Introduction
Primary hepatic carcinoma (PHC) is a common
malignant tumor, and its incidence rate ranks 6th in the world
(1). Among nearly 60 million patients
who succumb to PHC each year, patients from the developing
countries account for over 80%, of which Chinese patients account
for 55% (2,3). At present, the treatment methods of PHC
mainly include the surgical resection, chemotherapy and
radiotherapy, albeit with poor efficacy, resulting in frequent
side-effects. Biotherapy, with advantages such as high efficiency,
slight side effects and precision, may be able to serve as another
effective method for the treatment of PHC (4).
MicroRNAs (miRNAs or miRs), conserved in evolution,
non-coding, small RNA molecules 20 and 25 bp in length are
characterized by the regulation of the genetic expression at the
level of translation (5,6). The results of some experiments showed
that miR-143 is expressed at a low level in many malignant tumors,
such as colorectal (7), lung
(8), esophageal (9), and breast (10) cancers. However, few studies have
focused on the correlation between miR-143 and the occurrence and
development of liver cancer, and the effect of miR-143 on the
mechanism is also still unknown. In this study, artificially
synthesized miR-143 mimics were used to transfect the liver
carcinoma SMMC-7721 cell line to observe the effect of miR-143 on
the genetic and protein expression of liver carcinoma cells, and we
inferred the potential action mechanism in the development and
occurrence of liver carcinoma.
Materials and methods
Materials
The materials obtained for the study were: SMMC-7721
human liver carcinoma cell line (Institute of Cell Biology,
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences, Shanghai, China); cell apoptosis detection kit (Run-well
Industrial Co., Ltd., Shanghai, China); Lipofectamine™ 2000
(Biotsith Bioscience Co., Ltd., Suzhou, China); miR-143 mimics
(Miaoling Bioscience and Technology Co., Ltd., Wuhan, China);
methyl thiazolyl tetrazolium (MTT; Beijing Huamaike Biotechnology
Co., Ltd., Beijing, China); dimethyl sulfoxide (DMSO; Tideradar
Science and Technology Co., Ltd., Beijing, China); TRIGene kit
(Abbiotechnology, Inc., Guangzhou, China); reverse transcription
kit (GeneCopoeia, Rockville, MD, USA); cell total protein
extraction kit (Biogenro Biotechnology Co. Ltd., Beijing, China);
NF-κB p65 TransAM™ enzyme-linked immunosorbent assay (ELISA) kit
(Jiamay Biotech Co., Ltd., Beijing, China). Primary and secondary
antibodies, all from (Cell Signaling Technology, Danvers, MA,
USA).
Cell culture and transfection
Total cell culture medium was prepared as follows:
Dulbecco's modified Eagle's medium (DMEM) + 10% fetal bovine serum
(FBS) + 1% double-antibody. SMMC-7721 liver carcinoma cell line was
cultured in an incubator (37°C, 5% CO2), and the
following transfection experiments were prepared according to the
instructions in Lipofectamine™ 2000, in which miR-143 mimics were
transfected into the SMMC-7721 liver carcinoma cell line, and the
transfected cells were divided into the mimics and negative control
groups.
Detection of cell proliferation
through MTT experiments
Under the microscope (Olympus Corporation, Tokyo,
Japan), we observed cell growth. After being digested sufficiently
and the cells were counted, the cell density was adjusted to
5×105/ml, and the cells were inoculated into a 96-well
plate (100 µl/well). These cells were cultured for 24, 48 and 72 h,
and the prepared 20 µl MTT solution was added into the plate in the
dark. Then, the plate was placed into an incubator for 4 h at 37°C,
and the supernatant was discarded. A 150 ml DMSO solution was added
into each well followed by vibration on a shaker, and after
approximately 10 min, the crystal-like substance was totally
dissolved in DMSO solution. A Sunrise microplate reader (Tecan
Group Ltd., Männedorf, Switzerland) was used to detect the optical
density of each well at the wavelength of 490 nm
(OD490), and the average of OD values was taken as the
result.
