Introduction
Melanoma is an aggressive malignancy with the
highest mortality rate among types of skin cancer, affecting
1.5/100,000 individuals in Central and Eastern Europe and
2.3/100,000 in Germany (2012) (1).
Melanoma is characterized by marked genetic and phenotypic
heterogeneity. Activating mutations in the B-Raf proto-oncogene,
serine/threonine kinase, NRAS proto-oncogene, GTPase or KIT
protocol-oncogene receptor tyrosine kinase genes in melanoma were
identified to be important targets for treatment by specific
inhibitors, although acquired resistance compromises their efficacy
(2,3).
The search for novel molecular therapeutic targets is one of the
most challenging objectives in melanoma studies.
MicroRNAs (miRNAs), endogenous single-stranded RNA
molecules 18–24 nucleotides in length, serve a regulatory role in
numerous physiological and pathological processes (4,5).
Therefore, miRNAs have great potential as targets for the
development of novel therapeutics (6).
miR-204-5p was previously identified to be
downregulated in human melanoma compared with melanocytic nevi
(7). This observation is in agreement
with several other studies investigating the involvement of
miR-204-5p in the regulation of cell proliferation in colorectal
(8) and thyroid cancer (9), and oral squamous cell carcinoma
(10). miR-204-5p is involved in
various signaling cascades, including Signal transducer and
activator of transcription 3 (11)
and Wnt/β-catenin signaling (12).
One of the co-partners in the mediation of the effects of
miR-204-5p is considered to be miR-3065-5p, a relatively
newly-identified miRNA, for which limited functional data are
available (13). Therefore, the aim
of the present study was to determine the effects of miR-204-5p and
miR-3065-5p on the biological behavior of melanoma cells, and to
investigate the possibility of targeting matched miRNAs for the
modulation of melanoma cell function.
Materials and methods
Ethical approval
The present study was approved by the Krasnoyarsk
State Medical University Local Ethics Committee (protocol no.
70/2016, issued on June 6, 2016, Krasnoyarsk, Russia).
Tissue samples
Samples from primary melanomas (n=12; 5 males and 7
females; median age, 56 years; age range, 32–81 years) and benign
melanocytic tumors (n=9, 4 males and 5 females; median age, 36
years; age range 14–65 years) were obtained from patients from the
Krasnoyarsk Regional Oncology Center (Krasnoyarsk, Russia). Samples
were taken during the period from June 2016 to October 2016. Skin
biopsies were obtained after getting written informed consent. The
tissues were resected and immediately immersed in
1xRNAlater® stabilization solution (cat. no. AM7020;
Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at room
temperature for 1 to 2 h for transportation. Subsequently, all
samples were stored at −20°C prior to use. The diagnosis was
confirmed by certified pathologists using a light microscope (Leica
DM2500; Leica Microsystems, Germany) of the Krasnoyarsk
Pathological Anatomy Bureau. Melanoma was staged in accordance with
the American Joint Committee on Cancer 2009 guidelines (14).
Cell culture
The human melanoma BRO cell line was obtained from
the Research Institute of Fundamental and Clinical Immunology
(Novosibirsk, Russia). The melanoma SK-MEL1 cell line
(ATCC® HTB-67™) was obtained from the A.N. Sysin
Research Institute of Human Ecology and Environmental Health at the
Ministry of Health of the Russian Federation (Moscow, Russia). The
cells were cultured in RPMI-1640 with L-glutamine (Gibco; Thermo
Fisher Scientific, Inc.) and 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2 in a
CO2-incubator (Sanyo MSO-5AC; Sanyo Electric Co., Ltd.,
Osaka, Japan).
RNA isolation
For total RNA isolation, tissue samples were
homogenized with liquid nitrogen and then suspended in 200 µl
digestion buffer of the RecoverAll™ Total Nucleic Acid
Isolation kit (cat. no. AM1975; Ambion; Thermo Fisher Scientific,
Inc.). RNA was isolated according to the manufacturer's protocol,
and then eluted with 50 µl nuclease-free water. miRNA concentration
was estimated by a Qubit® 2.0 fluorimeter (Invitrogen;
Thermo Fisher Scientific, Inc.) with the use of Qubit®
microRNA Assay kit (ref. Q32880; Thermo Fisher Scientific,
Inc.).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
miRNA expression assay
RT-qPCR was performed using the TaqMan™
miRNA Assay (cat. no. 4427975; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The reaction mixture for RT, which consisted of
4 µl RNA from each sample, 0.4 µl 5X primers from the
TaqMan™ miRNA assay, 2 µl 50X OT buffer, 1 µl DTT, 1 µl
dNTPs mix and 0.5 µl revertase from the Moloney Murine Leukemia
Virus (MMLV) RT kit (cat. no. SK021, Evrogen, Moscow, Russia), was
incubated at 37°C for 30 min. Subsequently, 2 µl cDNA was added to
the PCR cocktail containing 8 µl 2.5-fold RT-PCR reaction mixture
with ROX reference dye (Syntol, Moscow, Russia), 1 µl 20X
primers/probe mix from the TaqMan™ miRNA Assay and 9 µl
de-ionized water. U6 small nuclear RNA and RNU6B were used as
endogenous controls (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The reaction was performed on a Step One™
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the following temperature cycling protocol: Preheating
at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles:
Denaturation step at 95°C for 15 sec and annealing and elongations
step at 60°C for 1 min with 6-carboxyfluorescein/ROX detection.
Data were analyzed using the ΔΔCq method, as previously described
(15).
Target gene expression assay
To estimate the expression levels of target genes,
RT of total RNA was performed using the MMLV RT kit (Evrogen) in a
mixture containing 4 µl RNA sample, 0.5 µl random primers, 2 µl 50X
OT buffer, 1 µl DTT, 1 µl dNTPs mix and 0.5 µl revertase. The 2 µl
cDNA was used for PCR with primers specific to the genes under
investigation (cat. no. 4331182, Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the aforementioned cycling protocol
for the miRNA expression assay.
Cell transfection and efficiency of
detection
The BRO and SK-MEL1 cells were transfected 24 h
prior to starting the subsequent experiments with specific
anti-miR™ miRNA inhibitors for miR-204-5p (mature miRNA
sequence: UUCCCUUUGUCAUCCUAUGCCU) and miR-3065-5p (mature miRNA
sequence: UCAACAAAAUCACUGAUGCUGGA) (both 5 nmol lyophilized pellet,
Assay IDs, AM11116 and AM18328, respectively; cat. no. AM17000;
Ambion; Thermo Fisher Scientific, Inc.) and with
mirVana® miRNA mimics in an additional experimental
series (5 nmol lyophilized pellet, Assay IDs, MC11116 and MC18328,
respectively; cat. no. 4464066, Ambion; Thermo Fisher Scientific,
Inc.). When cells reached a final concentration of
1–6×105 cells/ml, transfection experiments were
performed using 1.5 µl Lipofectamine® RNAiMAX
Transfection Reagent (cat. no. 13778150; Invitrogen; Thermo Fisher
Scientific, Inc., USA)/500 µl cells. Anti-miR or mimic solutions
were added to the cells containing culture medium RPMI-1640 with
L-glutamine (Gibco; Thermo Fisher Scientific, Inc.) to a final
concentration of 25 nM. The medium was replaced every 24 h after
transfection. To estimate the gain- or loss-of-function effect of
microRNA, anti-miR™ miRNA Inhibitor Negative Control #1
(5 nmol lyophilized pellet, cat. no. AM17010, Ambion; Thermo Fisher
Scientific, Inc.) and mirVana™ miRNA Mimic Negative
Control #1 (5 nmol lyophilized pellet, cat. no. 4464058, Ambion;
Thermo Fisher Scientific, Inc.) negative controls were used.
To evaluate the transfection efficiency, the effects
of anti-miR™ hsa-let-7c miRNA inhibitor (cat. no. 4392431; Ambion;
Thermo Fisher Scientific, Inc.) and hsa-miR-1 mimic (cat. no.
4464062; Ambion; Thermo Fisher Scientific, Inc.) positive controls
were measured and compared with those of the respective negative
controls. For this purpose, the cells were transfected using 1.5 µl
Lipofectamine® RNAiMAX Transfection Reagent (cat. no.
13778150; Invitrogen; Thermo Fisher Scientific, Inc.) with anti-miR
or mimic positive controls; anti-miR™ hsa-let-7c miRNA Inhibitor
Positive Control (5 nmol lyophilized pellet, cat. no. 4392431,
Ambion; Thermo Fisher Scientific, Inc.) and mirVana™ miRNA Mimic
miR-1 Positive Control (5 nmol lyophilized pellet, cat. no.
4464063, Ambion; Thermo Fisher Scientific, Inc.) and incubated at
37°C with 5% CO2 for 48 h. Subsequently, RNA was
isolated from the cells and the changes in levels of the
High-mobility group AT-hook 2. (HMGA2, Taqman® Gene
Expression Assays, cat. no 4331182, Applied Biosystems™; Thermo
Fisher Scientific, Inc.) for the inhibitors and Twinfilin
actin-binding protein 1 (TWF1, Taqman® Gene Expression
Assays, cat. no. 4331182, Applied Biosystems™; Thermo Fisher
Scientific, Inc.) for the mimics were estimated by PCR using a 2.5х
reaction mixture for PCR (cat. no. M431, Syntol, Russia).
