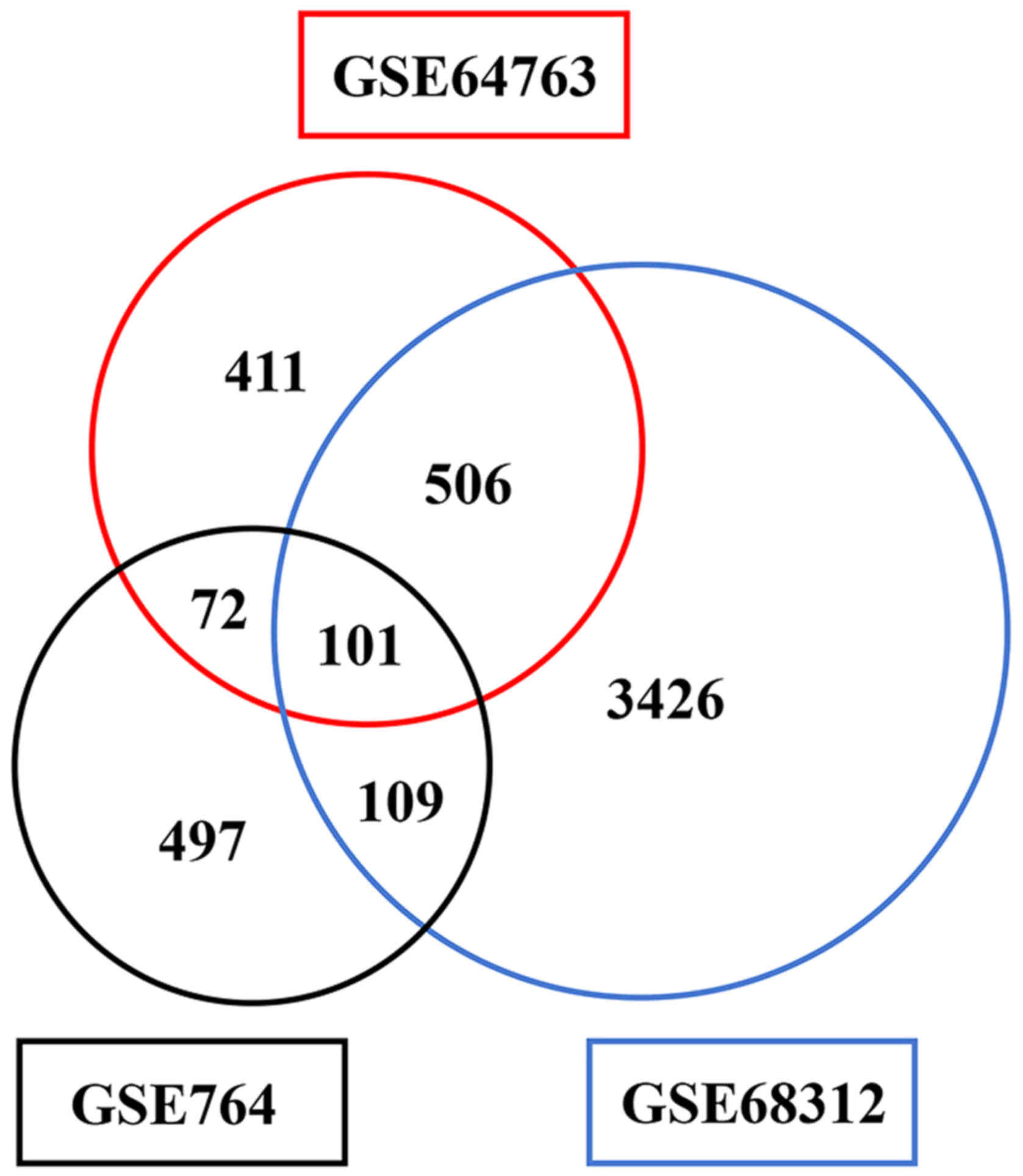
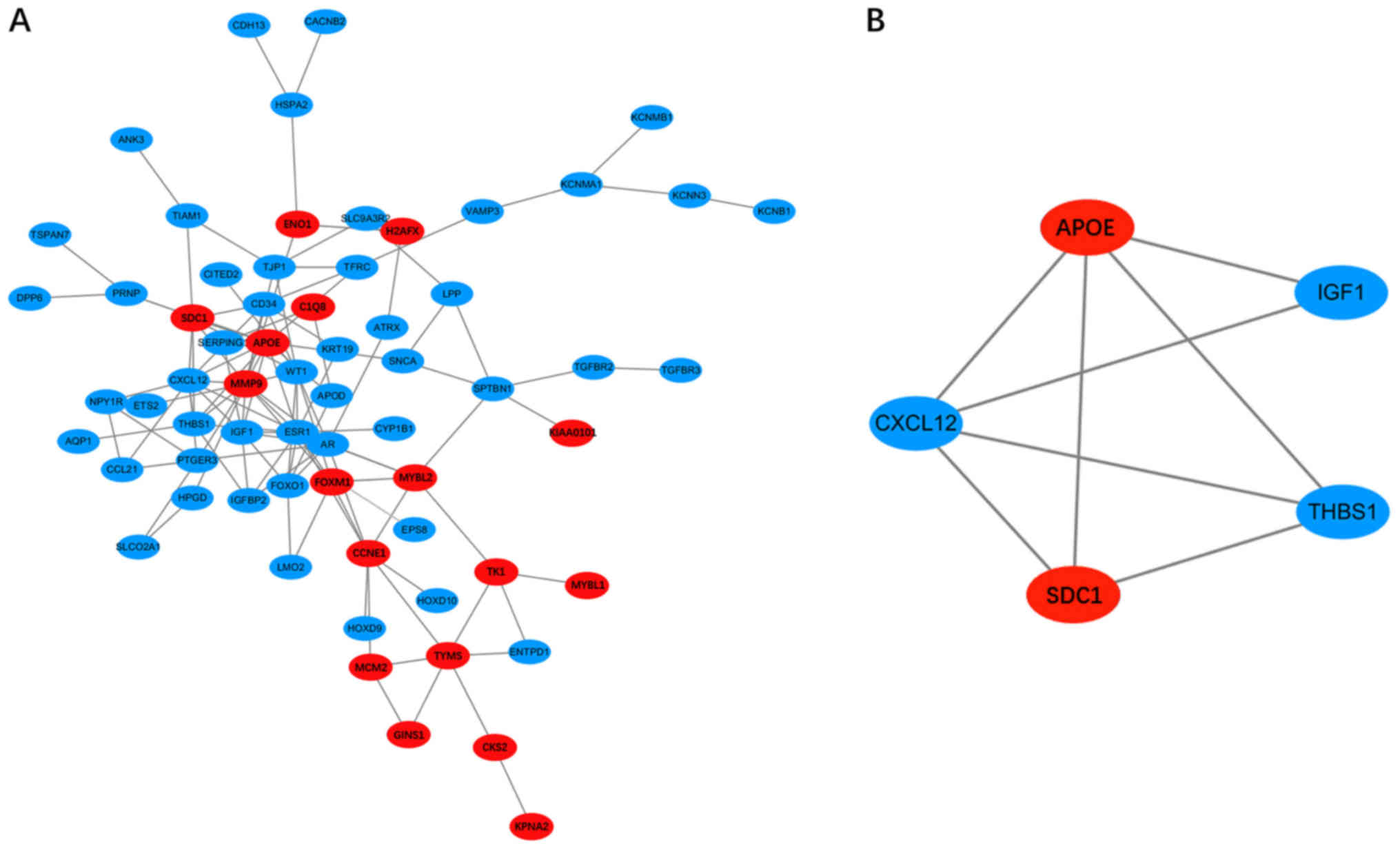
|
1
|
Ricci S, Stone RL and Fader AN: Uterine
leiomyosarcoma: Epidemiology, contemporary treatment strategies and
the impact of uterine morcellation. Gynecol Oncol. 145:208–216.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park JY, Lee JW, Lee HJ, Lee JJ, Moon SH,
Kang SY, Cheon GJ and Chung HH: Prognostic significance of
preoperative 18F-FDG PET/CT in uterine leiomyosarcoma. J
Gynecol Oncol. 28:e282017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ducie JA and Leitao MM Jr: The role of
adjuvant therapy in uterine leiomyosarcoma. Expert Rev Anticancer
Ther. 16:45–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cui RR, Wright JD and Hou JY: Uterine
leiomyosarcoma: A review of recent advances in molecular biology,
clinical management and outcome. BJOG. 124:1028–1037. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barlin JN, Zhou QC, Leitao MM, Bisogna M,
Olvera N, Shih KK, Jacobsen A, Schultz N, Tap WD, Hensley ML, et
al: Molecular subtypes of uterine leiomyosarcoma and correlation
with clinical outcome. Neoplasia. 17:183–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee TI and Young RA: Transcriptional
regulation and its misregulation in disease. Cell. 152:1237–1251.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cuppens T, Moisse M, Depreeuw J, Annibali