Detection of cell apoptosis
The SMMC-7721 liver carcinoma cell line was
inoculated into the 6-well plate, and transfection intervention was
performed for cells in the logarithmic phase (procedures were the
same as mentioned earlier). After 48 h of transfection, the cells
in the mimics and negative control groups were digested with
trypsin, and collected into the centrifuge tubes. The tubes were
centrifuged for 5 min at 8,600 × g and the supernatant was
discarded. The cells were mixed with the pre-cooled phosphate
buffer, and the procedure was repeated 3 times followed by
resuspension of cells to prepare the cell suspension at a density
of 5×105/ml, in which we extracted 1 ml suspension for
centrifugation and then the supernatant was discarded. In the
sediment, 500 µl binding buffer, 5 µl Annexin V-FITC and 10 µl
propidium iodide (PI) were sequentially added. The suspension was
then incubated in the dark for 10 min. A flow cytometer (Attune
NxT; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to
detect cell apoptosis in the mimics and negative control
groups.
Detection of mRNA expression using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
According to the procedures of instructions in the
TRIGene kit, the total RNA was extracted from the mimics and
negative control groups, respectively, and a spectrophotometer
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to detect
the concentration and purification of total RNAs, in which
A260/A280 values were between 1.8 and 2.0. In accordance with the
instructions of reverse transcription kit (RevertAid Fist Strand
cDNA Synthesis kit, K1622; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), and the primer sequences were produced by Shanghai JiRan
Biotechnology Co., Ltd. (Shanghai, China) (Table I). The reverse transcription was
performed for preparation of cDNA in 20 µl reaction system that was
placed on the RT-PCR apparatus.
 | Table I.Primer sequence of NF-κB p65. |
Table I.
Primer sequence of NF-κB p65.
| Genes | Primers |
|---|
| NF-κB p65 | F:
5′-TACCCTGAGGCTATAACTC-3′ |
|
| R:
5′-GACACTTGATAAGGCTTTG-3′ |
| GAPDH | F:
5′-TGGGTGTGAACCACGAGAA-3′ |
|
| R:
5′-GGCATGGACTGTGGTCATGA-3′ |
The 25 µl reaction system was prepared as per the
protocol of the quantitative PCR kit (2X RealStar Green Power
Mixture, A311; GenStar Biosolutions Co., Ltd., Beijing, China), and
the reaction conditions were set as follows: denaturation at 95°C
for 10 min, annealing at 95°C for 30 sec, extension at 59.4°C for
30 sec, for a total of 40 cycles, with a final extension at 95°C
for 15 sec, and cooling down to 65°C. GAPDH served as an internal
reference, and the quantitative value was used in the automatic
calculation of the relative mRNA expression of NF-κB p65 using
RT-PCR apparatus.
Detection of the protein expression
via western blot analysis
Total proteins from the mimics and negative control
groups were extracted as per the protocol of the cell total protein
extraction kit, and the concentration of extracted proteins was
assayed. The proteins were preserved at −70°C for later use. Gel at
different concentrations was prepared for SDS-PAGE, and the
position of NF-κB p65 was verified with the reference marker
stripe. A gel of appropriate size was cut for 30 min of membrane
transferring, and blocked using 5% bovine serum albumin (BSA) for 1
h. The mouse anti-human NF-κB and GAPDH primary monoclonal antibody
(dilution, 1:1,000; cat. nos. 6956 and 97166) were added onto the
membrane for incubation overnight at 4°C. Tris-buffered saline and
Tween-20 (TBST) was used to wash the polyvinylidene fluoride (PVDF)
membrane 3 times (5 min/time). The membrane was incubated at room
temperature using the horse anti-mouse HRP secondary polyclonal
antibody (dilution, 1:2,000; cat. no. 7076) for 1 h, and TBST was
used to wash the PVDF membrane 3 times (5 min/time).