Thermocycling conditions were as follows: Preheating at 50°C for 2
min and 95°C for 10 min, followed by 40 cycles of, denaturation at
95°C for 15 sec and annealing and elongations at 60°C for 1 min.
Data were analyzed using the ΔΔCq method (15). The expression of HMGA2 is specifically
downregulated by hsa-let-7c (16);
therefore, effective inhibition of let-7c leads to a decrease in
HMGA2 mRNA level in the cells. Similarly, TWF1 expression is
regulated by miR-1 and its effective transfection into the cells
leads to downregulation of TWF1 (17). To perform qPCR, the isolated RNA was
first converted to cDNA by RT with the Reverta kit (cat. no.
K3-1-100, AmpliSens, Moscow, Russia) and then amplified with TaqMan
primers for HMGA-2 (assay ID Hs00171569_m1; cat. no. 1287202;
Applied Biosystems; Thermo Fisher Scientific, Inc.) or for TWF1
(assay ID Hs00702289_s1; cat. no. 4331182; Applied Biosystems;
Thermo Fisher Scientific, Inc.) and endogenous control β-actin
(assay ID Hs01060665_g1; cat. no. 4331182; Applied Biosystems;
Thermo Fisher Scientific, Inc.).
Cell apoptosis detection
The melanoma SK-MEL1 and BRO cell lines were
transfected with relevant inhibitors or mimics and incubated in
24-well plates at 37°C in 5% CO2 for 48 h. The cells
were then detached with 0.25% trypsin solution in Hank's Balanced
Salt Solution (HBSS; Gibco; Thermo Fisher Scientific, Inc.), washed
twice with cold PBS and stained with the Annexin V-fluorescein
isothiocyanate/7-Aminoactinomycin D (7-AAD) kit (cat. no. IM3614,
Immunotech, Inc.; Beckman Coulter, Inc., Quebec, Canada) according
to the manufacturer's protocol. The proportion of viable (Annexin
V−/7-AAD−), early apoptotic (Annexin
V+/7-AAD−), apoptotic (Annexin
V+/7-AAD+) and necrotic (Annexin
V−/7-AAD+) cells was detected on Cytomics
FC-500 (Beckman Coulter, Inc., Brea, CA, USA) using CXP Software
(version 2.2; Beckman Coulter, Inc.). The experiment was performed
in triplicate.
Analysis of cell cycle by flow
cytometry
Cell cycle analysis was performed using propidium
iodide staining. Transfected SK-MEL1 and BRO cells were incubated
in 24-well plates at 37°C in 5% CO2 for 48 h.
Thereafter, the cells were washed twice with PBS, then treated with
RNase A (100 µg/ml) for 30 min, and stained with propidium iodide
(100 µg/ml) for 30 min at 37°C in the dark. The proportion of cells
in each phase was detected using a Cytomics FC-500 flow cytometer
(Beckman Coulter, Inc.) using CXP Software (version 2.2).
Cell viability/cell proliferation
assay
MTT assay
Melanoma cells transfected with miRNA inhibitors or
mimics were cultured at 37°C with 5% CO2 for 24 h, then
detached with 0.25% trypsin solution in HBSS (Gibco; Thermo Fisher
Scientific, UK), and placed in 96-well plates at a final
concentration of 3×104 cells/ml. An MTT assay was
performed at 24, 48, 72 and 96 h following transfection. For this
purpose, the culture medium was replaced and 10 µl MTT
solution/well was added. The cells with MTT were incubated for 4 h
at 37°C with 5% CO2. The cells were then washed, and 200
µl DMSO/well was added followed by incubation at room temperature
for 10 min. Absorbance of stained supernatants was measured with
Efos-9305 spectrophotometer (Shvabe Photosystems, Moscow, Russia)
at a wavelength of 560 nm. Cell viability/proliferation was
directly proportional to the absorbance rate. The experiment was
performed in triplicate.
Fluorescence microscopy
A total of 150 µl transfected cells at a
concentration of 5×104 cells/ml were placed in 96-well
plates and incubated at 37°C in 5% CO2. The cells were
stained using the CyQUANT® Direct Cell Proliferation
Assay (cat. no. 35011; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol, followed by addition of
NucBlue® Live reagent from Ready Probes® Cell
Viability Imaging kit (Blue/Red) (cat. no. R37610, Molecular
Probes; Thermo Fisher Scientific, Inc.). Following 30 min of
incubation at room temperature, fluorescence microscopy
(magnification, ×460) was performed using the Floid®
Cell Imaging Station (Floid Software, version no. 22809; Thermo
Fisher Scientific, Inc.). The nuclei of proliferating cells were
stained green and blue, while non-proliferating live cell nuclei
were stained dark blue.
Migration and invasion assay
Migration and invasion experiments were performed
with CytoSelect™ 24-Well Cell Migration and Invasion
assay kit (cat. no. CBA-100-C, Colorimetric Format; Cell Biolabs,
Inc., San Diego, CA, USA). The 24-well migration and invasion
plates contained polycarbonate membrane inserts with 8 µm pores
(kit component, item no. 10001). The inserts of the invasion assay
had an additional uniform layer of dried basement membrane matrix
solution on the upper membrane surface (kit component, item no.
11001). Melanoma cells transfected with miRNA mimic or inhibitor
were incubated at 37°C in 5% CO2 and then detached with
0.25% trypsin solution in HBSS (Gibco; Thermo Fisher Scientific,
Inc., UK), washed with PBS and suspended in FBS-free medium
RPMI-1640 to a final concentration of 1×105 cells/ml.
Following this, the cells were placed inside each insert, while the
lower well of the migration plate was filled by the medium
containing 10% FBS. Following a 22-h incubation at 37°C in 5%
CO2, the cells in the interior of the inserts were
mechanically removed and the rest were dissolved in the extraction
solution (kit component, item no. 11003-C) of the assay for 10 min
and transferred to a 96-well plate for optical density evaluation
at a wavelength 560 nm using the Efos-9305 spectrophotometer
(Shvabe Photosystems). Relative migration and invasion activity was
determined by the ratio of the mean absorbance in the test cells
vs. the mean absorbance in the negative control cells. The
experiments were performed in triplicate.
Colony formation analysis
The ability for colony formation was examined 24 h
following transfection of the cells with miR-204-5p and miR-3065-5p
inhibitors and mimics. Melanoma cells were detached with 0.25%
trypsin solution and placed in a 6-well culture plate containing 2
ml culture medium RPMI-1640 with L-glutamine (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS at a concentration of 103
cells/well. The cells were then incubated for 7–10 days at 37°C
with a 5% CO2 until they formed visible colonies
containing ≥50 tumor cells. The culture medium was changed every 3
days. Following the formation of the colonies, they were washed
twice with PBS, then fixed with 70% ethanol for 10 min at room
temperature, then stained with 0.05% crystal violet solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at room
temperature. The stained cells were washed four times with running
water, air-dried at room temperature and the number of colonies in
each well was counted by the naked eye. The experiment was
performed in triplicate.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis of miRNA target genes
DIANA-mirPath v.3.0 was used for the KEGG pathway
analysis (http://www.genome.jp/kegg;
01/01/2018) of miRNA signature. miRNA targets were predicted based
on DIANA-microT-coding sequence. The threshold (P≤0.05) was
calculated using the Fisher's exact test. To predict the gene
potential targets of miR-3065-5p and miR-204-5-p, three different
algorithms [TargetScan (version 7.0; http://www.targetscan.org), miRDB (version 5.0;
http://mirdb.org/miRDB) and miRTarBase (version
4.5; http://mirtarbase.mbc.nctu.edu.tw/)] were applied to
identify the validated targets of hsa-miR-3065-5p and miR-204-5-p.
Subsequently, genes associated with carcinogenesis for each of the
miRNAs were selected.
Enrichment analysis
A Gene Ontology term enrichment analysis (http://www.geneontology.org; 14/12/2017) was performed
to identify the biological processes, molecular functions and
cellular components associated with the target genes of miR-3065-5p
and miR-204-5p. The KEGG pathway enrichment analysis was used to
identify the miRNA targets associated with signaling pathways in
melanoma. P<0.05 was considered to indicate a statistically
significant difference. The PANTHER™ software
classification system (version 10.0; Protein ANalysis THrough
Evolutionary Relationships; www.pantherdb.org) was used to provide interpretation
of the biological functions of the validated targets of miR-204-5p
and miR-3065-5p.
Statistical analysis
Statistical analysis for all experiments was
performed using non-parametric Mann-Whitney U-tests for two
independent groups (negative control group and experimental group)
with Statistica 6.1 software (StatSoft, Inc., Dell, Round Rock, TX,
USA). The data are presented as the mean ± standard error of mean.
miR expression levels data in melanoma, melanocytic nevi and
melanoma cell lines were compared using Kruskal-Wallis H test.
