Introduction
Anaplastic (ATC) and poorly differentiated thyroid
cancer (PDTC) take origin from the follicular cells but during the
process of tumoral transformation, they lose progressively the
typical features of these cells, being the ATC completely
undifferentiated while PDTC showing an intermediate spectrum of
differentiation (1,2). These tumors, although rare (about 2 and
5% of thyroid cancer cases, respectively) (3), represent the most aggressive thyroid
tumors but their median survival time is rather different being of
6–12 months and 3–5 years for ATC and PDTC, respectively (4,5).
The histological diagnosis of ATC and PDTC is not
always straightforward (6) and some
controversial issues regarding the differential diagnosis among ATC
and PDTC are not yet fully resolved by pathologists (7). However, at least two or three
histological classifications to better distinguish these two types
of advanced thyroid tumors have been proposed and the one chosen by
the pathologist for the diagnosis should be indicated in the
pathological report (8,9).
Molecular alterations causative of these tumors have
been studied over the years by Sanger analysis and/or other
techniques (10,11). Very recently the genomic and
transcriptomic landscape of ATC and PDTC has been studied with a
Next Generation Sequencing (NGS) approach (12). This study demonstrated that ATC, and
to a lesser extent PDTC, are characterized by the accumulation of
several different oncogenic alterations. In particular, so far, no
genetic mutations or chromosomal rearrangements have been
demonstrated to be unique for ATC and/or PDTC but, it is worth to
note, that the majority of these studies are affected by the lack
of using common histologic criteria to define either ATC or PDTC
(13). In both cases the two most
frequently altered genes are TERT oncogene and TP53
tumor suppressor gene. However, while the prevalence of the
mutations of the two genes is very high in ATC (i.e., 70–75%) they
are much lower, especially for TP53, in PDTC (i.e., 40 and
8%, for TERT and TP53, respectively). Mutations of
other genes that impair the MAPK and PI3K-AKT pathways including
BRAF, RAS, AKT, PTEN and PIK3CA have been also
reported with a prevalence higher than 10% at least in one of the
two hystotypes (12). Other novel and
rare (i.e., prevalence <10%) gene's alterations have been also
reported in NGS studies on ATC (14,15) but
their tumorigenic driving role is still undefined.
Tumor associated macrophages (TAMs) have been
described in human cancer and their presence in tumoral tissues has
been correlated with an advanced stage of the disease and a worse
prognosis (16–18). A high density of TAM has been
previously reported also in thyroid carcinomas with a higher
density in ATC and PDTC with respect to well differentiated
papillary thyroid cancer (PTC) (19,20).
The aim of the present study was to verify if, in
the clinical practice, ATC and PDTC diagnosed by our pathologist
according to the WHO (21) and Turin
classification (6) respectively, show
a different clinical-pathological profile at diagnosis, a different
oncogenic profile, and a different pattern of macrophage
infiltration. The correlation between the somatic alterations and
the survival time of the patients has been also explored.
Materials and methods
Patients and tissue samples
Forty-two patients affected with ATC (n=21) or PDTC
(n=21) were diagnosed and followed at the Endocrine Unit of the
Department of Clinical and Experimental Medicine, University
Hospital of Pisa (Pisa, Italy), from 2006 to 2016. We analyzed the
thyroid tumoral DNA from all included patients. Thirty-one tissues
were obtained at surgery, immediately frozen in liquid nitrogen and
stored at −80°C until extraction; 4 samples derived from formalin
fixed paraffin embedded (FFPE) tissues. Seven samples were derived
from fine-needle aspiration (FNA) biopsies; FNA was performed under
echo guidance by a skilled endocrinologist and using a 23-gauge
needle attached to a 10-ml syringe. The aspirated material was used
to prepare slides for cytological analysis. The left over cells
were recovered in an appropriate buffer for DNA extraction and
stored at −80°. In 6 cases normal thyroid tissue and/or blood was
available. Clinical data were recorded in a computerized data base
and analyzed according to the presence of genetic alterations.
A signed informed consent was obtained from all
patients and the study received the approval of the Institutional
Reviewing Board.
Pathological diagnosis
Patients underwent total or subtotal thyroidectomy
at the Department of Surgery of the University of Pisa, Italy. A
pre-surgical cytological diagnosis of ATC or PDTC was available for
all patients. Hematoxylin-eosin stained sections of all patients
from the archives of the section of Pathology of the University of
Pisa were re-evaluated independently by two pathologists (C.U. and
F.B.). According to the ‘Turin proposal’ (6) PDTC have been defined as a neoplasm
derived from follicular epithelial cells with specific
characteristics, that are: i) a solid/trabecular/insular growth
pattern; ii) the absence of conventional nuclear features of PTC;
and iii) the presence of other features, such as convoluted nuclei,
mitotic activity (≥3 mitoses per 10 HPF), or necrosis. Compared to
ATC, PDTC shows a monotonous cell population and lack of
significant pleomorphism or marked atypia of the nuclei. Moreover,
necrosis is typically less prominent in PDTC, usually in focal
areas, as compared with the geographic distribution in ATC. Since
the immunophenotype is fundamental for the distinction between ATC
and PDTC, the immunohistochemistry (IHC) for keratins,
thyroglobulin (Tg) and thyroid transcription factor-1 (TTF-1) was
performed following standard procedures (22) and the diagnosis took into account the
evidence that keratins, Tg and TTF-1 are strongly expressed in
well-differentiated carcinomas but typically less intense in PDTC,
with Tg and TTF-1 that are completely lost in ATC, that retain only
focal and weak expression of keratins.
Genomic DNA and RNA extraction, PCR
amplification and sequencing
About 30–40 mg of frozen tissue and the left over
cells of FNA were used for genomic DNA extraction using the
Maxwell® 16 Tissue DNA Purification Kit (Promega
Corporation, Madison, WI, USA). Genomic DNA was eluted in 300 µl of
water. For FFPE tissues, one or two 5 µm sections were
deparaffinized and digested with proteinase K overnight. DNA
extraction was performed using the Maxwell® 16 FFPE
Tissue LEV DNA Purification Kit (Promega Corporation). DNA was
finally diluted in 50–100 µl of water. DNA concentration was
measured using an UV spectrophotometer (SmartSpec Plus
Spectrophotometer; Bio-Rad Laboratories, Inc., Hercules, CA, USA),
and 30 ng of genomic DNA was used for PCR in a final volume of 20
µl using a master mix (Kapa2G fast hs ready mix; Resnova S.r.l.,
Genzano di Roma, Italy).