Electrochemiluminescence (ECL) color-development solution was added
onto the membrane for exposure in the dark, and the ChemiDoc™ MP
imaging system was used for scanning. Images imported into the
system were analyzed using ImageJ professional image analytic
software, and the optical density was recorded.
Assay of NF-κB activity
The NF-κB p65 TransAM™ ELISA kit was used for assay
of NF-κB activity in SMMC-7721 cells. The cell proteins were
extracted from the mimics and negative control groups, and then
added into the 96-well plate containing oligonucleotides in the
homologous region of NF-κB. The plate was agitated slightly and
incubated at room temperature for 1 h followed by washing using
distilled water 3 or 4 times. Thereafter, the antibody of NF-κB was
added for 1 h of incubation at room temperature. Again, the plate
was washed 3 or 4 times using distilled water. Horse anti-mouse HRP
secondary polyclonal antibody (dilution, 1:1,000; cat. no. 7076)
was added and the proteins were incubated for 1 h at room
temperature followed by the color development reaction and
terminating reaction. The Sunrise microplate reader was used to
detect the activity of NF-κB at a wavelength of 450 nm.
Statistical analysis
Statistical Product and Service Solutions (SPSS)
16.0 (SPSS, Inc., Chicago, IL, USA) software was used for
statistical analysis of the data. Measurement data are presented as
mean ± standard deviation, and one-way analysis of variance and
paired t-test were applied in the analysis in comparison between
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
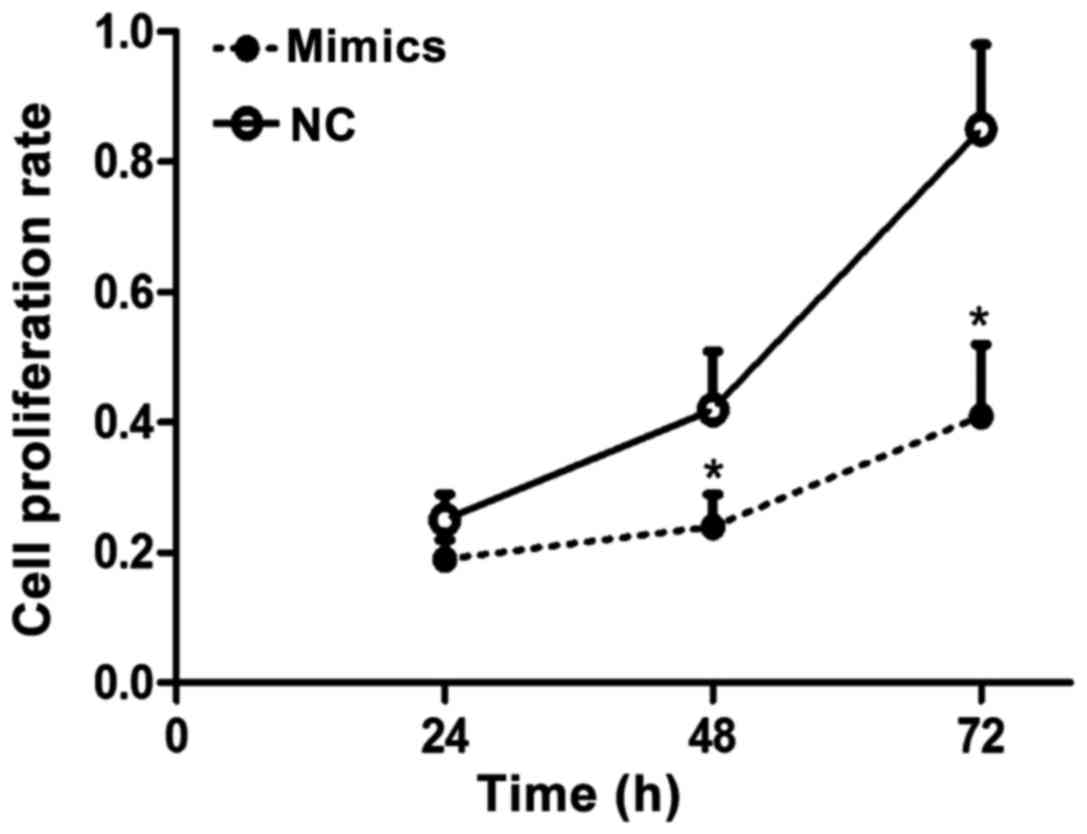
Results of the MTT assay
Results of the MTT assay for proliferation of liver
carcinoma cells showed that after SMMC-7721 liver carcinoma cell
line was transfected by mimics for 24, 48 and 72 h, the cell
proliferation rate in the negative control group was significantly
higher than that in the mimics group, and the difference had
statistical significance (p<0.05), suggesting that miR-143 can
obviously inhibit the proliferation of liver carcinoma cells
(Fig. 1).