Multiple comparisons of mean ranks were conducted for all groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-204-5p and miR-3065-5p expression
levels in melanocytic lesions and melanoma cells
miR-204-5p and miR-3065-5p expression was evaluated
in skin samples obtained from patients with melanoma and
melanocytic nevi, and in the melanoma BRO and SK-MEL1 cell lines.
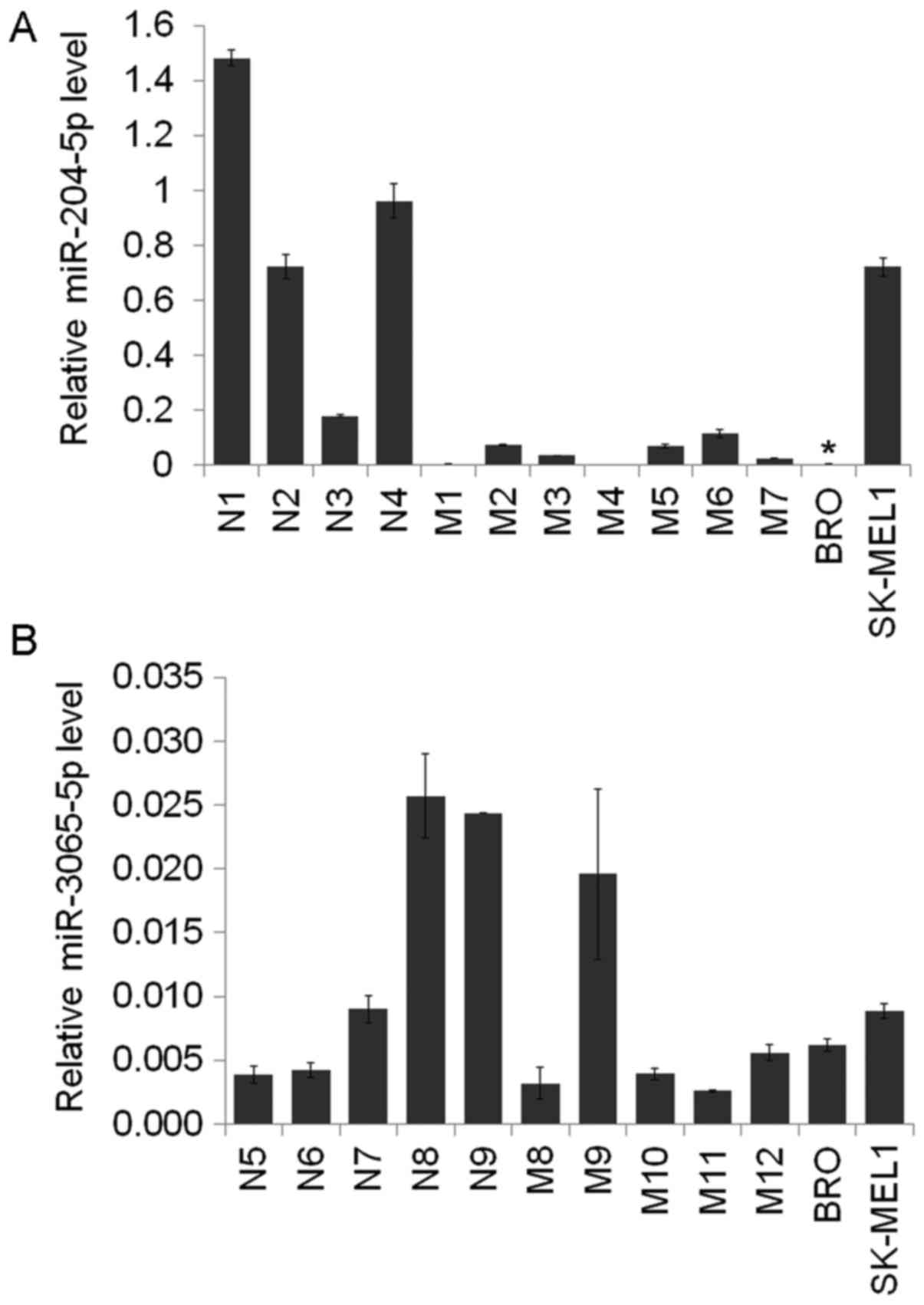
The miR-204-5p levels were identified to be decreased in the BRO
melanoma cells compared with those in the melanocytic nevi
(P=0.023, respectively; Fig. 1). The
expression level of miR-3065-5p did not differ between the groups
analyzed.
Bioinformatics analysis of miR-204-5p
and miR-3065-5p targets and associated pathways
To determine the functional role of miR-204-5p and
miR-3065-5p in melanoma cells, bioinformatics analysis was
performed using the aforementioned databases. A total of 235 target
genes were identified for miR-204-5p. Among those, 36 were selected
as being associated with carcinogenesis (Table I). Of those, the predicted targets
were: Negative apoptosis regulator B-cell lymphoma 2 (Bcl-2),
transcription factor Sex Determining Region Y-Box 4 (SOX4), and
Forkhead box C1 (FOXC1). These genes were predicted using three
databases: TargetScan, miRDB, miRTarBase. The primary signaling
pathways associated with miR-204-5p were steroid biosynthesis
(hsa00100), cytochrome P450 xenobiotic metabolism and apoptosis
(hsa04210). The 184 genes corresponding with miR-3065-5p were
detected to be associated with 48 altered biological processes,
among which 13 target genes were associated with tumor growth and
progression (Table II). One of those
is Homeodomain-interacting protein kinase 1 gene (HIPK1), which
participates in apoptosis, angiogenesis and cell proliferation
(18,19). The other miR-3065-5p gene target was
ITGA1 (integrin receptor subunit α1), which may regulate tumor
growth and angiogenesis (20).
miR-3065-5p was associated with the following signaling pathways in
accordance with DIANA-mirPath v.3.0: Ubiquitin-mediated proteolysis
(hsa04120), protein processing in the endoplasmic reticulum
(hsa04141), lysine degradation (hsa00310), transcriptional
dysregulation in cancer (hsa05202) and adherens junctions
(hsa04520).
 | Table I.microRNA-204-5p target genes and
their biological functions. |
Table I.
microRNA-204-5p target genes and
their biological functions.
| Target gene
symbol | Biological
processes involved |
|---|
| AKAP1 | Protein
binding |
| ANKRD13A | Cell migration |
| AP1S1 | Protein transporter
activity |
| AP1S2 | Protein
binding |
| ARHGAP29 | Rho protein signal
transduction |
| ATP2B1 | Calcium ion
transmembrane transport |
| Bcl-2 | Apoptosis |
| Bcl-2L2 | Negative regulation
of apoptosis |
| BDNF | Collateral
sprouting, nervous system development, synapse assembly |
| CDH2 | Cell migration,
cell adhesion, β-catenin binding, adherent junction
organization |
| CDX2 | Negative regulation
of transcription from RNA polymerase II promoter |
| COL5A3 | Collagen binding,
cell-matrix adhesion |
| CREB5 | Protein
binding |
| EDEM1 | Protein
binding |
| ELOVL6 | Fatty acid
metabolism |
| EZR | Cadherin binding
involved in cell-cell adhesion |
| FARP1 | Negative regulation
of phosphatase activity |
| FOXC1 | Cell migration,
cell proliferation, negative regulation of mitotic cycle |
| HAS2 | Hyaluronic acid
biosynthesis |
| HMX1 | Negative regulation
of transcription, DNA-templated |
| ITPR1 | Calcium ion
transport, response to hypoxia, signal transduction |
| M6PR | Transmembrane
signaling receptor activity |
| MAP1LC3B | Non-motor
microtubule binding protein |
| MEIS1 | Protein
binding |
| MEIS2 | Protein binding,
negative regulation of neuron differentiation |
| RAB22A | Protein binding,
endosome organization |
| RUNX2 | Protein
binding |
| SERINC3 | Innate immune
response, defense response to virus, L-serine transport |
| SIRT1 | Protein binding,
p53 binding, metal ion binding, keratin filament binding, cellular
response to tumor necrosis factor, necrosis factor, cellular
triglyceride homeostasis, cholesterol homeostasis, chromatin
organization, angiogenesis, UV-damage excision repair, positive
regulation of apoptotic process, regulation of mitotic cell
cycle |
| SOX4 | Negative regulation
of cell proliferation |
| TCF12 | Protein
binding |
| TCF4 | Positive regulation
of transcription, DNA-templated transcription, positive regulation
of neuron differentiation |
| TGFBR1 | Transforming growth
factor beta-activated receptor activity, angiogenesis, apoptotic
process, epithelial to mesenchymal transition, positive regulation
of cell migration, apoptotic process, epithelial to mesenchymal
transition, positive regulation of cell migration, positive
regulation of cell proliferation, transforming growth factor beta
receptor signaling pathway, protein serine/threonine kinase
activity |
| TGFBR2 | Transforming growth
factor beta-activated receptor activity |
| TRPM3 | Cation
transport |
| USP47 | Proliferation, cell
growth |
 | Table II.microRNA-3065-5p target genes and
their biological functions. |
Table II.
microRNA-3065-5p target genes and
their biological functions.