Primers and PCR conditions were specially defined to
the purpose of the present study (Table
I). The amplified DNA was analyzed on 2% agarose gels and
purified using a commercial kit (Jetquick purification kit; Celbio,
Milan, Italy). Sequence analysis was performed with an automated
system employing fluorescent dye terminators (ABI Prism 3110;
PerkinElmer, Inc., Waltham, MA, USA). Hot spot positions of
NRAS, HRAS, KRAS, BRAF, AKT, PIK3CA, PTEN, TP53, and
TERT oncogenes have been sequenced in this study.
 | Table I.PCR conditions and primers sequence
used for the amplification of putative oncogenes involved in
thyroid cancer. |
Table I.
PCR conditions and primers sequence
used for the amplification of putative oncogenes involved in
thyroid cancer.
| Gene | Exons | 5′→3′ Sequence | Tm | Lenght | Gene | Exons | 5′→3′ Sequence | Tm | Lenght |
|---|
| CTNNB1 | EX3 |
TCACTGAGCTAACCCTGGCTAT | 60 | 497 | TERT | EX2 |
CTACTCCTCAGGCGACAAGG | 60 | 197 |
|
|
|
TCTCTTTTCTTCACCACAACATTT |
|
|
|
|
CAAGCAGCTCCAGAAACAGG |
|
|
| PIK3CA | EX10 |
TGCTTTTTCTGTAAATCATCTGTGA | 61 | 285 | AKT | EX1 |
TAGAGTGTGCGTGGCTCTCA | 60 | 171 |
|
|
|
TGCTGAGATCAGCCAAATTC |
|
|
|
|
CGCCACAGAGAGAAGTTGTTGA |
|
|
|
| EX20 |
TGACATTTGAGCAAAGACCTG | 60 | 387 | H-RAS | EX2 |
GTGGGTTTGCCCTTCAGAT | 60 | 325 |
|
|
|
TGTGGAATCCAGAGTGAGCTT |
|
|
|
|
CCTATCCTGGCTGTGTCCTG |
|
|
| TP53 | EX5/6 |
CACTTGTGCCCTGACTTTCA | Touch | 464 | H-RAS | EX3 |
AGAGGCTGGCTGTGTGAACT | 60 | 344 |
|
|
|
CTTAACCCCTCCTCCCAGAG | 70/55 |
|
|
|
ACATGCGCAGAGAGGACAG |
|
|
|
| EX7 |
CTTGGGCCTGTGTTATCTCC | Touch | 228 | N-RAS | EX2 |
GAAAGCTTTAAAGTACTGTAGATGTGG | 60 | 240 |
|
|
|
TGGAAGAAATCGGTAAGAGGTG | 70/55 |
|
|
|
CCGACAAGTGAGAGACAGGA |
|
|
|
| EX8/9 |
CAAGGGTGGTTGGGAGTAGA | Touch | 545 | N-RAS | EX3 |
TTGCATTCCCTGTGGTTTTT | 60 | 325 |
|
|
|
ACCAGGAGCCATTGTCTTTG | 70/55 |
|
|
|
TGGTAACCTCATTTCCCCATA |
|
|
| PTEN | ex5 |
TGCAACATTTCTAAAGTTACCTACTTG | 60 | 397 | K-RAS | EX2 |
TTAACCTTATGTGTGACATGTTCTAA | 60 | 230 |
|
|
|
AAACCCAAAATCTGTTTTCCAA |
|
|
|
|
ATCAAAGAATGGTCCTGCAC |
|
|
|
| ex6 |
GGCTACGACCCAGTTACCAT | 60 | 347 | K-RAS | EX3 |
TCAAGTCCTTTGCCCATTTT | 60 | 375 |
|
|
|
CCTGCATAAATTTCAAATGTGG |
|
|
|
|
TGCATGGCATTAGCAAAGAC |
|
|
|
| ex7 |
TCCATATTTCGTGTATATTGCTGA | 60 | 397 | ALK | EX23 |
TTCCTCCCAGTTTAAGATTTGC | 60 | 298 |
|
|
|
GCAAAACACCTGCAGATCTAA |
|
|
|
|
CATCGAGGAACTTGCTACCC |
|
|
|
| ex8 |
TGATAGTTTATTTTGTTGACTTTTTGC | 60 | 452 | EML4/ | EX23/0 |
GTGCAGTGTTTAGCATTCTTGGGG | 60 | 244 |
|
|
|
GTCAAGCAAGTTCTTCATCAGC |
|
| ALK | EX2 |
TGCCAGCAAAGCAGTAGTTG |
|
|
| BRAF | EX15 |
TTGACTCTAAGAGGAAAGATGAAGTACT | 60 | 320 | STRN/ | EX3/ |
CGGGACAGAATTGAATCAGGG | 60 | 244 |
|
|
|
AGCATCTCAGGGCAAAAAT |
|
| ALK | EX20 |
TGCCAGCAAAGCAGTAGTTG |
|
|
| TERT | Promoter |
CTGGCGTCCCTGCACCCTGG | 62 | 474 | ALK | EX27/ |
ACCCAAGAACTGCCCTGG | 60 | 233 |
|
|
|
ACGAACGTGGCCAGCGGCAG | Q-SOL |
|
| EX29 |
GAGAGACCAGGAGAGGAGGA |
|
|
Cloning of the deletions
The PCR product of the sample characterized by the
presence of heterozigous deletions has been subjected to direct
cloning by using the TA cloning Kit (TOPO®-TA cloning;
Invitrogen; Thermo Fisher Scientific, Inc.) according to the
instruction of the manufacturer.