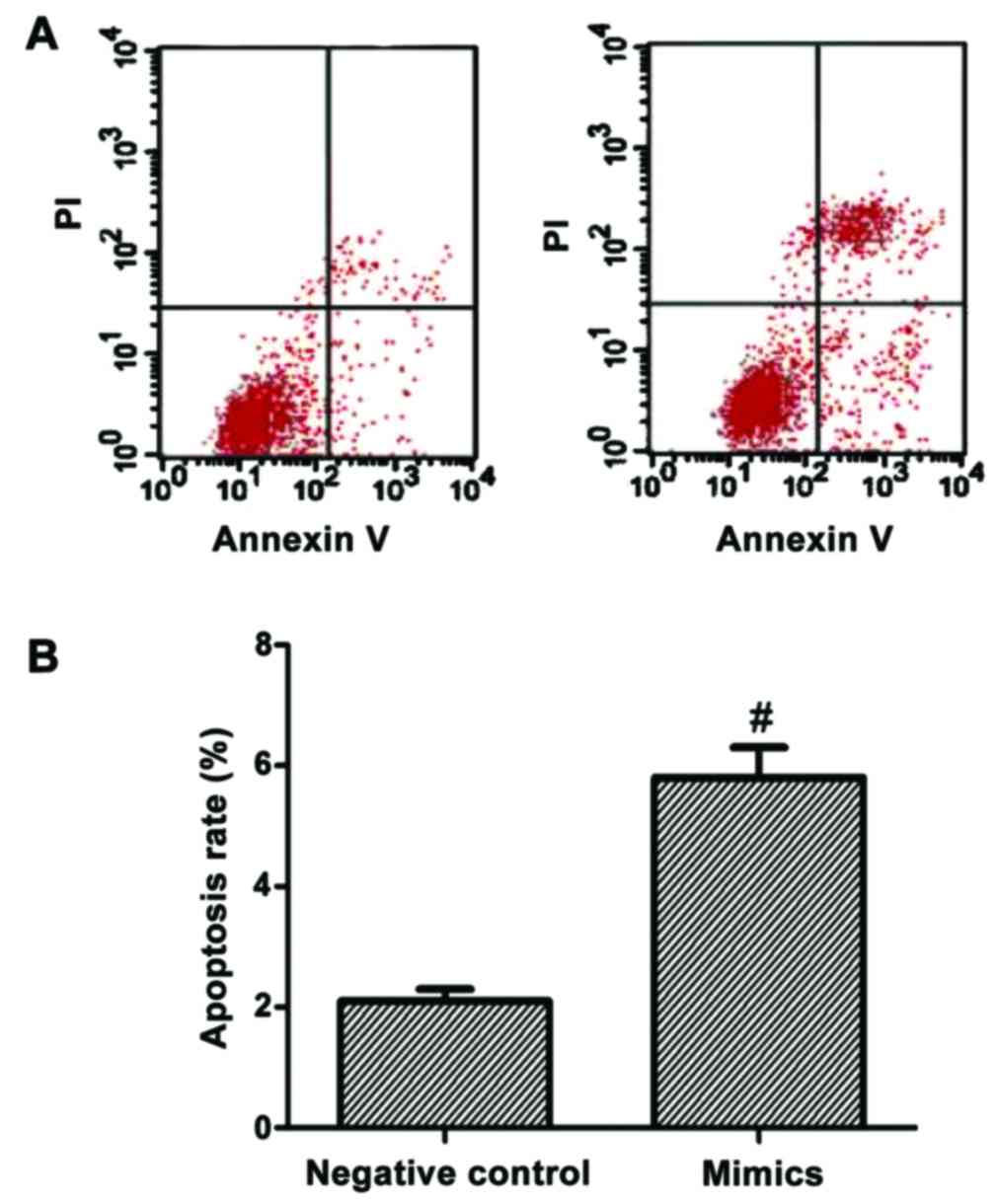
Results of cell apoptosis
experiment
After the SMMC-7721 liver carcinoma cell line was
transfected by mimics for 48 h, cell apoptotic rates of the mimics
and negative control groups were detected using a flow cytometer.
The results showed that the cell apoptotic rate in the mimics group
was significantly higher than that in the negative control group,
with a statistically significant difference (p<0.05; Fig. 2).