D, Colas E, Gil-Moreno A, Huvila J, Carpén O, Zikán M, Matias-Guiu
X, et al: Integrated genome analysis of uterine leiomyosarcoma to
identify novel driver genes and targetable pathways. Int J Cancer.
142:1230–1243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iozzo RV and Sanderson RD: Proteoglycans
in cancer biology, tumor microenvironment and angiogenesis. J Cell
Mol Med. 15:1013–1031. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Purushothaman A, Babitz SK and Sanderson
RD: Heparanase enhances the insulin receptor signaling pathway to
activate extracellular signal-regulated kinase in multiple myeloma.
J Biol Chem. 287:41288–41296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brule S, Charnaux N, Sutton A, Ledoux D,
Chaigneau T, Saffar L and Gattegno L: The shedding of syndecan-4
and syndecan-1 from HeLa cells and human primary macrophages is
accelerated by SDF-1/CXCL12 and mediated by the matrix
metalloproteinase-9. Glycobiology. 16:488–501. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Asuthkar S, Velpula KK, Nalla AK, Gogineni
VR, Gondi CS and Rao JS: Irradiation-induced angiogenesis is
associated with an MMP-9-miR-494-syndecan-1 regulatory loop in
medulloblastoma cells. Oncogene. 33:1922–1933. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wen N, Wang Y, Wen L, Zhao SH, Ai ZH, Wang
Y, Wu B, Lu HX, Yang H, Liu WC and Li Y: Overexpression of FOXM1