IHC for CD68
In order to investigate the presence of macrophage
in tumoral samples, CD68 expression was evaluated by IHC on
FFPE tumor sections using mouse monoclonal anti-human antibody
(Ventana Medical Systems, Inc., Tucson, AZ, USA). Sections were
stained using the Ventana automated slide stainer (Ventana Medical
Systems, Inc.). Expression of CD68 was evaluated independently by
two of the authors (C.U. and F.B.) who were both blinded to the
clinicopathologic data. Discrepancies between the two observers
were discussed with a third pathologist (L.T.). The authors
evaluated the CD68 expression in macrophages. The samples were
evaluated as positive if staining was present. The intensity of the
staining was evaluated in 5 high power fields (×40 magnification)
and the ratio between the number of CD68 macrophages positive cells
and the number of tumoral cells of the samples (i.e., 1:1 and 1:10)
was used to create a graduated score: Low (1:100, 1:200), Medium
(1:50, 1:20), and High (1:1, 1:2, 1:3, 1:5).
Statistical analysis
The statistical analysis was performed with the
Chi-square test according to the studied variables. Survival curves
were analyzed using the Kaplan-Meier method and the statistical
significance was assessed by log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinico-pathological features of the
studied patients
The mean age at diagnosis of the 21 ATC patients (10
males, 11 females) was 65.7 years (median 67, range 45–84).
Clinico-pathological data were available in 17/21 patients.
According to the TNM classification (23), 2 patients had a stage IVA disease
(T4aN0-1M0), 4 a stage IVB (T4bN0-1M0) and 11 a stage IVC
(T1-4N0-1M1). Among the 21 patients, 4 were lost at follow-up, 2
were free of disease, 1 had persistent disease and 14 were dead for
the tumor with a median survival time of 6 months.
The mean age at diagnosis of the 21 PDTC patients (9
males, 12 females) was 60.2 years (median 58, range 41–83).
Clinico-pathological data were available in 17/21 patients;
According to the TNM classification (23), 2 patients had a stage II disease
(T2N0M0), 1 a stage III (T1-3N0-1M0), 11 a stage IVA (T4aN0-1M0)
and 1 a stage IVC (T1-4N0-1M1). For 2 patients that were not
surgically treated at our hospital, the presence of node and
distant metastases at diagnosis was not known. Among the 21
patients, 4 were lost at follow-up, 4 had persistent disease and 13
were dead for the tumor and the median survival was 47 months.
When comparing the clinical and pathological
features of the 2 groups we found that, although not significantly,
ATC patients were older than PDTC patients (median 67 vs. 58 years,
respectively). Moreover ATC patients had distant metastases at
diagnosis much more frequently than PDTC patients (stage IVC 11/17
vs. 1/17, respectively). Finally, although there was no difference
in the final outcome in terms of deaths, the median time of
survival was significantly shorter in ATC than in PDTC (median
survival time 6 vs. 47 months, respectively; P=0.0014).
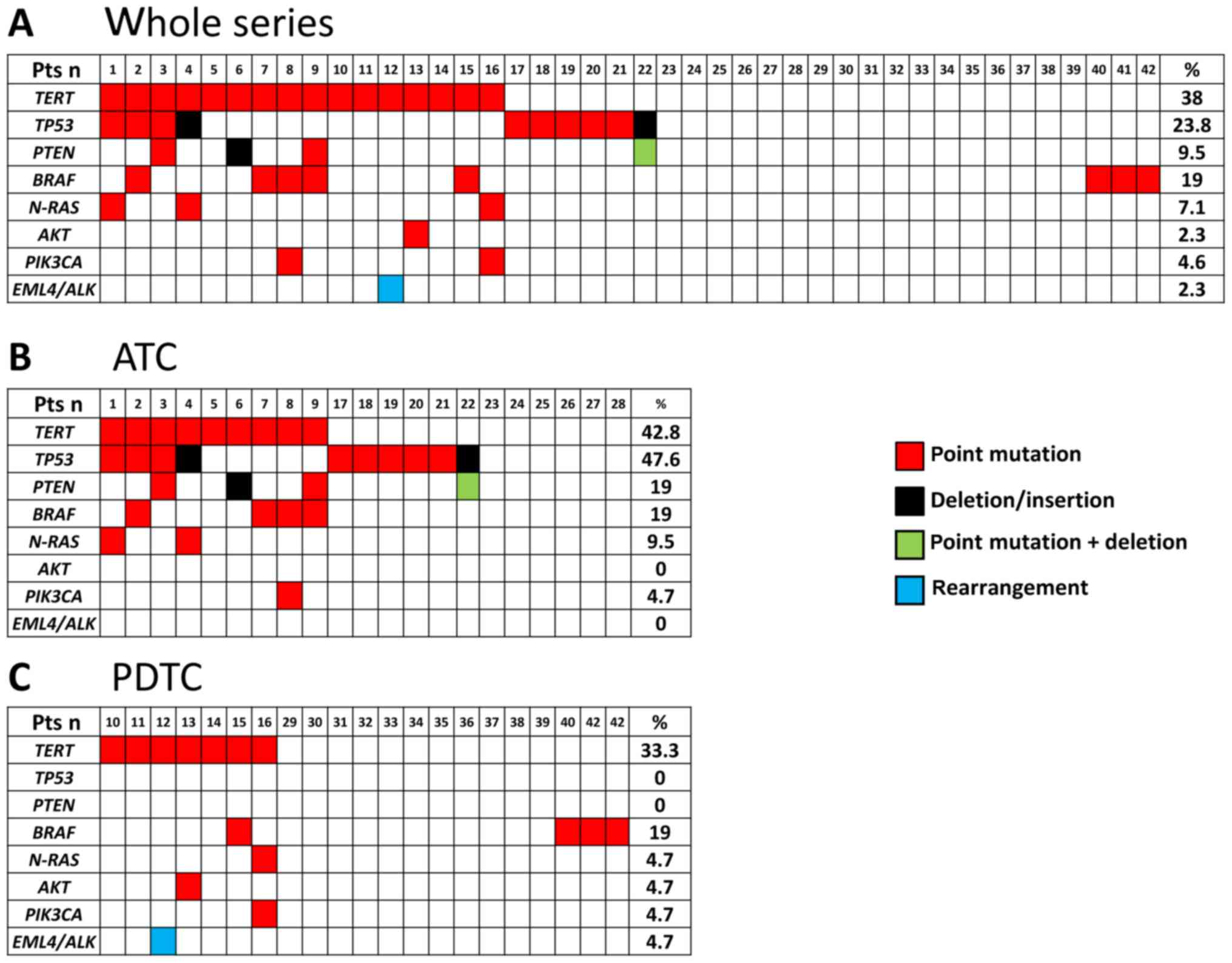
Analysis of somatic mutations
All cases, 21 ATC and 21 PDTC, were screened for
gene alterations in 10 selected genes. Overall 44 somatic
alterations (40 point mutations and 4 deletions/insertions) were
identified in the whole series. Table
II shows detailed information on the type of mutations and
their prevalence in our series.
 | Table II.Genetic alterations in our series
(n=42) of ATC and PDTC cases. |
Table II.