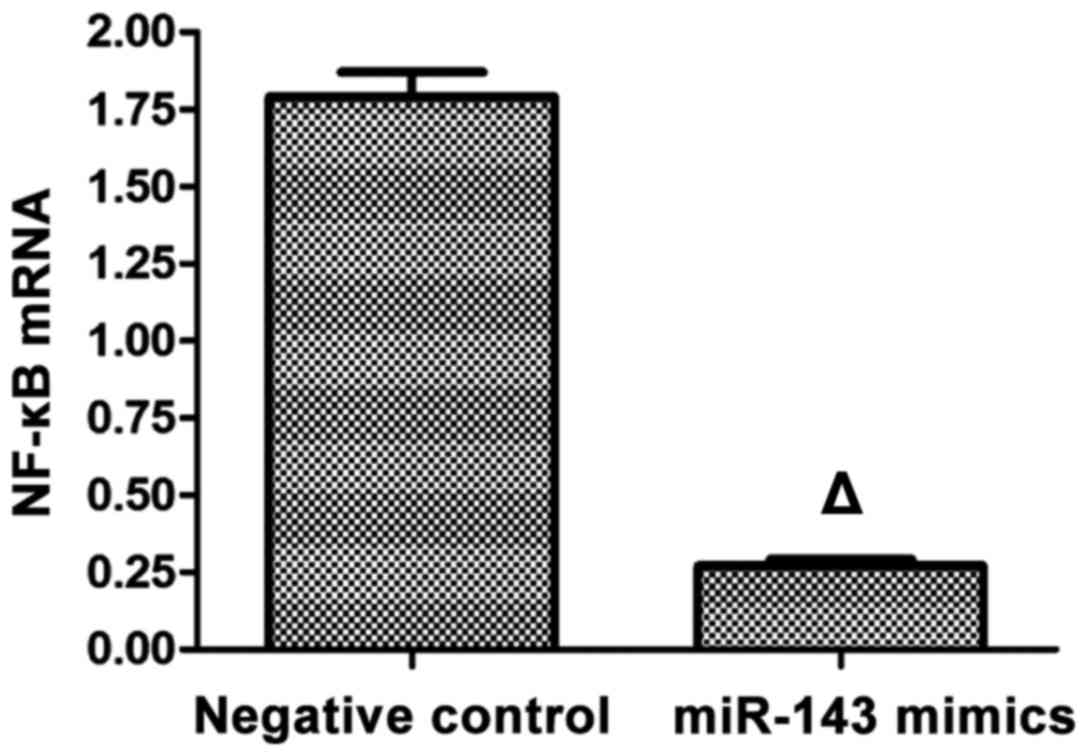
Detection of mRNA expression of NF-κB
using RT-qPCR
RT-qPCR was applied in the detection of mRNA
expression of NF-κB, and the results showed that after the
SMMC-7721 liver carcinoma cell line was transfected with mimics for
48 h, the mRNA expression of NF-κB in the mimics group was
significantly lower than that in the negative control group with a
statistically significant difference (p<0.05; Fig. 3).