| Target gene
symbol | Biological
processes involved |
|---|
| EPT1 |
Phosphatidylethanolamine biosynthesis |
| HIPK1 | Apoptosis, positive
regulation of angiogenesis, positive regulation of cell
proliferation |
| ITGA1 | Negative regulation
of cell proliferation, negative regulation of epidermal growth
factor receptor signaling pathway, cellular extravasation,
cell-matrix adhesion, activation of MAPK activity |
| MYBL1 | Positive regulation
of transcription, DNA-templated |
| MYEF2 | DNA binding |
| NUFIP2 | RNA binding |
| PDP2 | [Pyruvate
dehydrogenase (lipomide)] phosphatase activity, metal ion
binding |
| RAB1A | Cell-cell adhesion,
positive regulation of glycoprotein metabolic process |
| SPATA13 | Guanyl-nucleotide
exchange factor activity |
| SYPL1 | Chemical synaptic
transmission |
| WASF3 | Actin binding |
| ZMYM6 | Multicellular
organism development |
| PCDH9 | Regulation of
adhesion molecules, calcium ion binding |
Effect of the application of
miR-204-5p inhibitors and mimics on melanoma cell apoptosis,
migration, invasion and colony formation
To investigate the functional role of miR-204-5p in
melanoma, its specific inhibitors or mimics were transfected in BRO
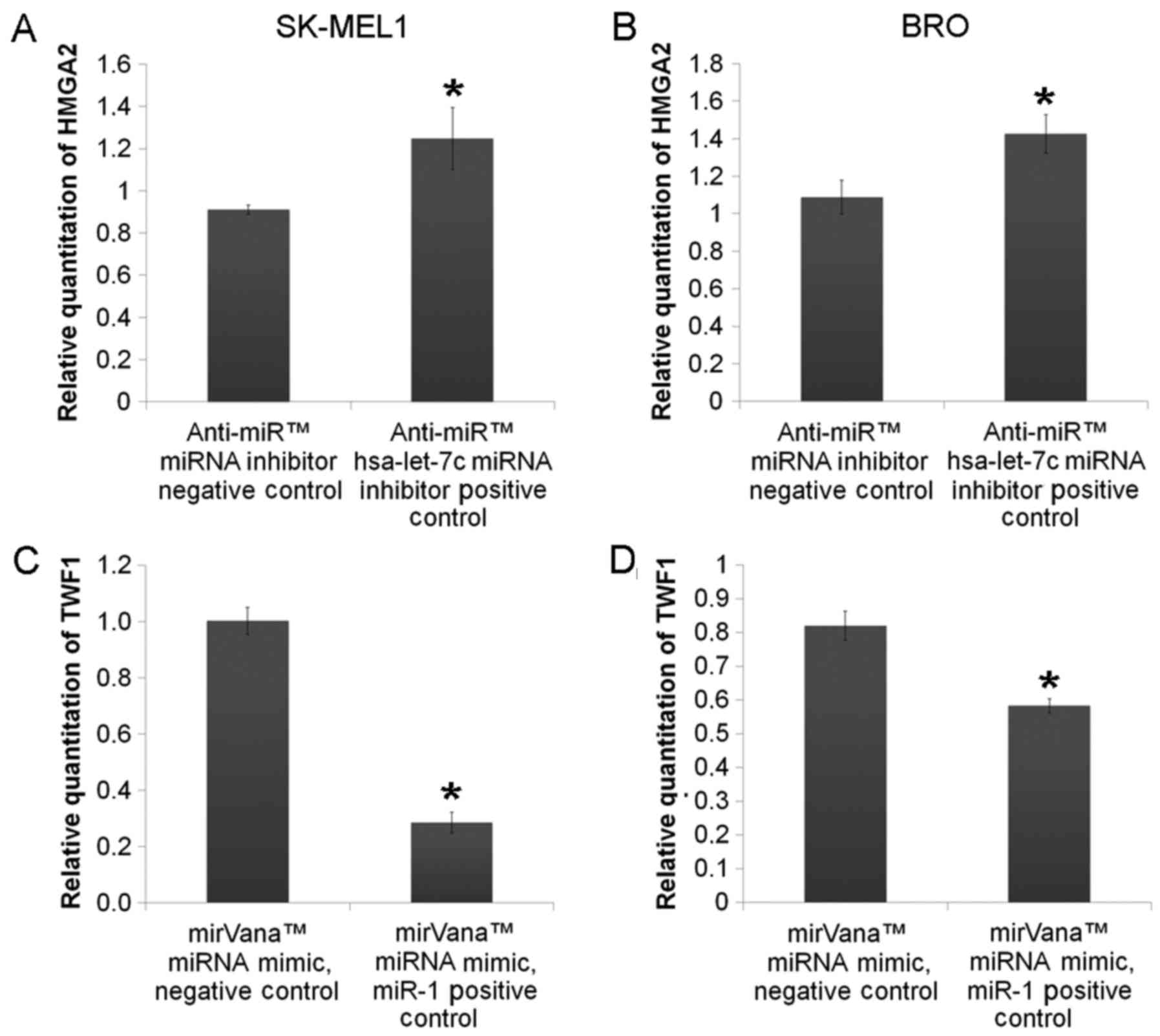
and SK-MEL1 melanoma cells. Transfection efficiency analysis
