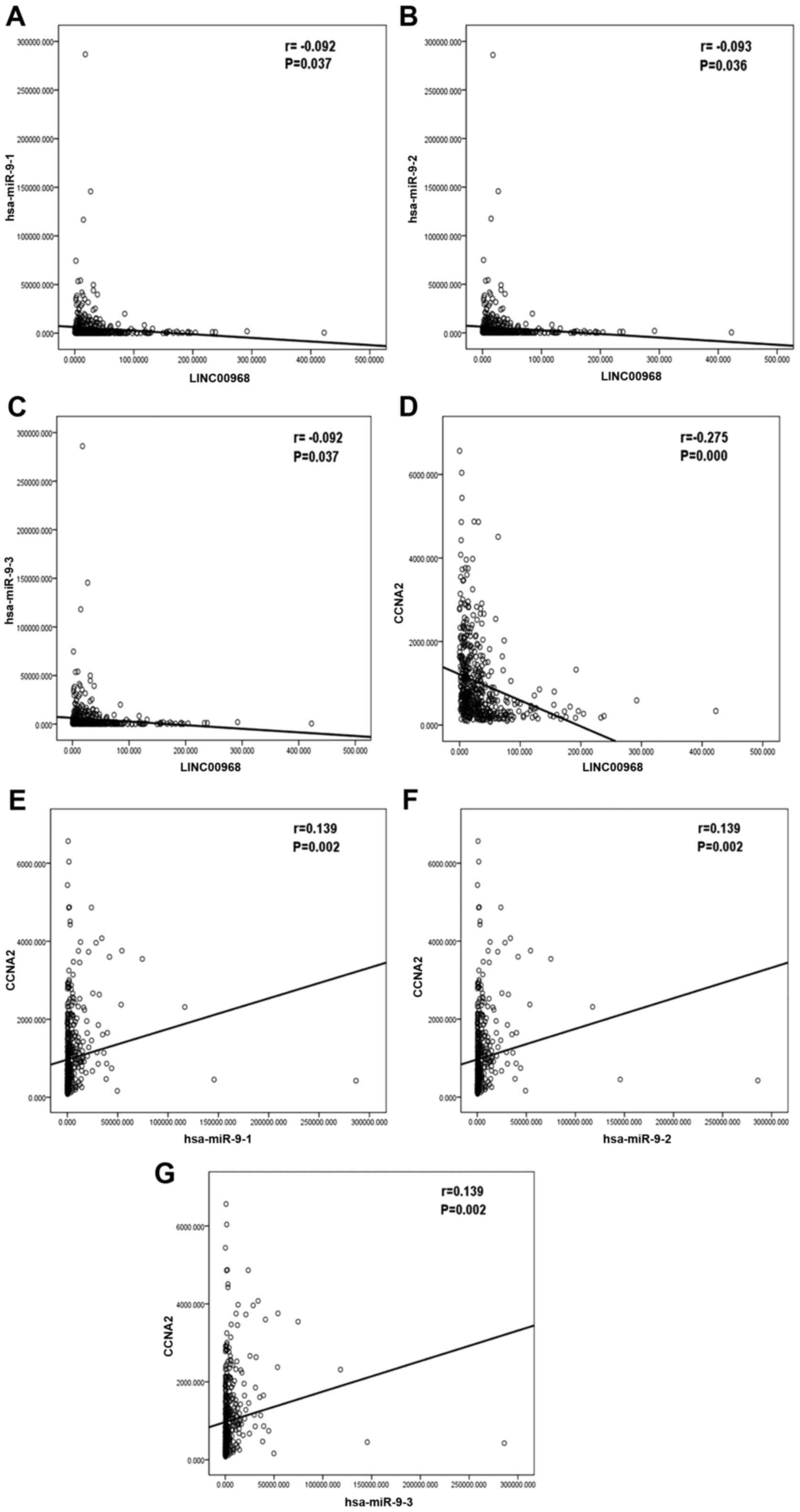
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD: The 2015 WHO classification of
lung tumors. Pathologe. 35 Suppl 2:S1882014. View Article : Google Scholar
|
|
3
|
Yang R, Li P, Zhang G, Lu C, Wang H and
Zhao G: Long non-coding RNA XLOC_008466 functions as an oncogene in
human non-small cell lung cancer by targeting miR-874. Cell Physiol
Biochem. 42:126–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Lett. 344:20–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wan L, Sun M, Liu GJ, Wei CC, Zhang EB,
Kong R, Xu TP, Huang MD and Wang ZX: Long noncoding RNA PVT1
promotes non-small cell lung cancer cell proliferation through
epigenetically regulating LATS2 expression. Mol Cancer Ther.