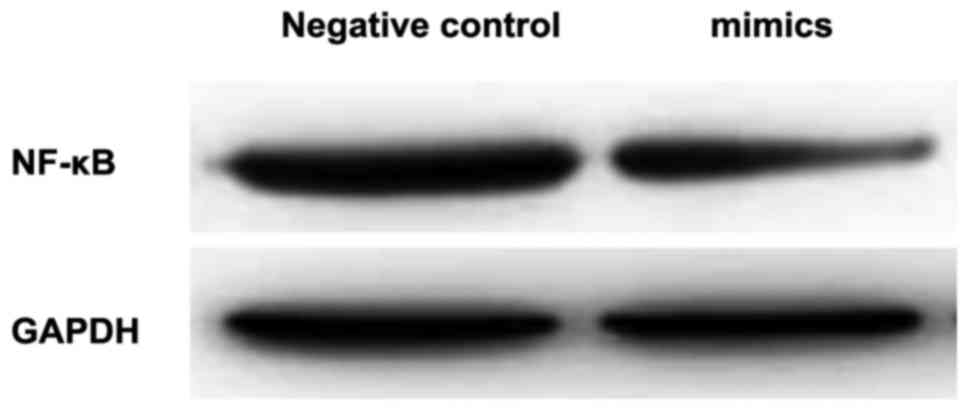
Detection of the expression of NF-κB
p65 via western blot analysis
Western blot analysis revealed that the protein
expression level of NF-κB p65 in the intervention group was
significantly lower than that in the control group, and the
difference was of statistical significance (p<0.05; Table II and Fig.
4).
 | Table II.Protein expression of NF-κB p65 in the
negative control and mimics groups (mean ± SD). |
Table II.
Protein expression of NF-κB p65 in the
negative control and mimics groups (mean ± SD).
| Variable | Negative control
group | Mimics group |
|---|
| NF-κB p65 | 1.1582±0.0319 |
0.4376±0.0428a |
Result of NF-κB activity assay
After the cells were transfected with miR-143 mimics
for 48 h, the NF-κB activity in the SMMC-7721 liver carcinoma cell
line was 0.22±0.04, which was significantly lower than that in the
control group (p<0.01; Table
III).
 | Table III.NF-κB activity assay. |
Table III.
NF-κB activity assay.
| Group | NF-κB activity | F-value | P-value |
|---|
| Negative control | 0.51±0.05 | 341.374 | <0.01 |
| miR-143 mimics |
0.22±0.04a |
|
|
Discussion
Liver carcinoma is usually characterized by high
malignancy, and due to the difficulties in diagnosis at early or
middle stage, patients with liver carcinoma are usually diagnosed
at the late stage, making it difficult for radical resection.
Consequently, conventional surgeries, chemotherapy and radiotherapy
have been greatly limited in clinical practice as treatment for
liver carcinoma (11). In recent
years, the newly emerged biotherapy has been viewed as a new method
for the treatment of malignant tumors, and, therefore, searching
for the new targets is of great clinical and practical significance
in the treatment of malignant tumors. In humans, there are a
variety of oncogenes and anti-oncogenes, which can act alone, or
together to form a network system to perform a synergistic effect,
thereby promoting or inhibiting the occurrence and development of
tumors (12). miRNAs, the important
genetic regulatory factors in humans, have been reported to exert
regulatory effects on the occurrence and development of liver
carcinoma (13). Nevertheless, the
mechanism of miR-143 involved in the development of liver carcinoma
remains to be determined (14). Thus,
in this study, we preliminarily investigated the mechanism how
miR-143 affects the development of liver carcinoma.