revealed that the mimics and inhibitors of the studied miRNAs
resulted in respective up- or downregulation of the target
molecules (Fig. 2). miR-204-5p
expression alterations caused by application of either inhibitors
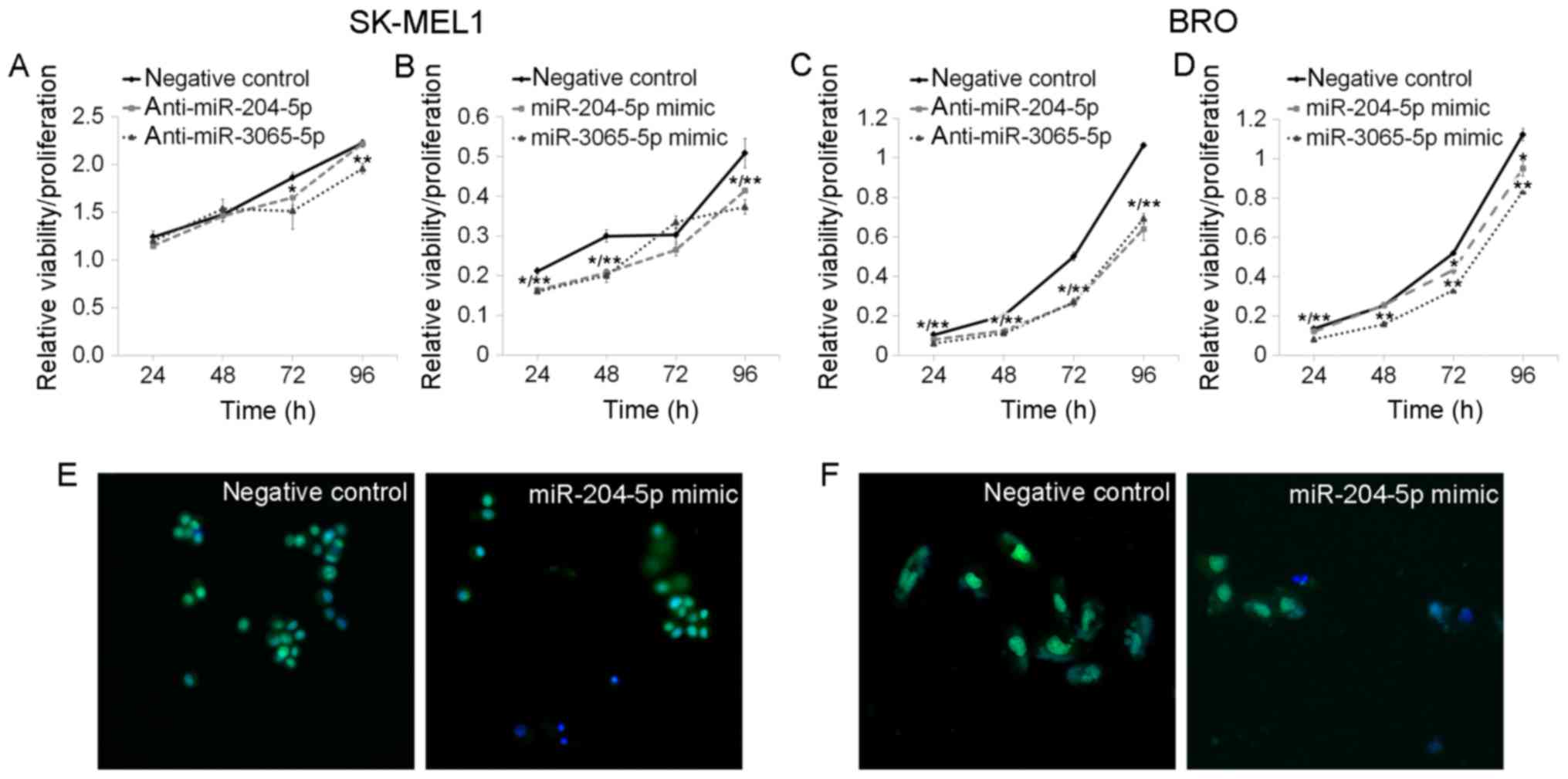
or mimics resulted in decreased cell viability/proliferation, as
measured by the MTT assay in BRO and SK-MEL1 melanoma cells
(Fig. 3). The miR-204-5p inhibitors
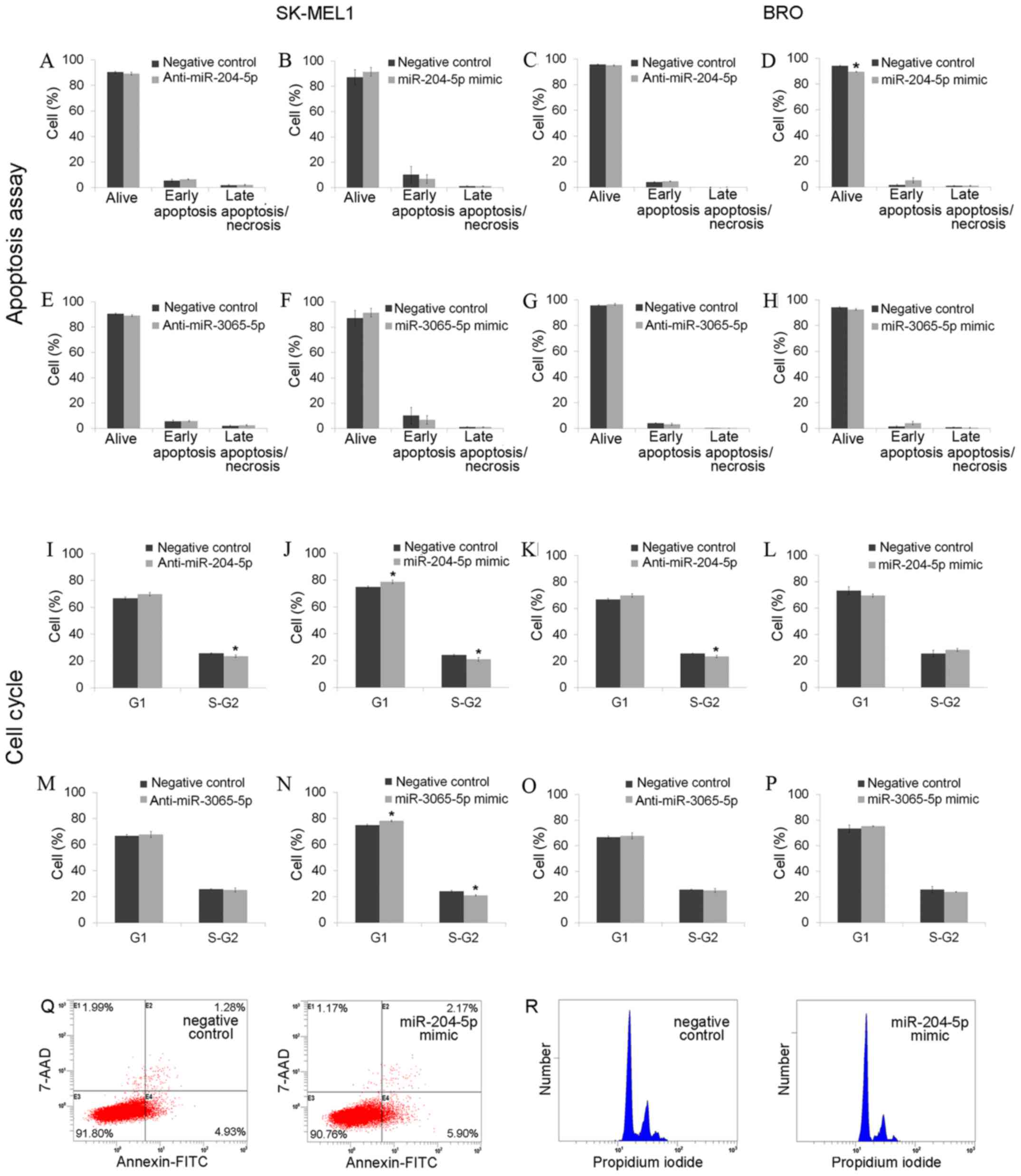
exerted no effect on the rate of BRO melanoma cell apoptosis.
Application of miR-204-5p mimics led to a decrease in the number of
surviving BRO melanoma cells, but did not affect the ratio of
apoptotic and necrotic cells (Fig.
4).
Application of either miR-204-5p inhibitors or
mimics resulted in the increase of the percentage of cells at the
G1 stage and the decrease of those at stage S-G2 among SK-MEL1
melanoma cells (Fig. 4), and
miR-204-5p inhibitor significantly decreased S-G2 stage in BRO
melanoma cells (Fig. 4). miR-204-5p
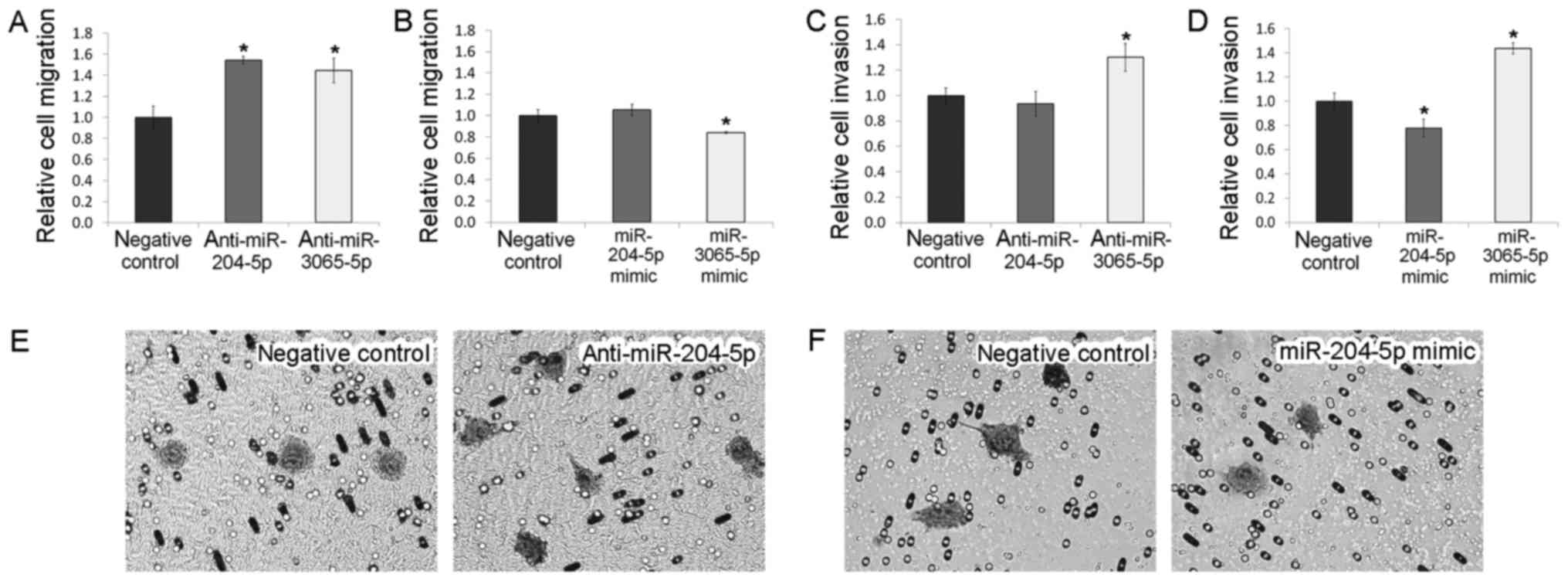
downregulation resulted in 1.54-fold increase of cell migration but
did not affect the invasion of BRO melanoma cells (Fig. 5). Transfection of the cells by mimics
did not alter these characteristics (P>0.05) but reduced their
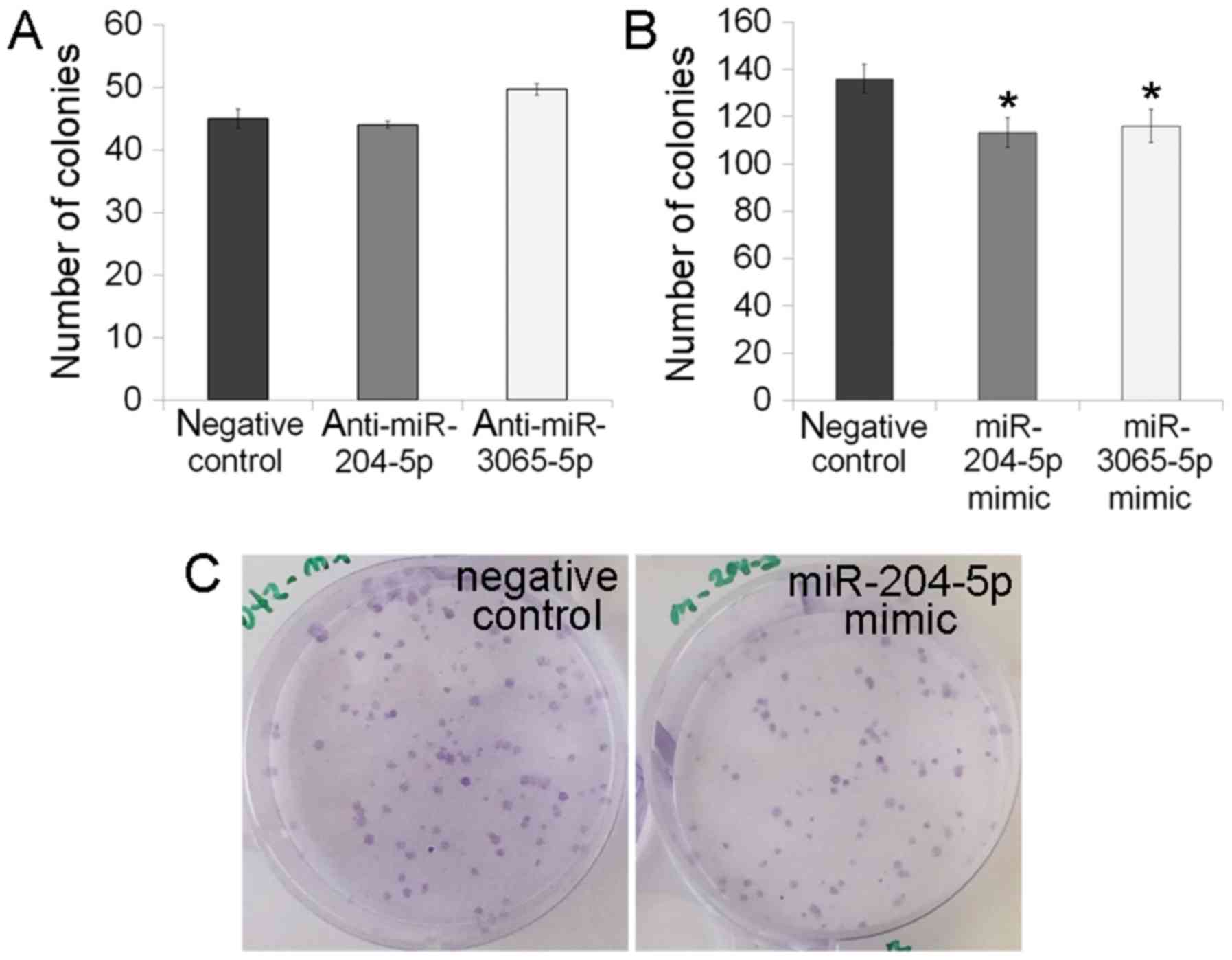
colony formation ability (Fig. 6).