15:1082–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arima C, Kajino T, Tamada Y, Imoto S,
Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T,
et al: Lung adenocarcinoma subtypes definable by lung
development-related miRNA expression profiles in association with
clinicopathologic features. Carcinogenesis. 35:2224–2231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fujita Y, Yagishita S, Hagiwara K,
Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H,
Tamura T, et al: The clinical relevance of the
miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant
non-small-cell lung cancer. Mol Ther. 23:717–727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Liu J and Wang W: Construction and
investigation of breast-cancer-specific ceRNA network based on the
mRNA and miRNA expression data. IET Syst Biol. 8:96–103. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
16
|
Yang J, Lin J, Liu T, Chen T, Pan S, Huang
W and Li S: Analysis of lncRNA expression profiles in non-small
cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer.
85:110–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu X, Wang X, Fu B, Meng L and Lang B:
Differentially expressed genes and microRNAs in bladder carcinoma
cell line 5637 and T24 detected by RNA sequencing. Int J Clin Exp
Pathol. 8:12678–12687. 2015.PubMed/NCBI
|
|
18
|
Li Q, Ge X, Xu X, Zhong Y and Qie Z:
Comparison of the gene expression profiles between gallstones and
gallbladder polyps. Int J Clin Exp Pathol. 7:8016–8023.
2014.PubMed/NCBI
|
|
19
|
Jiang CM, Wang XH, Shu J, Yang WX, Fu P,
Zhuang LL and Zhou GP: Analysis of differentially expressed genes
based on microarray data of glioma. Int J Clin Exp Med.
8:17321–17332. 2015.PubMed/NCBI
|
|
20
|
Chen L, Zhuo D, Chen J and Yuan H:
Screening feature genes of lung carcinoma with DNA microarray
analysis. Int J Clin Exp Med. 8:12161–12171. 2015.PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
R Core Team: R: A language and environment
for statistical computing, R foundation for statistical computing.
Vienna, Austria: 2013, https://www.R-project.org
|
|
24
|
Naemura M, Murasaki C, Inoue Y, Okamoto H
and Kotake Y: Long noncoding RNA ANRIL regulates proliferation of
non-small cell lung cancer and cervical cancer cells. Anticancer
Res. 35:5377–5382. 2015.PubMed/NCBI
|
|
25
|
Liu YY, Chen ZH, Peng JJ, Wu JL, Yuan YJ,
Zhai ET, Cai SR, He YL and Song W: Up-regulation of long non-coding
RNA XLOC_010235 regulates epithelial-to-mesenchymal transition to
promote metastasis by associating with Snail1 in gastric cancer.
Sci Rep. 7:24612017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng Q, Ren M, Li Y and Song X:
LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One.
11:e01648452016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao S, Wang Y, Li J, Lv M, Niu H and Tian
Y: Tumor-suppressive function of long noncoding RNA MALAT1 in
glioma cells by suppressing miR-155 expression and activating FBXW7
function. Am J Cancer Res. 6:2561–2574. 2016.PubMed/NCBI
|
|
29
|
Chou J, Wang B, Zheng T, Li X, Zheng L, Hu
J, Zhang Y, Xing Y and Xi T: MALAT1 induced migration and invasion
of human breast cancer cells by competitively binding miR-1 with
cdc42. Biochem Biophys Res Commun. 472:262–269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang G, Wu X, Li S, Xu X, Zhu H and Chen
X: The long noncoding RNA CASC2 functions as a competing endogenous
RNA by sponging miR-18a in colorectal cancer. Sci Rep. 6:265242016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang CG, Yin DD, Sun SY and Han L: The
use of lncRNA analysis for stratification management of prognostic
risk in patients with NSCLC. Eur Rev Med Pharmacol Sci. 21:115–119.