miR-143 exists in various cells in humans, and
miR-143 has been reported to have lower expression levels in many
kinds of tumor cells, such as cervical (15) and prostate (16) cancers. Currently, there are few
studies reporting the correlations between miR-143 and the
occurrence and development of liver carcinoma. In addition, there
is controversy regarding this correlation in the published
literature. Some scholars have found that miR-143 is expressed at
low levels in liver carcinoma tissues (17). However, the findings of other authors
indicated that miR-143 is highly expressed in liver carcinoma, and
acts as an anti-oncogene to effectively suppress the invasion and
metastasis of liver carcinoma cells (18).
In this study, we performed an MTT assay after the
SMMC-7721 liver carcinoma cell line was transfected with miR-143
mimics, and found that the proliferation rate of liver carcinoma
cells in the mimics group was significantly lower than that in the
negative control group, and the difference was of statistical
significance. The subsequent cell apoptosis experiment revealed
that after cells were transfected with miR-143 mimics, the cell
apoptotic rate in the mimics group was significantly higher than
that in the negative control group, with a statistically
significant difference. To further determine the potential
mechanism regarding miR-143 promotion of apoptosis of liver
carcinoma cells, we assayed the signal transduction pathway of
NF-κB.
The NF-κB signal transduction pathway is known to be
involved in the development of various cancers including lung
(18), colorectal (19) and liver cancers (20), in which it plays important roles. In
the present study, we detected the protein and genetic expression
of NF-κB in the mimics and the negative control groups, and the
western blotting showed that after the expression of miR-143 in
SMMC-7721 liver carcinoma cell line was upregulated, the protein
and genetic expression of NF-κB were significantly
downregulated.
In conclusion, our results show that the
upregulation in miR-143 expression in the SMMC-7721 liver carcinoma
cell line can suppress the activity of NF-κB signal transduction
pathway to a certain degree by inducing apoptosis of liver
carcinoma cells, resulting in the significant inhibition of the
cell proliferation rate. Since miRNAs can act on various target
genes, the process in which miR-143 promotes apoptosis of liver
carcinoma cells and inhibits the proliferation of liver carcinoma
cells may be caused by the effect of other target genes, or the
synergistic effect of several signal pathways. Thus, to clarify the
mechanism regarding how miR-143 induces apoptosis of liver
carcinoma cells requires in-depth investigation in future
studies.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Padhya KT, Marrero JA and Singal AG:
Recent advances in the treatment of hepatocellular carcinoma. Curr
Opin Gastroenterol. 29:285–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Groulx JF, Giroux V, Beauséjour M,
Boudjadi S, Basora N, Carrier JC and Beaulieu JF: Integrin α6A
splice variant regulates proliferation and the Wnt/β-catenin
pathway in human colorectal cancer cells. Carcinogenesis.
35:1217–1227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paul I, Bhattacharya S, Chatterjee A and
Ghosh MK: Current understanding on EGFR and Wnt/β-catenin signaling
in glioma and their possible crosstalk. Genes Cancer. 4:427–446.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen P, Zhao X and Ma L: Downregulation of
microRNA-100 correlates with tumor progression and poor prognosis
in hepatocellular carcinoma. Mol Cell Biochem. 383:49–58. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Dysregulated
microRNAs in the pathogenesis and progression of cervical neoplasm.
Cell Cycle. 11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bae HJ, Jung KH, Eun JW, Shen Q, Kim HS,
Park SJ, Shin WC, Yang HD, Park WS, Lee JY, et al: MicroRNA-221
governs tumor suppressor HDAC6 to potentiate malignant progression
of liver cancer. J Hepatol. 63:408–419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bai JW, Xue HZ and Zhang C:
Down-regulation of microRNA-143 is associated with colorectal
cancer progression. Eur Rev Med Pharmacol Sci. 20:4682–4687.
