Introduction
Even though, the incidence of breast cancer (BC) in
China is lower compered to Western countries, since the 1990s it
has increased twice as fast as the global rate (1) with very early onset (2,3). In China,
BC patients <50 years old account for 46% of the total cases
(4), with the peak prevalence
observed in the 45 to 55 years old age (5–8).
Meanwhile, the highest mortality is observed in even younger age
i.e. in the 30 to 44 years old age (5). Therefore, in order to reduce the number
of BC patients, and the growing burden of this disease, it is
urgent to promote BC prevention in China.
BRCA (Breast Cancer Susceptibility Gene) is
associated with the majority of hereditary BC, which accounts for
about 5 to 10% of all cases of breast cancers (9). Meanwhile, the resemblance in clinical
and pathologic features between sporadic triple-negative BC (TNBC)
and BRCA1 mutant BC implies that mechanism behind
BRCA germline mutant tumors is strongly associated with
somatic mutation of sporadic BC (10). However, a lack of basic information
about the prevalence and spectrum of BRCA mutations hinders
research progress on the etiology (6)
and risk evaluation of model of breast cancer prevention in China.
Additionally, because of the large presence of ethnic-specific
contexts (11–14), the Western risk evaluation models do
not apply well to China. Therefore, the following paper assembled a
wide-range clinic-based cross-sectional study of hereditary risk
among BC patients, who were representative for BRCA
mutations study. The patients were from different parts of mainland
China, which was important for determining the prevalence and types
of BRCA mutations, as well as to provide basic information
for further studies of BRCA and BC prevention models in
China.
Materials and methods
Study population
The present study has been approved by center
medical ethics committee (Ethics Committee of the First Affiliated
Hospital of Third Military Medical University, PLA), and was
successfully registered with Chinese Clinical Trial Registry
(ChiCTR), which is the international clinical trial registration
platform.
In the time period from October 2015 to February
2016, 445 patients diagnosed with BC were recruited from 18
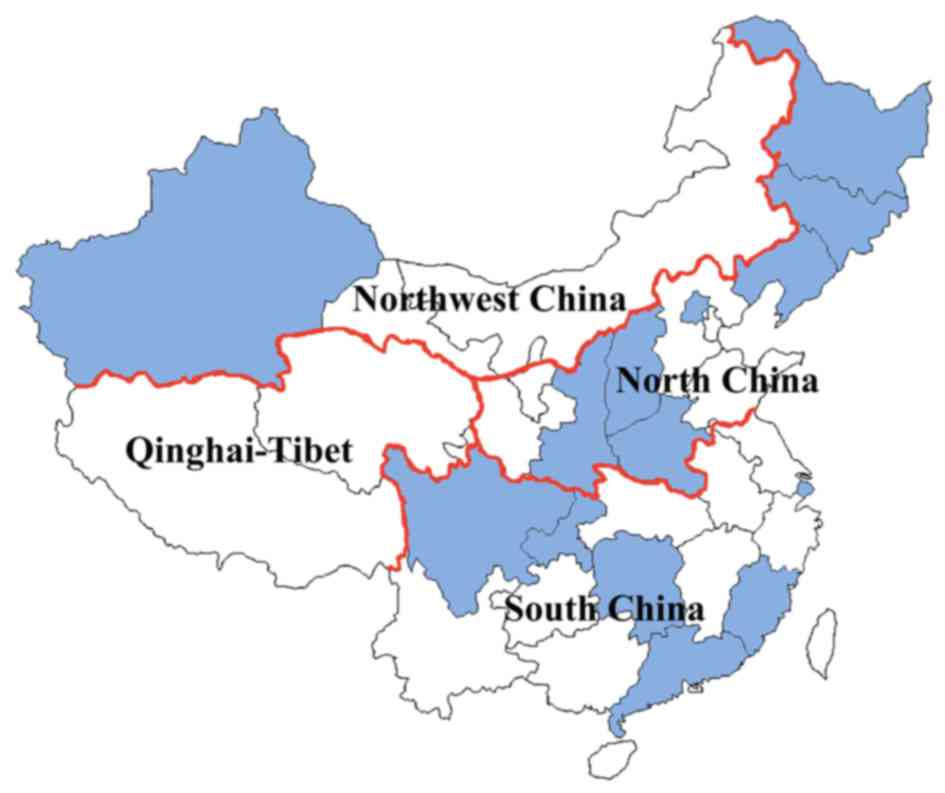
tertiary general clinics located in North, South and Northwest
China, which account for 3/4 of Chinese national territory
(Fig. 1). Among these patients, three
refused to participate in the study, while for other five the
recruitment criteria couldn't be confirmed. Pathological
examination was used to confirm the cancer diagnoses and breast
cancer subtype. Furthermore, twenty-seven participants were
additionally excluded from the clinical data analysis due to
incomplete or illegible clinical information. Where it was
possible, family members of probands with confirmed BC were
recruited to participate.
After reading and signing the informed consent,
participants were interviewed in order to provide medical details
including past medical history, present BC diagnosis age, tumor
size, states of axillary lymph nodes and metastasis, and details of
family cancer history. Consequently, fresh peripheral venous blood
(5 ml) was collected from each participant, and transferred into a
coded Ethylene Diamine Tetraacetie Acid (EDTA) tube at 4°C. During
the same day, blood samples were sent to Annoroad Gene Technology
(Beijing) Co., Ltd, for further analyses (BRCA genetic
testing). Each participant received a sealed file filled with
BRCA test result (excluding their clinical records). Genetic
consultation was provided in order to better explain the
BRCA test results. The participants did not encounter any
financial costs (including genetic consultation) for their
participation in the present study.