predicts poor prognosis and promotes cancer cell proliferation,
migration and invasion in epithelial ovarian cancer. J Transl Med.
12:1342014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tzeng HT, Tsai CH, Yen YT, Cheng HC, Chen
YC, Pu SW, Wang YS, Shan YS, Tseng YL, Su WC, et al: Dysregulation
of Rab37-mediated cross-talk between cancer cells and endothelial
cells via thrombospondin-1 promotes tumor neovasculature and
metastasis. Clin Cancer Res. 23:2335–2345. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Limoge M, Safina A, Beattie A, Kapus L,
Truskinovsky AM and Bakin AV: Tumor-fibroblast interactions
stimulate tumor vascularization by enhancing cytokine-driven
production of MMP9 by tumor cells. Oncotarget. 8:35592–35608. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lorenc Z, Waniczek D, Lorenc-Podgórska K,
Krawczyk W, Domagała M, Majewski M and Mazurek U: Profile of
expression of genes encoding matrix metallopeptidase 9 (MMP9),
matrix metallopeptidase 28 (MMP28) and TIMP metallopeptidase
inhibitor 1 (TIMP1) in colorectal cancer: Assessment of the role in
diagnosis and prognostication. Med Sci Monit. 23:1305–1311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roomi MW, Kalinovsky T, Niedzwiecki A and
Rath M: Modulation of u-PA, MMPs and their inhibitors by a novel
nutrient mixture in adult human sarcoma cell lines. Int J Oncol.