Melanoma SK-MEL1 is a low-adhesion cell line, which becomes
slightly invasive in vitro (21); therefore, its migration, invasion and
colony formation were not estimated.
Effect of miR-3065-5p inhibitor and
mimic application on melanoma cell apoptosis, migration, invasion
and colony formation
miR-3065-5p expression modulation by inhibitors or
by mimics led to an apparent decrease of cell
viability/proliferation in BRO and SK-MEL1 melanoma cells (Fig. 3). Apoptosis analysis demonstrated that
all transfected cells had live and apoptotic cell ratios similar to
negative controls (P>0.05). miR-3065-5p inhibition did not
affect the cell cycle of either cell line, but miR-3065-5p mimics
reduced the number of SK-MEL1 cells at the S-G2 phase (from
24.06±0.64 to 20.95±0.57%; P=0.0495) and increased the cell
population at the G1-phase (from 74.87±0.72 to 78.21±0.54%;
P=0.0495; Fig. 4). miR-3065-5p
inhibition stimulated BRO melanoma cell migration, whereas
miR-3065-5p upregulation exerted the opposite effect (Fig. 5). It was also identified that
miR-3065-5p inhibitor or mimic application promoted invasion of BRO
melanoma cells, whereas miR-204-5p mimics induced suppression of
BRO melanoma cell invasive ability (Fig.
5). Upregulation of miR-3065-5p caused the change in the colony
number of BRO cells (Fig. 6).
Effects of miR-204-5p and miR-3065-5p
on target gene expression
To elucidate the molecular mechanisms underlying the
involvement of miR-204-5p and miR-3065-5p in melanoma cell
biological behavior, the effect of these miRNAs on the expression
of their target genes was investigated. Bcl-2, Transforming growth
factor β receptor 1 (TGFβR1) and SOX4 gene expression levels were
evaluated following performing gain- and loss-of-function
experiments for miR-204-5p, and HIPK1 and ITGA1 for miR-3065-5p.
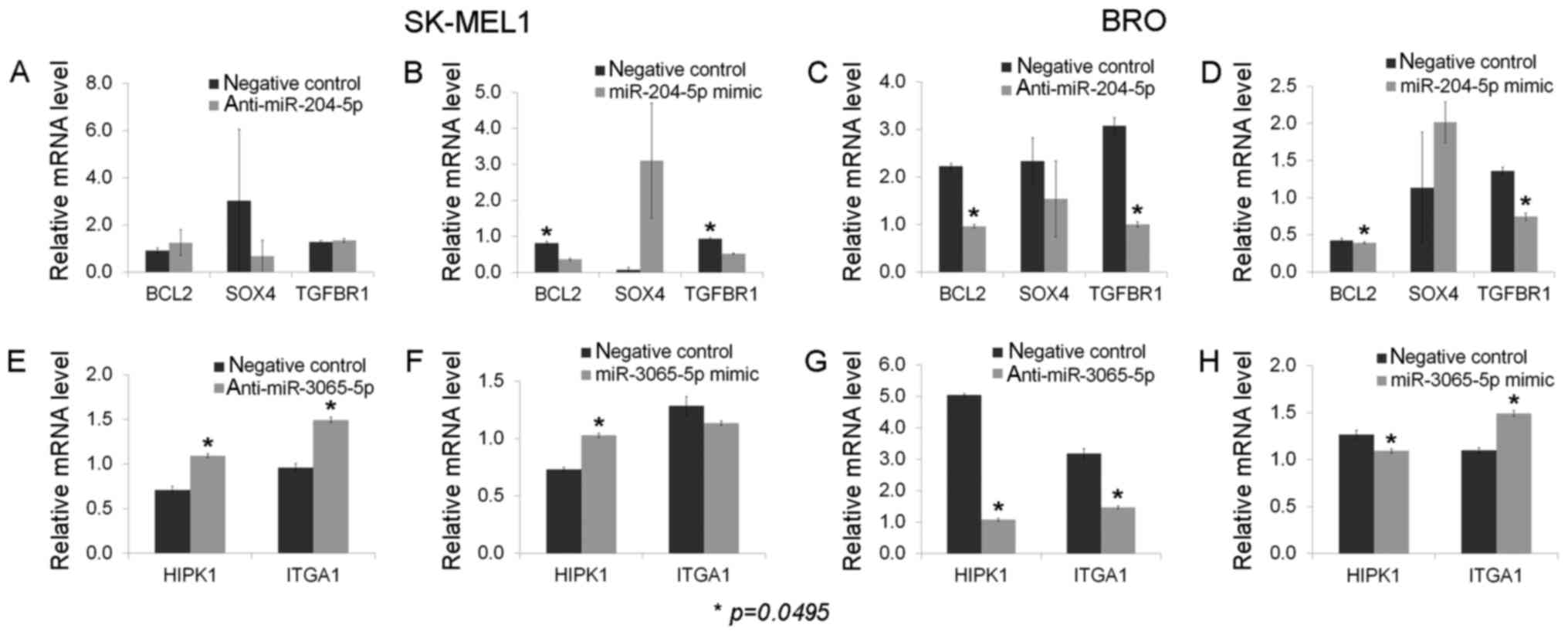
The inhibition of miR-204-5p in BRO melanoma cells was identified
to decrease the level of Bcl-2, while stimulation of miR-204-5p
exhibited no effect on Bcl-2 expression. Conversely, Bcl-2
expression was decreased in melanoma SK-MEL1 cells following
miR-204-5p mimic transfection, and remained stable following
specific miR-204-5p inhibitor application. The mRNA levels of
TGFβR1 were downregulated following the application of the
inhibitor and mimic of miR-204-5p in BRO melanoma cells, and
following miR-204-5p mimic transfection in SK-MEL1 melanoma cells.
miR-204-5p inhibition did not affect TGFβR1 expression in SK-MEL1
cells. No alterations in SOX4 expression were observed following
miR-204-5p inhibitor and mimic application in either cell line
(Fig. 7).
Melanoma cell transfection by miR-3065-5p inhibitors
and mimics led to a downregulation of HIPK1 in BRO melanoma cells
and upregulation in SK-MEL1 melanoma cells. The ITGA1 level was
downregulated by miR-3065-5p inhibition and upregulated by its
mimics in BRO melanoma cells. On the contrary, overexpression of
ITGA1 was observed following miR-3065-5p knockdown in SK-MEL1
cells, while transfection with miR-3065-5p mimics did not affect
ITGA1 expression (Fig. 7).