2017.PubMed/NCBI
|
|
32
|
Xue Y, Ni T, Jiang Y and Li Y: Long
noncoding RNA GAS5 inhibits tumorigenesis and enhances
radiosensitivity by suppressing miR-135b expression in non-small
cell lung cancer. Oncol Res. 25:1305–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Mu Y, Cui H, Liang Y and Su X:
MiR-9-3p augments apoptosis induced by H2O2 through down regulation
of Herpud1 in glioma. PLoS One. 12:e01748392017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Higashi T, Hayashi H, Ishimoto T, Takeyama
H, Kaida T, Arima K, Taki K, Sakamoto K, Kuroki H, Okabe H, et al:
miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in
hepatocellular carcinoma cells. Br J Cancer. 113:252–258. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meng Q, Xiang L, Fu J, Chu X, Wang C and
Yan B: Transcriptome profiling reveals miR-9-3p as a novel tumor
suppressor in gastric cancer. Oncotarget. 8:37321–37331. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuva-Aydemir Y, Simkin A, Gascon E and Gao
FB: MicroRNA-9: Functional evolution of a conserved small
regulatory RNA. RNA Biol. 8:557–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Zhu L, Ma Z, Sun G, Luo X, Li M,
Zhai S, Li P and Wang X: Oncogenic miR-9 is a target of erlotinib
in NSCLCs. Sci Rep. 5:170312015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mitra R, Edmonds MD, Sun J, Zhao M, Yu H,
Eischen CM and Zhao Z: Reproducible combinatorial regulatory
networks elucidate novel oncogenic microRNAs in non-small cell lung
cancer. RNA. 20:1356–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Illiano M, Nigro E, Sapio L, Caiafa I,
Spina A, Scudiero O, Bianco A, Esposito S, Mazzeo F, Pedone PV, et
al: Adiponectin down-regulates CREB and inhibits proliferation of
A549 lung cancer cells. Pulm Pharmacol Ther. 45:114–120. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo XF, Zhu XF, Cao HY, Zhong GS, Li L,
Deng BG, Chen P, Wang PZ, Miao QF and Zhen YS: A bispecific
enediyne-energized fusion protein targeting both epidermal growth
factor receptor and insulin-like growth factor 1 receptor showing
enhanced antitumor efficacy against non-small cell lung cancer.
Oncotarget. 8:27286–27299. 2017.PubMed/NCBI
|
|
41
|
Troncone M, Cargnelli SM, Villani LA,
Isfahanian N, Broadfield LA, Zychla L, Wright J, Pond G, Steinberg
GR and Tsakiridis T: Targeting metabolism and AMP-activated kinase
with metformin to sensitize non-small cell lung cancer (NSCLC) to
cytotoxic therapy: Translational biology and rationale for current
clinical trials. Oncotarget. 8:57733–57754. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yim NH, Hwang YH, Liang C and Ma JY: A
platycoside-rich fraction from the root of Platycodon grandiflorum
enhances cell death in A549 human lung carcinoma cells via mainly
AMPK/mTOR/AKT signal-mediated autophagy induction. J
Ethnopharmacol. 194:1060–1068. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Li ZY, Hou XX, Wang X, Luo YH,
Ying YP and Chen G: Clinical significance and effect of AEG-1 on
the proliferation, invasion, and migration of NSCLC: A study based
on immunohistochemistry, TCGA, bioinformatics, in vitro and in vivo
verification. Oncotarget. 8:16531–16552. 2017.PubMed/NCBI
|
|
44
|
Praveen P, Hülsmann H, Sültmann H, Kuner R
and Fröhlich H: Cross-talk between AMPK and EGFR dependent
signaling in non-small cell lung cancer. Sci Rep. 6:275142016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang X, Zheng Q, Wang C, Zhou H, Jiang G,
Miao Y, Zhang Y, Liu Y, Li Q, Qiu X and Wang E: CCDC106 promotes
non-small cell lung cancer cell proliferation. Oncotarget.
8:26662–26670. 2017.PubMed/NCBI
|
|
46
|
Chen R, Zhang Y, Zhang C, Wu H and Yang S:
miR-137 inhibits the proliferation of human non-small cell lung
cancer cells by targeting SRC3. Oncol Lett. 13:3905–3911. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wen XP, Ma HL, Zhao LY, Zhang W and Dang
CX: MiR-30a suppresses non-small cell lung cancer progression
through AKT signaling pathway by targeting IGF1R. Cell Mol Biol
(Noisy-le-grand). 61:78–85. 2015.PubMed/NCBI
|
|
48
|
Mei Y, Yang JP and Qian CN: For robust big
data analyses: A collection of 150 important pro-metastatic genes.
Chin J Cancer. 36:162017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Van Laere S, Dirix L and Vermeulen P:
Molecular profiles to biology and pathways: A systems biology
approach. Chin J Cancer. 35:532016. View Article : Google Scholar : PubMed/NCBI
|