnav R,
Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC,
et al: Activation of Notch-1 signaling maintains the neoplastic
phenotype in human Ras-transformed cells. Nat Med. 8:979–986. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singletary SE, Allred C, Ashley P, Bassett
LW, Berry D, Bland KI, Borgen PI, Clark GM, Edge SB, Hayes DF, et
al: Staging system for breast cancer: Revisions for the 6th edition
of the AJCC cancer staging manual. Surg Clin North Am. 83:803–819.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei XL, Dou XW, Bai JW, Luo XR, Qiu SQ, Xi
DD, Huang WH, Du CW, Man K and Zhang GJ: ERα inhibits
epithelial-mesenchymal transition by suppressing Bmi1 in breast
cancer. Oncotarget. 6:21704–21717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al: American society of clinical Oncology/College of American
pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. Arch Pathol Lab Med.
131:18–43. 2007.PubMed/NCBI
|
|
21
|
Hao L, Zhang C, Qiu Y, Wang L, Luo Y, Jin
M and Zhang Y, Guo TB, Matsushima K and Zhang Y: Recombination of
CXCR4, VEGF, and MMP-9 predicting lymph node metastasis in human
breast cancer. Cancer Lett. 253:34–42. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shawber CJ, Funahashi Y, Francisco E,
Vorontchikhina M, Kitamura Y, Stowell SA, Borisenko V, Feirt N,
Podgrabinska S, Shiraishi K, et al: Notch alters VEGF
responsiveness in human and murine endothelial cells by direct
regulation of VEGFR-3 expression. J Clin Invest. 117:3369–3382.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parr C, Watkins G and Jiang WG: The
possible correlation of Notch-1 and Notch-2 with clinical outcome
and tumour clinicopathological parameters in human breast cancer.
Int J Mol Med. 14:779–786. 2004.PubMed/NCBI
|
|
25
|
Zardawi SJ, Zardawi I, McNeil CM, Millar
EK, McLeod D, Morey AL, Crea P, Murphy NC, Pinese M, Lopez-Knowles
E, et al: High Notch1 protein expression is an early event in
breast cancer development and is associated with the HER-2
molecular subtype. Histopathology. 56:286–296. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rizzo P, Miao H, D'Souza G, Osipo C, Song
LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, et al:
Cross-talk between notch and the estrogen receptor in breast cancer
suggests novel therapeutic approaches. Cancer Res. 68:5226–5235.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yao K, Rizzo P, Rajan P, Albain K, Rychlik
K, Shah S and Miele L: Notch-1 and notch-4 receptors as prognostic
markers in breast cancer. Int J Surg Pathol. 19:607–613. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Magnifico A, Albano L, Campaner S, Delia
D, Castiglioni F, Gasparini P, Sozzi G, Fontanella E, Menard S and
Tagliabue E: Tumor-initiating cells of HER2-positive carcinoma cell
lines express the highest oncoprotein levels and are sensitive to
trastuzumab. Clin Cancer Res. 15:2010–2021. 2009. View Article : Google Scholar : PubMed/NCBI
|