Discussion
miR-204-5p was identified to be downregulated in
melanoma BRO cells compared with that in melanocytic nevi. This
result is concurrent with previous studies, which demonstrated that
this miRNA functioned as a tumor suppressor and had diminished
expression in oral squamous cell carcinoma (10), colorectal cancer (8) and prostate cancer (22). The miR-204-5p mimic/inhibitor and
miR-3065-5p mimic/inhibitor application lead to a decrease of
melanoma cell viability. This observation may be explained by the
numerous targets of miRNA modulation that are presently under
study, which may ultimately trigger several signaling pathways.
More prominent cell viability inhibition in comparison with the
negative control was observed in melanoma BRO cells following
anti-miR-204-5p/anti-miR-3065-5p inhibition, as demonstrated by a
>3-fold decrease in the MTT assay results, which also allows
exclusion of unspecific results. The up- and downregulation of
miR-204-5p resulted in downregulation of the expression of its
target gene TGFβR1 in the BRO melanoma cell line, and
downregulation of TGFβR1 following miR-204-5p mimics application in
SK-MEL1 cells. TGFβR1 is a key component of TGFβ signaling, which
is implicated in several physiological and pathological processes,
including wound healing, epithelial-to-mesenchymal transition and
tumor-suppressor activities in pre-malignant melanocytes (23). TGFβ specifically interacts with its
type I receptor, promoting advanced melanoma growth and progression
(24). Therefore, the decrease in
cell proliferation observed following miR-204-5p expression level
modulation in the metastatic melanoma cell lines BRO and SK-MEL1
may be mediated by TGFβ signaling. The colony assay indicated that
the miR-204-5p and miR-3065-5p mimics reduced the colony formation,
corresponding with the cell cycle assay results. The colony forming
assay was based on the number of colonies counted, which was
possible when the number of cells was ≥50. Therefore, the cells,
depending on the type, were grown from 7–10 days up to 2–3 weeks.
Transient transfection with mimics and inhibitors was performed
using Lipofectamine® RNAiMAX. The application of a
transfection reagent allowed the sustenance of mimics or inhibitor
molecules in cells for several days, usually for 3–5 days in
accordance with the manufacturer's protocol. Within 3–5 days of the
effect of the inhibitors/mimics, the colony formation was analyzed.
Cells continued to grow when there was no effect of the
mimic/inhibitor, but if the modulators evidently affected cells
growth, a difference between control and experimental groups was
observed.
We performed similar experiments with BRO melanoma
cells following miR-106a inhibitor application, and indicated that
evaluation of the number of colonies was possible on the 6–7th day
of experimentation (25). The same
approaches were described in several previous studies examining
miRNAs (26,27).
miR-204-5p acts as a tumor suppressor by targeting
SOX4 in renal cell carcinoma (28)
and T-cell acute lymphoblastic leukemia (29). In the present study, the SOX4 mRNA
level was not altered in the miR-204-5 gain- and loss-of-function
experiments, while certain biological effects, which may be
associated with SOX4, were observed in melanoma cells. This may be
due to the repressive action of miRNA on translation, but not on
mRNA degradation (30,31). It may also be hypothesized that SOX4
was not triggered by miR-204-5p in the melanoma cells studied.
Bcl-2, which was identified as an additional miR-204-5p target
gene, is a crucial negative apoptosis regulator (32). Its downregulation following the
application of miR-204-5p inhibitors was identified in BRO cells,
and following the application of miR-204-5p mimics in SK-MEL1
cells. As apoptosis was not triggered by miRNA modulation, the
observed Bcl-2 dysregulation may be implicated in non-apoptotic
functions, including cell survival, in melanoma cells (33).
miR-3065 mimic and inhibitor application modulated
HIPK1 levels in BRO and SK-MEL1 cells, but in different ways: The
miR-3065 mimics and inhibitors upregulated HIPK1 levels in SK-MEL1
cells, but downregulated HIPK1 levels in BRO cells. HIPK1 has been
identified as one of the nuclear protein kinases, participating in
the regulation of several processes, including cell growth and
apoptosis (18,19). In colorectal cancer cell growth, the
suppressive action of HIPK1 was mediated through the p53 signaling
pathway (34). HIPK1 is considered to
possess both pro-tumorigenic and onco-suppressive properties
(34), explained by the possible
bidirectional role of this protein, depending on the oxidative
status of the cell. This may explain the oppositely directed
alterations of HIPK1 observed in BRO and SK-MEL1 cells, which may
differ in oxygen supply, due to their distinct origins: SK-MEL1
cells originated from melanoma metastases in regional lymph nodes
and are subjected to less oxidative stress compared with BRO
melanoma cells, which are derived from melanoma metastases to
visceral organs (35).
miR-3065 modulation altered the migration and
invasion levels and modulated ITGA1 gene expression in BRO melanoma
cells. This glycoprotein belongs to the integrin family and
regulates cell-cell adhesion (36);
therefore, alterations of its expression may facilitate cell
contact and migration. It was demonstrated that ITGA1 polymorphisms
are associated with a high risk of gastric cancer development and
progression (37). ITGA1 is a key
component of Ras/mitogen-activated protein kinase
kinase/extracellular signal-related kinase signaling activation,
which regulates cell proliferation in melanoma, as previously
described (38). In the present
study, ITGA1 expression was modulated by miR-3065-specific mimic
and inhibitor application in BRO melanoma cells: Anti-miR-3065-5p
led to ITGA1 downregulation, whereas miR-3065-5p mimics increased
ITGA1 expression. ITGA1 expression levels were the most clearly
modulated among the target genes evaluated in the present study,
with >2-fold downregulation following specific miR-3065-5p
inhibitor application, suggesting that this gene is the most likely
target of miR-3065-5p.
Among other miR-204-5p target genes, CDH2 and FOXC1
should also be under consideration. CDH2 is a gene that codes
N-cadherin, which is broadly expressed in melanoma cells, serving
as a promotor of melanoma cell invasion and metastasis via
triggering several signaling cascades and transcription factors
(39). N-cadherin-overexpressing
melanoma cells lose contacts with neighboring keratinocytes and
demonstrate increased adhesion and motility features (40). FOXC1 is a member of Forkhead box
family transcription factors. FOXC1 overexpression was previously
identified to activate proliferation, migration, invasion and
colony formation of melanoma cells by targeting PI3K/AKT signaling
pathway (41). Therefore, we
hypothesized that the inhibition of melanoma cell migration
following miR-204-5p inhibitor application and the decrease of the
invasion rate by melanoma cells following miR-204-5p mimic
application may be accounted for by miR targeting FOXC1, which
requires additional clarification. Our observation is in line with
an additional miR-204-5p functional study on laryngeal squamous
cell carcinoma cells, where miR-204-5p mimic and FOXC1 siRNA
application suppressed cancer cell migration and invasion (42). miR-204-5p upregulation in glioma cells
suppresses glioma cells growth, migration and invasion by
triggering RAB22A, which belongs to RAS proteins superfamily
(43). miR-204-5p was demonstrated to
be downregulated in several malignancies including renal cell
carcinoma and gastric cancer (44,45). Our
results correspond to the aforementioned data that suggest that
miR-204-5p functions as cancer suppressor in various malignancies,
including melanoma, with the targeting of several genes implicated
in carcinogenesis.
Target genes for microRNAs investigated in the
present study were selected by application of three computational
tools for microRNA target prediction and estimated functionally by
qPCR. However, the luciferase assay may have provided the correct
determination of more specific interactions between the microRNA
and its targeting site in the mRNA. The absence of luciferase assay
data is a limitation of the present study.
miR-204-5p and miR-3065-5p were previously described
as miRNAs with tumor-suppressor properties, although their effects
are exerted through triggering different signaling pathways and
target genes (13). In the present
study, their suppressor effects on cell viability and
proliferation, and migration and invasion capacity of melanoma
cells was observed, to determine whether they may be of value as an
anticancer treatment option.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Russian
Science Foundation (grant no. 14-15-00074).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
NP, AK, MA, and AM performed the research. TR
designed the study. NP, AK, MA, AS and TR analyzed the data. NP,
AK, MA and TR wrote the paper.
Ethics approval and consent to
participate
The present study was approved by the Krasnoyarsk
State Medical University Local Ethics Committee (protocol no.
70/2016, issued on June 6, 2016, Krasnoyarsk, Russia). Written
informed consent was obtained from all patients.
Consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forsea AM, Del Marmol V, de Vries E,
Bailey EE and Geller AC: Melanoma incidence and mortality in
Europe: New estimates, persistent disparities. Br J Dermatol.
167:1124–1130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carvajal RD, Antonescu CR, Wolchok JD,
Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ,
Chmielowski B, Lutzky J, et al: KIT as a therapeutic target in
metastatic melanoma. JAMA. 305:2327–2334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chakraborty R, Wieland CN and Comfere NI:
Molecular targeted therapies in metastatic melanoma.
Pharmacogenomics Pers Med. 6:49–56. 2013.