43:39–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lusby K, Savannah KB, Demicco EG, Zhang Y,
Ghadimi MP, Young ED, Colombo C, Lam R, Dogan TE, Hornick JL, et
al: Uterine leiomyosarcoma management, outcome, and associated
molecular biomarkers: A single institution's experience. Ann Surg
Oncol. 20:2364–2372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu XQ, Shi YF, Cheng XD and Wu YZ: The
differential diagnosis between uterine leiomyosarcoma and the
special subtypes of leiomyoma. Zhonghua Yi Xue Za Zhi.
83:1419–1421. 2003.(In Chinese). PubMed/NCBI
|
|
20
|
Koivisto-Korander R, Butzow R, Koivisto AM
and Leminen A: Immunohistochemical studies on uterine
carcinosarcoma, leiomyosarcoma, and endometrial stromal sarcoma:
Expression and prognostic importance of ten different markers.
Tumour Biol. 32:451–459. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Garcia C, Kubat JS, Fulton RS, Anthony AT,
Combs M, Powell CB and Littell RD: Clinical outcomes and prognostic
markers in uterine leiomyosarcoma: A population-based cohort. Int J
Gynecol Cancer. 25:622–628. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
George S, Feng Y, Manola J, Nucci MR,
Butrynski JE, Morgan JA, Ramaiya N, Quek R, Penson RT, Wagner AJ,
et al: Phase 2 trial of aromatase inhibition with letrozole in
patients with uterine leiomyosarcomas expressing estrogen and/or
progesterone receptors. Cancer. 120:738–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho YL, Bae S, Koo MS, Kim KM, Chun HJ,
Kim CK, Ro DY, Kim JH, Lee CH, Kim YW and Ahn WS: Array comparative
genomic hybridization analysis of uterine leiomyosarcoma. Gynecol
Oncol. 99:545–551. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uluer ET, Inan S, Ozbilgin K, Karaca F,
Dicle N and Sanci M: The role of hypoxia related angiogenesis in
uterine smooth muscle tumors. Biotech Histochem. 90:102–110. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bodner-Adler B, Nather A, Bodner K,
Czerwenka K, Kimberger O, Leodolter S and Mayerhofer K: Expression
of thrombospondin 1 (TSP 1) in patients with uterine smooth muscle
tumors: An immunohistochemical study. Gynecol Oncol. 103:186–189.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hulsurkar M, Li Z, Zhang Y, Li X, Zheng D
and Li W: Beta-adrenergic signaling promotes tumor angiogenesis and
prostate cancer progression through HDAC2-mediated suppression of
thrombospondin-1. Oncogene. 36:1525–1536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Adams JC, Kureishy N and Taylor AL: A role
for syndecan-1 in coupling fascin spike formation by
thrombospondin-1. J Cell Biol. 152:1169–1182. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alexander CM, Reichsman F, Hinkes MT,
Lincecum J, Becker KA, Cumberledge S and Bernfield M: Syndecan-1 is
required for Wnt-1-induced mammary tumorigenesis in mice. Nat
Genet. 25:329–332. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marinkovich MP: Tumor microenvironment:
Laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 7:370–380.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jary M, Lecomte T, Bouché O, Kim S, Dobi
E, Queiroz L, Ghiringhelli F, Etienne H, Léger J, Godet Y, et al:
Prognostic value of baseline seric Syndecan-1 in initially
unresectable metastatic colorectal cancer patients: A simple
biological score. Int J Cancer. 139:2325–2335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wilsie LC, Gonzales AM and Orlando RA:
Syndecan-1 mediates internalization of apoE-VLDL through a low
density lipoprotein receptor-related protein (LRP)-independent,
non-clathrin-mediated pathway. Lipids Health Dis. 5:232006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang YA, Zhou B, Wernig M and Südhof TC:
ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription
and Aβ secretion. Cell. 168:427–441.e21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo J, Song J, Feng P, Wang Y, Long W, Liu
M and Li L: Elevated serum apolipoprotein E is associated with
metastasis and poor prognosis of non-small cell lung cancer. Tumour
Biol. 37:10715–10721. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Wan J, Yuan L, Ba J, Feng P, Long W,
Huang H, Liu P, Cai Y, Liu M, et al: Serum levels of apolipoprotein
E correlates with disease progression and poor prognosis in breast
cancer. Tumour Biol. Oct 5–2016.(Epub ahead of print). View Article : Google Scholar
|
|
35
|
Shi X, Xu J, Wang J, Cui M, Gao Y, Niu H
and Jin H: Expression analysis of apolipoprotein E and its
associated genes in gastric cancer. Oncol Lett. 10:1309–1314. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Roy I, Zimmerman NP, Mackinnon AC, Tsai S,
Evans DB and Dwinell MB: CXCL12 chemokine expression suppresses
human pancreatic cancer growth and metastasis. PLoS One.