Genetic alterations in our series
(n=42) of ATC and PDTC cases.
| Gene | Mutation | Exon | Mutated cases in
ATC (n=21) | Mutated cases in
PDTC (n=21) | P
(χ2) |
|---|
| NRAS | Q61R | 3 | 2
(9.5%) | 1 (4.7%) | NS |
| BRAF | V600E | 15 | 4 (19%) | 4 (19%) | NS |
| AKT | E17K | 1 | 0 | 1 (4.7%) | NS |
| TP53 | C176Y | 5 | 1 (4.75%) | 0 | 0.0003 |
|
| H179R |
| 1 (4.75%) | 0 |
|
|
| c.572_574delCTC,
P191delP | 6 | 1 (4.75%) | 0 |
|
|
| M246L | 7 | 1 (4.75%) | 0 |
|
|
| R248W |
| 1 (4.75%) | 0 |
|
|
| c.702_703 del AC,
p. H233 fs*1 |
| 1 (4.75%) | 0 |
|
|
| R267W | 8 | 1 (4.75%) | 0 |
|
|
| R273C |
| 1 (4.75%) | 0 |
|
|
| R273H |
| 1 (4.75%) | 0 |
|
|
| K320N | 9 | 1 (4.75%) | 0 |
|
| PIK3CA | E545K | 10 | 1 (4.75%) | 0 | NS |
|
| H1047R | 20 | 0 | 1 (4.7%) |
|
| PTEN | c.355delG, p.
V119Lfs*15 |
|
|
|
|
|
| Q97R | 5 | 1 (4.75%) | 0 | 0.03 |
|
| D115N |
| 1 (4.75%) | 0 |
|
|
| S170I | 6 | 1 (4.75%) | 0 |
|
|
| c.741_742insA, p.
P248fs*5 | 7 | 1 (4.75%) | 0 |
|
| TERT | C250T | Promoter | 1 (4.75%) | 0 | NS |
|
| C228T |
| 7 (33%) | 9 (43%) |
|
|
| P376A | 2 | 0 | 1
(4.7%) |
|
| ALK | EML4/ALK
rearrangement | 14/20 | 0 | 1/9 (11.1%) | NS |
| n. mutations | >2 |
| 6
(28.6%) | 1
(4.7%) | 0.03 |
|
| ≤2 |
| 15 (71.4%) | 20 (95.3%) |
|
In the whole series 25/42 (59.5%) cases were found
to harbor at least 1 somatic alteration (Fig. 1A) and 17/42 (40.5%) were found to be
negative for the tested mutations. Moreover, in this series,
multiple alterations in the same sample were found in several cases
(n=13). In the ATC series (Fig. 1B) a
total of 15/21 (71.4%) cases were found to harbor at least 1
somatic alteration and 6/21 (28.6%) were found to be negative for
the tested mutations. In this group, mutations of TP53 were
the most frequently found (10/21, 47.6%). In addition to
TP53 mutations, we found mutations in TERT (9/21,
42.8%), PTEN (4/21, 19%), BRAF (4/21, 19%),
N-RAS (2/21, 9.5%), and PIK3CA (1/21, 4.7%). In the
PDTC series a total of 10/21 (47.6%) cases were found to harbor at
least 1 somatic alteration and 11/21 (52.4%) were found to be
negative for the tested mutations. Mutations were found in
TERT (7/21, 33.3%), BRAF (4/23, 19%), N-RAS
(1/23, 4.7%), AKT (1/23, 4.7%) and PIK3CA (1/23,
4.7%) (Fig. 1C). No mutations were
found in TP53 tumor suppressor gene in our PDTC.
Four ATC cases, already included in the above
mentioned mutated cases, harbored complex alterations. Two of them
were found in the PTEN tumor suppressor gene, the first in
exon 5 that was characterized by a 1 bp interstitial deletion
(c.355delG, p.V119Lfs*15; COSM28903) and a missense mutation at
codon 97 (Q97R) on the same allele; the second was in exon 7
(c.741_742insA, p.P248fs*5; COSM4986). In 2 cases we found
deletions in the TP53 tumor suppressor gene: an in frame 3
bp deletion in the exon 6 (c.572_574delCTC, P191delP, COSM111721)
and a never reported deletion in exon 7 (c.702_703 del AC, p. H233
fs*1).
When we compared the prevalence of somatic mutations
in the group of ATC and PDTC cases we found that TP53
mutations, as well as PTEN mutations, were present in ATC
but not in PDTC. Moreover, it is worth to note that complex
mutations were found only in ATC. At variance, no major differences
were observed in the prevalence of other gene alterations between
the two tumoral histotypes. Finally, the prevalence of
heterogeneity (i.e., cases with >2 mutations) was significantly
higher in the ATC group (6/21, 28.6%) than in the PDTC group (1/21,
4.7%) (P=0.03).
The presence of a germline mutation was excluded in
6/6 cases whose normal thyroid tissue and/or blood was
available.