2016.PubMed/NCBI
|
|
8
|
Zhang HB, Sun LC, Ling L, Cong LH and Lian
R: miR-143 suppresses the proliferation of NSCLC cells by
inhibiting the epidermal growth factor receptor. Exp Ther Med.
12:1795–1802. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mao Y, Liu J, Zhang D and Li B: miR-143
inhibits tumor progression by targeting FAM83F in esophageal
squamous cell carcinoma. Tumour Biol. 37:9009–9022. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johannessen C, Moi L, Kiselev Y, Pedersen
MI, Dalen SM, Braaten T and Busund LT: Expression and function of
the miR-143/145 cluster in vitro and in vivo in human breast
cancer. PLoS One. 12:e01866582017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Takahashi S, Tasaka A, Yoshima T,
Ochi H and Chayama K: Involvement of microRNA-224 in cell
proliferation, migration, invasion, and anti-apoptosis in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:565–575.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang FT, Peng JF, Cheng WJ, Zhuang YY,
Wang LY, Li CQ, Tang J, Chen WY, Li YH and Zhang SN: miR-143
targeting TAK1 attenuates pancreatic ductal adenocarcinoma
progression via MAPK and NF-κB pathway in vitro. Dig Dis Sci.
62:944–957. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao N, Wang R, Zhou L, Zhu Y, Gong J and
Zhuang SM: MicroRNA-26b suppresses the NF-κB signaling and enhances
the chemosensitivity of hepatocellular carcinoma cells by targeting
TAK1 and TAB3. Mol Cancer. 13:352014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khafaei M, Samie S, Mowla SJ, Alvanegh AG,
Mirzaei B, Chavoshei S, Dorraj GS, Esmailnejad M, Tavallaie M and
Nourani M: Evaluation of miR-9 and miR-143 expression in urine
specimens of sulfur mustard exposed patients. Interdiscip Toxicol.
8:169–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deftereos G, Corrie SR, Feng Q, Morihara
J, Stern J, Hawes SE and Kiviat NB: Expression of mir-21 and
mir-143 in cervical specimens ranging from histologically normal
through to invasive cervical cancer. PLoS One. 6:e284232011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z,
Li P, Zhang W, Wu H, Feng N, et al: miR-143 decreases prostate
cancer cells proliferation and migration and enhances their
sensitivity to docetaxel through suppression of KRAS. Mol Cell
Biochem. 350:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Borralho PM, Simões AE, Gomes SE, Lima RT,
Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE and Rodrigues
CM: miR-143 overexpression impairs growth of human colon carcinoma
xenografts in mice with induction of apoptosis and inhibition of
proliferation. PLoS One. 6:e237872011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He J, Qian X, Carpenter R, Xu Q, Wang L,
Qi Y, Wang ZX, Liu LZ and Jiang BH: Repression of miR-143 mediates
Cr (VI)-induced tumor angiogenesis via IGF-IR/IRS1/ERK/IL-8
pathway. Toxicol Sci. 134:26–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szele E, Gombos K, Juhász K, Wohler V,
Kovács A and Ember I: Effects of purified glycerol from biodiesel
on miRNAs compared to the expression profile of selected mRNAs in
Balb/c mice. In Vivo. 27:107–111. 2013.PubMed/NCBI
|
|
20
|
Mosquera JM, Sboner A, Zhang L, Chen CL,
Sung YS, Chen HW, Agaram NP, Briskin D, Basha BM, Singer S, et al:
Novel MIR143-NOTCH fusions in benign and malignant glomus tumors.
Genes Chromosomes Cancer. 52:1075–1087. 2013. View Article : Google Scholar : PubMed/NCBI
|