9:e904002014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang YP and Wu XH: Correlations of
chemokine CXCL12 and its receptor to tumor metastasis. Ai Zheng.
26:220–224. 2007.(In Chinese). PubMed/NCBI
|
|
38
|
Wu Y, Guo X, Brandt Y, Hathaway HJ and
Hartley RS: Three-dimensional collagen represses cyclin E1 via β1
integrin in invasive breast cancer cells. Breast Cancer Res Treat.
127:397–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nakayama N, Nakayama K, Shamima Y,
Ishikawa M, Katagiri A, Iida K and Miyazaki K: Gene amplification
CCNE1 is related to poor survival and potential therapeutic target
in ovarian cancer. Cancer. 116:2621–2634. 2010.PubMed/NCBI
|
|
40
|
Kanska J, Zakhour M, Taylor-Harding B,
Karlan BY and Wiedemeyer WR: Cyclin E as a potential therapeutic
target in high grade serous ovarian cancer. Gynecol Oncol.
143:152–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin SC, Kao CY, Lee HJ, Creighton CJ,
Ittmann MM, Tsai SJ, Tsai SY and Tsai MJ: Dysregulation of
miRNAs-COUP-TFII-FOXM1-CENPF axis contributes to the metastasis of
prostate cancer. Nat Commun. 7:114182016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gormally MV, Dexheimer TS, Marsico G,
Sanders DA, Lowe C, Matak-Vinković D, Michael S, Jadhav A, Rai G,
Maloney DJ, et al: Suppression of the FOXM1 transcriptional
programme via novel small molecule inhibition. Nat Commun.
5:51652014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leitao MM, Soslow RA, Nonaka D, Olshen AB,
Aghajanian C, Sabbatini P, Dupont J, Hensley M, Sonoda Y, Barakat
RR and Anderson S: Tissue microarray immunohistochemical expression
of estrogen, progesterone, and androgen receptors in uterine
leiomyomata and leiomyosarcoma. Cancer. 101:1455–1462. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nachtigal MW, Hirokawa Y,
Enyeart-VanHouten DL, Flanagan JN, Hammer GD and Ingraham HA:
Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1
in sex-specific gene expression. Cell. 93:445–454. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Patil DT, Laskin WB, Fetsch JF and
Miettinen M: Inguinal smooth muscle tumors in women-a dichotomous
group consisting of Müllerian-type leiomyomas and soft tissue
leiomyosarcomas: An analysis of 55 cases. Am J Surg Pathol.
35:315–324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Coosemans A, Van Calster B, Verbist G,
Moerman P, Vergote I, Van Gool SW and Amant F: Wilms tumor gene 1
(WT1) is a prognostic marker in high-grade uterine sarcoma. Int J
Gynecol Cancer. 21:302–308. 2011.PubMed/NCBI
|