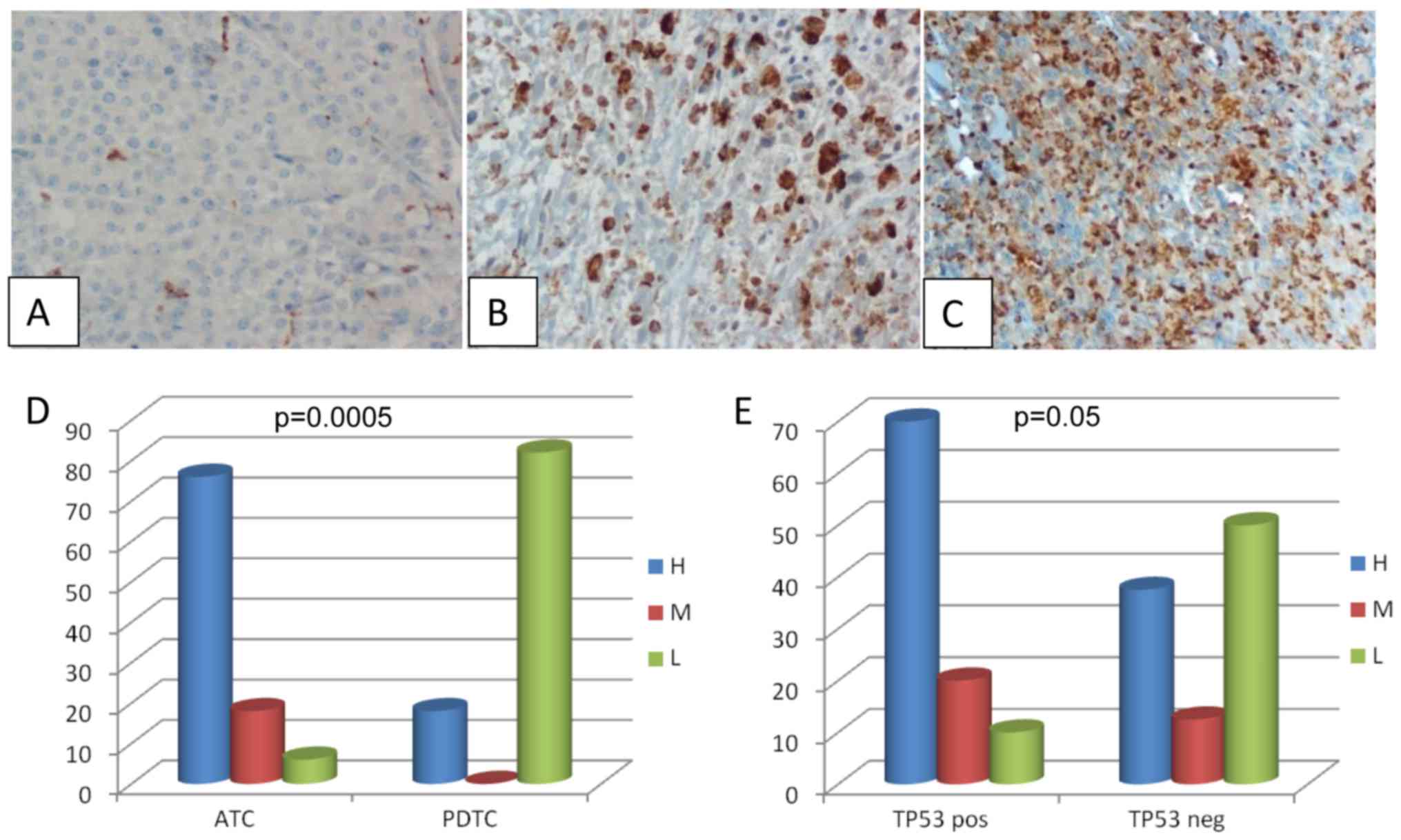
Analysis of macrophagic infiltration
in ATC and PDTC
Analysis of TAM was performed by evaluating the CD68
expression by IHC. Twenty-six cases (15 ATC and 11 PDTC) whose
paraffin embedded tissues were available, were evaluated. All
samples turned out to be positive for CD68 although at a variable
degree (Fig. 2A-C). In particular
among PDTC the CD68 staining was high in 2 cases, and low in the
remaining 9 cases; among ATC the CD68 staining was high in 11
cases, medium in 3 cases and low in 1 case. When comparing the
intensity of CD68 staining in ATC and PDTC we found that ATC were
characterized by a higher macrophagic infiltration (P=0.0005)
(Fig. 2D). Moreover, in ATC the
intensity of CD68 staining correlated with the presence of
TP53 mutations even if, although very close, this
correlation did not reach a formal statistical significance
(Fig. 2E, P=0.05).
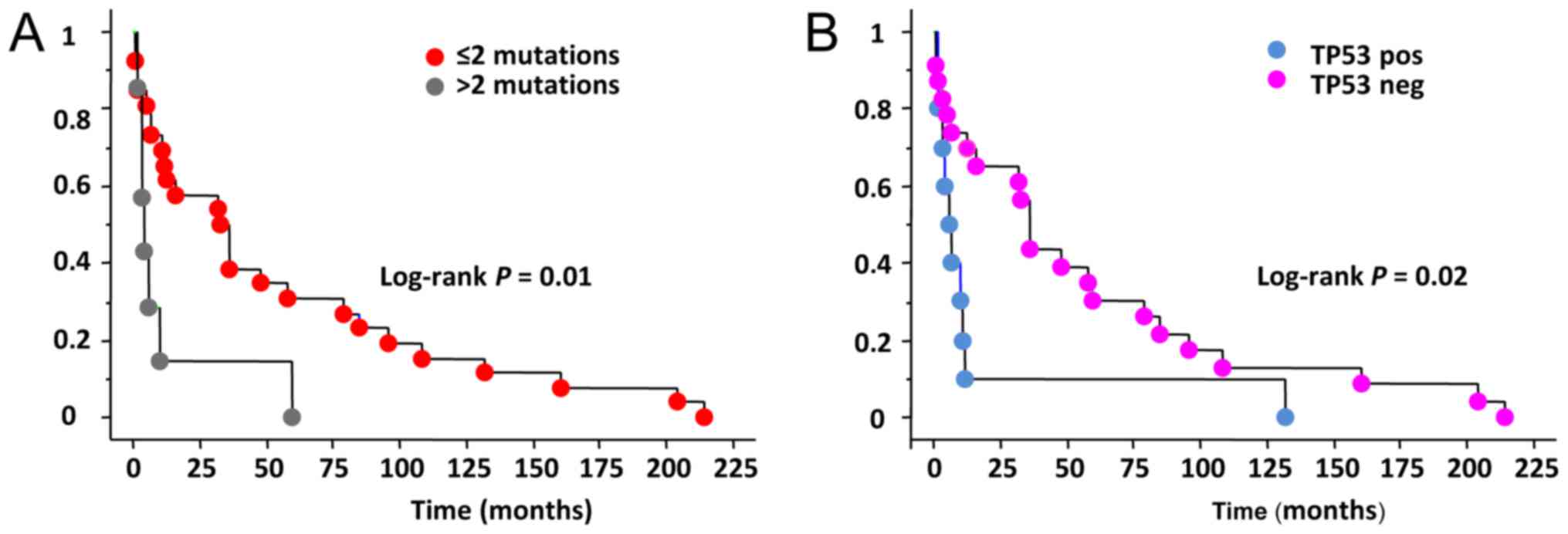
Correlation of somatic alterations
with the outcome
We correlated the presence of the somatic
alterations with the outcome of PDTC and ATC patients. When
considering the whole group, data on the outcome of patients were
available in 34 cases (17 PDTC and 17 ATC). Among them, 28 patients
were died for the tumor, 4 have persistent disease and 2 have been
cured and still alive. It is worth to note that these latter
patients were both affected by ATC (confirmed by 3 different
pathologists) but negative for all studied mutations.