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Z: MicroRNA: A matter of life or
death. World J Biol Chem. 1:41–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Costa PM and de Lima Pedroso MC: MicroRNAs
as molecular targets for cancer therapy: On the modulation of
microRNA expression. Pharmaceuticals. 6:1195–1220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komina A, Palkina N, Aksenenko M,
Tsyrenzhapova S and Ruksha T: Antiproliferative and pro-apoptotic
effects of miR-4286 inhibition in melanoma cells. PLoS One.
11:e01682292016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Li F and Zhou X: MiR-204-5p
regulates cell proliferation and metastasis through inhibiting
CXCR4 expression in OSCC. Biomed Pharmacother. 82:202–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao W, Wang HH, Tian FJ, He XY, Qiu MT,
Wang JY, Zhang HJ, Wang LH and Wan XP: A TrkB-STAT3-miR-204-5p
regulatory circuitry controls proliferation and invasion of
endometrial carcinoma cells. Mol Cancer. 12:1552013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He H, Chen K, Wang F, Zhao L, Wan X, Wang
L and Mo Z: miR-204-5p promotes the adipogenic differentiation of
human adipose-derived mesenchymal stem cells by modulating DVL3
expression and suppressing Wnt/β-catenin signaling. Int J Mol Med.
35:1587–1595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Philipone E, Yoon AJ, Wang S, Shen J, Ko
YC, Sink JM, Rockafellow A, Shammay NA and Santella RM:
MicroRNAs-208b-3p, 204–5p, 129-2-3p and 3065-5p as predictive
markers of oral leukoplakia that progress to cancer. Am J Cancer
Res. 6:1537–1546. 2016.PubMed/NCBI
|
|
14
|
Balch CM, Gershenwald JE, Soong S,
Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG,
Ding S, et al: Final version of 2009 AJCC melanoma staging and
classification. J Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kinoshita T, Nohata N, Yoshino H, Hanazawa
T, Kikkawa N, Fujimura L, Chiyomaru T, Kawakami K, Enokida H,
Nakagawa M, et al: Tumor suppressive microRNA-375 regulates lactate
dehydrogenase B in maxillary sinus squamous cell carcinoma. Int J
Oncol. 40:185–193. 2012.PubMed/NCBI
|
|
18
|
Isono K, Nemoto K, Li Y, Takada Y, Suzuki
R, Katsuki M, Nakagawara A and Koseki H: Overlapping roles for
homeodomain-interacting protein kinases Hipk1 and Hipk2 in the
mediation of cell growth in response to morphogenetic and genotoxic
signals. Mol Cell Biol. 26:2758–2771. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Zhang R, Luo D, Park SJ, Wang Q, Kim
Y and Min W: Tumor necrosis factor alpha-induced desumoylation and
cytoplasmic translocation of homeodomain-interacting protein kinase
1 are critical for apoptosis signal-regulating kinase 1-JNK/p38
activation. J Biol Chem. 280:15061–15070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gouon V, Tucker GC, Kraus-Berthier L,
Atassi G and Kieffer N: Up-regulated expression of the beta3
integrin and the 92-kDa gelatinase in human HT-144 melanoma cell
tumors grown in nude mice. Int J Cancer. 68:650–662. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin YC, Lin JF, Tsai TF, Chou KY, Chen HE
and Hwang TI: Tumor suppressor miRNA-204-5p promotes apoptosis by
targeting BCL2 in prostate cancer cells. Asian J Surg. 40:396–406.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishimura EK, Suzuki M, Igras V, Du J,
Lonning S, Miyachi Y, Roes J, Beermann F and Fisher DE: Key roles
for transforming growth factor beta in melanocyte stem cell
maintenance. Cell Stem Cell. 6:130–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Principe DR, Doll JA, Bauer J, Jung B,
Munshi HG, Bartholin L, Pasche B, Lee C and Grippo PJ: TGF-β:
Duality of function between tumor prevention and carcinogenesis. J
Natl Cancer Inst. 106:djt3692014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Palkina NV, Komina AV, Aksenenko MB and
Ruksha TG: The pro-oncogenic effect of miR-106a microRNA inhibition
in melanoma cells in vitro. Cell Tissue Biol. 11:pp1–8. 2017.
View Article : Google Scholar
|
|
26
|
Lee H, Lee S, Bae H, Kang HS and Kim SJ:
Genome-wide identification of target genes for miR-204 and miR-211
identifies their proliferation stimulatory role in breast cancer
cells. Sci Rep. 6:252872016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mao A, Zhao Q, Zhou X, Sun C, Si J, Zhou
R, Gan L and Zhanga H: MicroRNA-449a enhances radiosensitivity by
downregulation of c-Myc in prostate cancer cells. Sci Rep.
6:273462016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu D, Pan H, Zhou Y, Zhang Z, Qu P, Zhou J
and Wang W: Upregulation of microRNA-204 inhibits cell
proliferation, migration and invasion in human renal cell carcinoma
cells by downregulating SOX4. Mol Med Rep. 12:7059–7064. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yin JJ, Liang B and Zhan XR: microRNA-204
inhibits cell proliferation in T-cell acute lymphoblastic leukemia
by down-regulating SOX4. Int J Clin Exp Pathol. 8:9189–9195.
2015.PubMed/NCBI
|
|
30
|
Hu W and Coller J: What comes first:
Translational repression or mRNA degradation? The deepening mystery
of microRNA function. Cell Res. 22:1322–1324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dalmay T: Mechanism of miRNA-mediated
repression of mRNA translation. Essays Biochem. 54:29–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Akl H, Vervloessem T, Kiviluoto S,
Bittremieux M, Parys JB, De Smedt H and Bultynck G: A dual role for
the anti-apoptotic Bcl-2 protein in cancer: Mitochondria versus
endoplasmic reticulum. Biochim Biophys Acta. 1843:2240–2252. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu C, Gong Y, Shi X, Sun Z, Niu M, Sang W,
Xu L, Zhu F, Wang Y and Xu K: Plumbagin reduces chronic lymphocytic
leukemia cell survival by downregulation of Bcl-2 but upregulation
of the Bax protein level. Oncol Rep. 36:1605–1611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rey C, Soubeyran I, Mahouche I, Pedeboscq
S, Bessede A, Ichas F, De Giorgi F and Lartigue L: HIPK1 drives p53
activation to limit colorectal cancer cell growth. Cell Cycle.
12:1879–1891. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lockshin A, Giovanella BC, De Ipolyi PD,
Williams LJ Jr, Mendoza JT, Yim SO and Stehlin JS Jr: Exceptional
lethality for nude mice of cells derived from a primary human
melanoma. Cancer Res. 45:345–350. 1985.PubMed/NCBI
|
|
36
|
Boudjadi S, Carrier JC, Groulx JF and
Beaulieu JF: Integrin α1β1 expression is controlled by c-MYC in
colorectal cancer cells. Oncogene. 35:1671–1678. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yim DH, Zhang YW, Eom SY, Moon SI, Yun HY,
Song YJ, Youn SJ, Hyun T, Park JS, Kim BS, et al: ITGA1
polymorphisms and haplotypes are associated with gastric cancer
risk in a Korean population. World J Gastroenterol. 19:5870–5876.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Boudjadi S and Beaulieu JF: MYC and
integrins interplay in colorectal cancer. Oncoscience. 3:50–51.
2016.PubMed/NCBI
|
|
39
|
Bagati A, Bianchi-Smiraglia A, Moparthy S,
Kolesnikova K, Fink EE, Lipchick BC, Kolesnikova M, Jowdy P,
Polechetti A, Mahpour A, et al: Melanoma suppressor functions of
the carcinoma oncogene FOXQ1. Cell Rep. 20:2820–2832. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Murtas D, Maxia C, Diana A, Pilloni L,
Corda C, Minerba L, Tomei S, Piras F, Ferreli C and Perra MT: Role
of epithelial-mesenchymal transition involved molecules in the
progression of cutaneous melanoma. Histochem Cell Biol.
148:639–649. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Li L, Liu S, Zhao Y, Wang L and Du
G: FOXC1 promotes melanoma by activating MST1R/PI3K/AKT.
Oncotarget. 7:84375–84387. 2016.PubMed/NCBI
|
|
42
|
Gao W, Wu Y, He X, Zhang C, Zhu M, Chen B,
Liu Q, Qu X, Li W, Wen S and Wang B: MicroRNA-204-5p inhibits
invasion and metastasis of laryngeal squamous cell carcinoma by
suppressing forkhead box C1. J Cancer. 8:2356–2368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xia Z, Liu F, Zhang J and Liu L: Decreased
expression of MiRNA-204-5p contributes to glioma progression and
promotes glioma cell growth, migration and invasion. PLoS One.
10:e01323992015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shu X, Hildebrandt MA, Gu J, Tannir NM,
Matin SF, Karam JA, Wood CG and Wu X: MicroRNA profiling in clear
cell renal cell carcinoma tissues potentially links tumorigenesis
and recurrence with obesity. Br J Cancer. 116:77–84. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang T, Liu C, Huang S, Ma Y, Fang J and
Chen Y: A downmodulated MicroRNA profiling in patients with gastric
cancer. Gastroenterol Res Pract. 2017:15269812017. View Article : Google Scholar : PubMed/NCBI
|