In our series 19/19 (100%) patients with at least 1
somatic mutation died for the disease (ATC n=12, PDTC n=7) while in
the group of non mutated cases only 9/15 (60%) were died (ATC n=3,
PDTC n=6). The remaining non mutated cases 6/15 (40%) patients were
either cured (2/15, 13.3%) (ATC n=2) or affected by a persistent
disease (4/15, 26.7%) (PDTC n=4). When analyzing the overall
survival length we observed that in the whole series a shorter
survival was significantly correlated with the presence of more
than 2 gene mutations in the same tissue (Fig. 3A) Moreover, the absence of TP53
mutation appears to be a good prognostic factor for a longer
survival (Fig. 3B).
Discussion
ATC and PDTC are rare dedifferentiated thyroid
tumors that are supposed to derive either by pre-existing well
differentiated thyroid carcinomas (24) or to originate directly from normal
follicular thyroid cells (25). Both
of them are very aggressive but the ATC have a worse prognosis in
terms of shortness of survival (4,5) and also
in the modality of death which is by suffocation in the majority of
cases (26). The differential
diagnosis is very important at clinical level since the medical
decisions to be taken are very depending from histology mainly
because PDTC can be treated with tyrosine kinase inhibitors, such
as sorafenib and lenvatinib (27,28), while
these drugs are not yet approved for the treatment of ATC.
In the present series we found that our ATC and
PDTC, which were classified and distinguished according to the WHO
and Turin classifications, respectively, were represented by two
groups of patients with a different clinical and pathological
profile already at the time of the diagnosis. Although mean age of
patients was already >60 in both groups, the PDTC group showed a
younger age at diagnosis than ATC (median age 58 vs. 67 years).
Moreover, the stage IVC at diagnosis, the only one comparable since
ATC are all stage IV by definition, was more frequent in ATC
(11/17; 64.7%) than in PDTC (1/17; 5.9%). Finally, although the
rate of death was high in both in ATC and PDTC (i.e., 82.3 vs.
76.5%, respectively), the length of survival of the dead patients
was significantly longer in PDTC (median 47 months) than in ATC
(median 6 months). Our findings are in keeping with those reported
in the literature since it is known that PDTC, although lethal in
the majority of cases, have a slower aggressiveness than ATC and
are characterized by a lower stage at presentation and longer
follow up (2,5). In our opinion, the fact that in our
series the two groups maintain these clinical and biological
differences among them indirectly supports the truthfulness of the
histological classifications used by our pathologists.
As far as the molecular profiles of ATC and PDTC are
concerned, both traditional techniques and NGS confirmed a high
prevalence of mutations of ‘classical’ thyroid tumors related
oncogenes and/or tumor suppression genes such as TP53, TERT and
BRAF (12,14,15,29,30).
We analysed the most frequent genetic alterations involved in ATC
and PDTC tumorigenesis and in particular we analysed those
alterations that were found to be prevalent at least in more than
10% of cases (12). Also in our
series, TERT and TP53 mutations are the most frequent
in ATC histotype followed by BRAF V600E and N-RAS
mutations with a prevalence that is almost the half of that found
in the Landa et al study. A likely reason to explain this
difference can be found in the fact that the NGS method used by
Landa et al is much more sensitive than the Sanger
sequencing method used by us and since, as we also observed, ATC
tissues are reach of macrophages, their presence can further reduce
the sensitivity in detecting gene mutations. However, at variance,
the prevalence of PTEN alterations in our ATC series (19%)
was very similar to that found by Landa et al (18%) thus
raising the question of whether we had a real problem of low
sensitivity or whether our series of ATC were somehow different
from those analysed by Landa et al, although in both series
the WHO 2004 classification was used.
Regarding the PDTC gene alterations we confirmed
that also in our series TERT alterations were the most
frequent followed by BRAFV600E and N-RAS with a
prevalence that was again about half of that found by Landa et
al. A peculiar data is that we did not find any alteration of
TP53 and PTEN in our series of PDTC and this finding
is in contrast with data reported by NGS (12) and other studies (29,31,32) that
showed the presence of TP53 mutations in 8–28% of PDTC
cases. One possible explanation to justify this difference is that
the differential diagnosis between PDTC and ATC is dependent from
pathologists and type of classifications. Italian PDTC cases were
diagnosed taking into consideration the Turin classification and
Landa's PDTC cases were classified taking into consideration either
the Turin classification or their own (MSK) criteria. As shown in
the Landa's paper (12) the genetic
profile of PDTC tumor is different according to the different
system of classification and this could also justify the different
prevalence of TP53 and PTEN mutations in our
cases.
Another difference that we observed between ATC and
PDTC genetic profile is related to the presence of both a higher
number of mutated cases and a higher number of heterogeneous cases
with >2 mutations in the same tumor. This finding is in keeping
with that of Landa and colleagues that also showed that the median
number of Single Nucleotide Variants in ATC is higher than in PDTC
(12). Cases with a number of
mutation >2 showed a worse prognosis in our overall series and
the evidence that the number of cases with multiple mutations is
higher in ATC confirmed that these are much more aggressive than
PDTC. This finding is in line with the evidence that human tumors
with a higher degree of heterogeneity, such as pancreatic tumors,
are more aggressive (33,34). As far as the correlation between the
mutations and the clinical course of the disease was concerned, we
found that patients with tumors positive for TP53 mutation,
that must be underlined were all ATC, had a shorter survival time,
as observed also in other human cancer (35–37).
However, their final outcome (i.e. dead vs. alive patients) did not
show any statistically significant differences in the TP53
positive vs. negative cases. It is worth noting that the 2 cured
patients, although affected by well ascertained ATC, were both
negative for any mutation.
A clear distinction between ATC and PDTC has also
been highlighted by differences in the level of TAM infiltrating
the tumors. In fact, although all cases, both ATC and PDTC, turned
out to be positive for CD68 immunostaining, a higher percentage of
CD68 positive cells was demonstrated in ATC with respect to PDTC.
The correlation between tumor progression and presence of TAM has
been previously reported in thyroid tumors (19) and a correlation between TAM, tumor
vascularization and metastases has been demonstrated also in other
human tumors (16,38). A significant correlation of higher
levels of TAM expression has been observed in our TP53
positive cases, that were only ATC, in keeping with the observation
that mutations in this gene may support a pro-inflammatory tumor
microenvironment promoting tumor malignancy (39). To our knowledge this is the first
study demonstrating a correlation between TP53 and presence
and intensity of TAM in ATC.
We are aware of the limits of this study due to the
low number of cases and the incompleteness of some data but,
unfortunately (or fortunately) these tumors are very rare also in
referral centers like ours and the collection of the samples was in
10 years during which the strategies of therapy (i.e., surgery vs.
non surgery) and follow up (i.e., periodical active controls vs.
palliative care) of these patients have been changing and impacting
on the outcome of these patients. Only a multicentric study with
shared protocols of therapy and follow up could overcome this
problem.
Despite these considerations, according to our
results and taking into account the limit above mentioned, we can
conclude that ATC and PDTC, although both of them still lethal, are
different tumors not only because characterized by a different
clinical and pathological profile at diagnosis but also, at least
in our series, because they show a different molecular profile in
terms of type and number of mutations and also a different spectrum
of positivity for TAM. New targeted therapies for both, and in
particular for PDTC, can provide a chance of prolonged survival.
Finally, according to our results, the WHO classification for ATC
and the Turin classification for PDTC are likely those that better
distinguish the two types of very aggressive thyroid cancer.
Acknowledgements
This study was supported by the Associazione
Italiana Ricerca sul Cancro (AIRC, Investigator grant 2014, project
code 15431).
References
|
1
|
Khairy G: Anaplastic transformation of
differentiated thyroid carcinoma. Int J Health Sci (Qassim).
3:93–96. 2009.PubMed/NCBI
|
|
2
|
Yu MG, Rivera J and Jimeno C: Poorly
differentiated thyroid carcinoma: 10-year experience in a southeast
asian population. Endocrinol Metab (Seoul). 32:288–295. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Molinaro E, Romei C, Biagini A, Sabini E,
Agate L, Mazzeo S, Materazzi G, Sellari-Franceschini S, Ribechini
A, Torregrossa L, et al: Anaplastic thyroid carcinoma: From
clinicopathology to genetics and advanced therapies. Nat Rev
Endocrinol. 13:644–660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagaiah G, Hossain A, Mooney CJ,
Parmentier J and Remick SC: Anaplastic thyroid cancer: A review of
epidemiology, pathogenesis and treatment. J Oncol. 2011:5423582011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siironen P, Hagström J, Mäenpää HO,
Louhimo J, Heikkilä A, Heiskanen I, Arola J and Haglund C:
Anaplastic and poorly differentiated thyroid carcinoma: Therapeutic
strategies and treatment outcome of 52 consecutive patients.
Oncology. 79:400–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Volante M, Collini P, Nikiforov YE,
Sakamoto A, Kakudo K, Katoh R, Lloyd RV, LiVolsi VA, Papotti M,
Sobrinho-Simoes M, et al: Poorly differentiated thyroid carcinoma:
The Turin proposal for the use of uniform diagnostic criteria and
an algorithmic diagnostic approach. Am J Surg Pathol. 31:1256–1264.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Volante M, Bussolati G and Papotti M: The
story of poorly differentiated thyroid carcinoma: From Langhans'
description to the Turin proposal via Juan Rosai. Semin Diagn
Pathol. 33:277–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Volante M and Papotti M: Poorly
differentiated thyroid carcinoma: 5 years after the 2004 WHO
classification of endocrine tumours. Endocr Pathol. 21:1–6. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Asioli S, Erickson LA, Righi A, Jin L,
Volante M, Jenkins S, Papotti M, Bussolati G and Lloyd RV: Poorly
differentiated carcinoma of the thyroid: Validation of the Turin
proposal and analysis of IMP3 expression. Mod Pathol. 23:1269–1278.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smallridge RC and Copland JA: Anaplastic
thyroid carcinoma: Pathogenesis and emerging therapies. Clin Oncol
(R Coll Radiol). 22:486–497. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi X, Liu R, Qu S, Zhu G, Bishop J, Liu
X, Sun H, Shan Z, Wang E, Luo Y, et al: Association of TERT
promoter mutation 1,295,228 C>T with BRAF V600E mutation, older
patient age and distant metastasis in anaplastic thyroid cancer. J
Clin Endocrinol Metab. 100:E632–E637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Landa I, Ibrahimpasic T, Boucai L, Sinha
R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP,
Xu B, et al: Genomic and transcriptomic hallmarks of poorly
differentiated and anaplastic thyroid cancers. J Clin Invest.
126:1052–1066. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nikiforov YE: Genetic alterations involved
in the transition from well-differentiated to poorly differentiated
and anaplastic thyroid carcinomas. Endocr Pathol. 15:319–327. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kunstman JW, Juhlin CC, Goh G, Brown TC,
Stenman A, Healy JM, Rubinstein JC, Choi M, Kiss N, Nelson-Williams
C, et al: Characterization of the mutational landscape of
anaplastic thyroid cancer via whole-exome sequencing. Hum Mol
Genet. 24:2318–2329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeon MJ, Chun SM, Kim D, Kwon H, Jang EK,
Kim TY, Kim WB, Shong YK, Jang SJ, Song DE and Kim WG: Genomic
alterations of anaplastic thyroid carcinoma detected by targeted
massive parallel sequencing in a BRAF (V600E) mutation-prevalent
area. Thyroid. 26:683–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lissbrant IF, Stattin P, Wikstrom P,
Damber JE, Egevad L and Bergh A: Tumor associated macrophages in
human prostate cancer: Relation to clinicopathological variables
and survival. Int J Oncol. 17:445–451. 2000.PubMed/NCBI
|
|
17
|
Ohno S, Ohno Y, Suzuki N, Kamei T, Koike
K, Inagawa H, Kohchi C, Soma G and Inoue M: Correlation of
histological localization of tumor-associated macrophages with
clinicopathological features in endometrial cancer. Anticancer Res.
24:3335–3342. 2004.PubMed/NCBI
|
|
18
|
Hanada T, Nakagawa M, Emoto A, Nomura T,
Nasu N and Nomura Y: Prognostic value of tumor-associated
macrophage count in human bladder cancer. Int J Urol. 7:263–269.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryder M, Ghossein RA, Ricarte-Filho JC,
Knauf JA and Fagin JA: Increased density of tumor-associated
macrophages is associated with decreased survival in advanced
thyroid cancer. Endocr Relat Cancer. 15:1069–1074. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caillou B, Talbot M, Weyemi U,
Pioche-Durieu C, Al Ghuzlan A, Bidart JM, Chouaib S, Schlumberger M
and Dupuy C: Tumor-associated macrophages (TAMs) form an
interconnected cellular supportive network in anaplastic thyroid
carcinoma. PLoS One. 6:e225672011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lloyd RV OR, Klöppel G and Rosai J: WHO
Classification of Tumours of Endocrine Organs. 4th edition. WHO;
Geneva: 2017
|
|
22
|
Bejarano PA, Nikiforov YE, Swenson ES and
Biddinger PW: Thyroid transcription factor-1, thyroglobulin,
cytokeratin 7, and cytokeratin 20 in thyroid neoplasms. Appl
Immunohistochem Mol Morphol. 8:189–194. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wreesmann VB, Ghossein RA, Patel SG,
Harris CP, Schnaser EA, Shaha AR, Tuttle RM, Shah JP, Rao PH and
Singh B: Genome-wide appraisal of thyroid cancer progression. Am J
Pathol. 161:1549–1556. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
O'Neill JP and Shaha AR: Anaplastic
thyroid cancer. Oral Oncol. 49:702–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shaha AR, Ferlito A, Owen RP, Silver CE,
Rodrigo JP, Haigentz M Jr, Mendenhall WM, Rinaldo A and Smallridge
RC: Airway issues in anaplastic thyroid carcinoma. Eur Arch
Otorhinolaryngol. 270:2579–2583. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brose MS, Nutting CM, Jarzab B, Elisei R,
Siena S, Bastholt L, de la Fouchardiere C, Pacini F, Paschke R,
Shong YK, et al: Sorafenib in radioactive iodine-refractory,
locally advanced or metastatic differentiated thyroid cancer: A
randomised, double-blind, phase 3 trial. Lancet. 384:319–328. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schlumberger M, Tahara M, Wirth LJ,
Robinson B, Brose MS, Elisei R, Habra MA, Newbold K, Shah MH, Hoff
AO, et al: Lenvatinib versus placebo in radioiodine-refractory
thyroid cancer. N Engl J Med. 372:621–630. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Donghi R, Longoni A, Pilotti S, Michieli
P, Della Porta G and Pierotti MA: Gene p53 mutations are restricted
to poorly differentiated and undifferentiated carcinomas of the
thyroid gland. J Clin Invest. 91:1753–1760. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fagin JA, Matsuo K, Karmakar A, Chen DL,
Tang SH and Koeffler HP: High prevalence of mutations of the p53
gene in poorly differentiated human thyroid carcinomas. J Clin
Invest. 91:179–184. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ho YS, Tseng SC, Chin TY, Hsieh LL and Lin
JD: p53 gene mutation in thyroid carcinoma. Cancer Lett. 103:57–63.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dobashi Y, Sugimura H, Sakamoto A, Mernyei
M, Mori M, Oyama T and Machinami R: Stepwise participation of p53
gene mutation during dedifferentiation of human thyroid carcinomas.
Diagn Mol Pathol. 3:9–14. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cros J, Raffenne J, Couvelard A and Pote
N: Tumor heterogeneity in pancreatic adenocarcinoma. Pathobiology.
Aug 5–2017.(Epub ahead of print). PubMed/NCBI
|
|
34
|
Riesco-Eizaguirre G and Santisteban P:
Endocrine Tumours: Advances in the molecular pathogenesis of
thyroid cancer: Lessons from the cancer genome. Eur J Endocrinol.
175:R203–R217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Girgin C, Tarhan H, Hekimgil M, Sezer A
and Gürel G: P53 mutations and other prognostic factors of renal
cell carcinoma. Urol Int. 66:78–83. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim HW, Lee HM, Hwang SH, Ahn SG, Lee KA
and Jeong J: Patterns and biologic features of p53 mutation types
in korean breast cancer patients. J Breast Cancer. 17:1–7. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Horio Y, Takahashi T, Kuroishi T, Hibi K,
Suyama M, Niimi T, Shimokata K, Yamakawa K, Nakamura Y, Ueda R, et
al: Prognostic significance of p53 mutations and 3p deletions in
primary resected non-small cell lung cancer. Cancer Res. 53:1–4.
1993.PubMed/NCBI
|
|
38
|
Bolat F, Kayaselcuk F, Nursal TZ,
Yagmurdur MC, Bal N and Demirhan B: Microvessel density, VEGF
expression and tumor-associated macrophages in breast tumors:
Correlations with prognostic parameters. J Exp Clin Cancer Res.
25:365–372. 2006.PubMed/NCBI
|
|
39
|
Ubertini V, Norelli G, D'Arcangelo D,
Gurtner A, Cesareo E, Baldari S, Gentileschi MP, Piaggio G, Nistico
P, Soddu S, et al: Mutant p53 gains new function in promoting
inflammatory signals by repression of the secreted interleukin-1
receptor antagonist. Oncogene. 34:2493–2504. 2015. View Article : Google Scholar : PubMed/NCBI
|