Recruitment criteria
The recruitment criteria adhered to the breast
cancer diagnosis and treatment guidelines and specifications
(Chinese Cancer Society, V2015). Participants who met one of the
following criteria were included in the study: i) BC with onset age
≤35 years old (early-onset BC group), ii) ≥1 relativea
(either sex) from the same side of the family as BC patient and
(or) ovarian cancer (BC/OC) diagnosed at any age (BC/OC family
history group), iii) two primary BC cancersb [bilateral
BC (BBC) group], iv) male BC patients (MBC group), v) BC with OC
(BC&OC group) and vi) meeting ≥2 criteria above simultaneously
(mixed group). aIncluding first, second and third degree
relative with no age limitation. bTwo primary BC,
bilateral BC excluding metastatic contralateral BC or unilateral BC
including two or more different types of cancers.
In case the patients failed to sign the informant
consent, they were excluded from the present study.
BRCA gene analysis
BRCA1 (MIM:113705) and BRCA2
(MIM:600185) testing was performed using Annoroad Gene Technology
(Beijing, China) on an Illumina HiSeq 2500 platform (Illumina, San
Diego, CA, USA). Genomic DNA was first extracted from peripheral
blood white cells using the QIAGEN DNeasy Blood and Tissue Kit
(Qiagen, Shanghai, China). The genomic DNA was then fragmented by a
bioruptor sonication device (Diagenode, Leige, Belgium). The
construction and capture of the DNA library followed the standard
protocols from Illumina (Illumina, San Diego, CA, USA) and Roche
(Roche, Shanghai, China). A Qubit 3.0 Fluorometer (Invitrogen, San
Francisco, CA, USA) and Bioanalyzer (Agilent, Santa Clara, CA, USA)
were used to determine the quantity of the library. Finally, the
library was sequenced on one lane using 100 paired-end
(2x100 bp) strategies.
Variant nomenclature
Reference sequences used for BRCA1 and
BRCA2 analyses were GenBank NM_007294.2 (BRCA1) and
NM_000059.3 (BRCA2). Mutation nomenclature was described
according to Human Genome Variation Society (v2.0) (15).
Statistical analysis
Medians were used with interquartile ranges of
abnormally distributed data for continuous variables (diagnosed
ages of patients) and rank test for analyses. Proportions were
shown for categorical variables. Comparisons of mutation rates and
proportions were analyzed by Chi-square test of unordered
categorical variable. Univariate and multivariate logistic analysis
were used to examine the relationships between hormone receptors
(HRs, including estrogen receptor ER, progesterone receptor PR) and
human epidermal growth factor receptor 2 (HER2) and BRCA
mutation states with odd ratios (OR) and 95% confidence intervals
(95% CI).
All P-values were two-sided. P<0.05 was
considered to indicate a statistically significant difference. All
data were analyzed using PASW Statistics 22.0 (SPSS, Inc., Chicago,
IL, USA).
Results
Study population
From a total of 437 BC patients enrolled in the
study, almost half came from north China (Table I). Forty percent of early-onset BCs
(173/437) and 30% of patients with BC/OC family history (163/437)
were observed in more than half of the participants (Table II). The median age of 437 BC patients
was 35.0 (31.0, 46.0) years. The median age of mutation carriers
was higher compared to non-carriers [41.0 (34.0, 47.0) vs. 35.0
(30.0, 46.0), P=0.002].
 | Table I.The geographic mutation rates
(n=437). |
Table I.
The geographic mutation rates
(n=437).
| Area | N | BRCA+a (%) | P-value |
|---|
| North China | 201 | 31 (15.4) | 0.5 |
| South China | 151 | 30 (19.9) |
|
| Northwest
China | 85 | 15 (17.6) |
|
| Total | 437 | 76 (17.4) |
|
 | Table II.The mutation rates in the recruited
groups (n=437). |
Table II.
The mutation rates in the recruited
groups (n=437).
| Recruit
Criteria | N | BRCA+a (%) | P-value |
|---|
| EOb | 173 | 16 (9.2) |
|
| BC/OC FH | 129 | 28 (21.7) |
|
| BBC | 50 | 12 (24.0) |
|
| MBC | 9 | 2 (22.2) | 0.007 |
| BC&OC | 6 | 1 (16.7) |
|
| Fixed | 70 | 17 (24.3) |
|
| Total | 437 | 76 (17.4) |
|
Mutation frequency
Seventy-six (17.4%) BRCA mutation carriers
were identified, 31 (15.4%) of which were from patients from North
China, 30 (19.9%) from patients from South China, and 15 (17.6%)
from patients from Northwest China. No significant difference in
gene mutation rates was found between different region areas
(Table I). Furthermore, the
early-onset patient rate (9.2%) was significantly different in
relation to remaining 5 groups (P=0.007) (Table II); while no significant difference
was found between the 5 groups (P>0.05).
BRCA mutation status
According to the American College of Medical
Genetics and Genomics (ACMG) (16), a
total of 61 deleterious mutation points (29 in BRCA1, 32 in
BRCA2) were observed in 76 carriers (Table III), and consequently classified
into the already ‘known’ (Table IV)
and the ‘novel’ (Table V) mutations.
Briefly, 72% of novel variations were found in BRCA2
(Tables IV and V). Moreover, five mutations of 61 were
observed in more than one unrelated patients from different areas
(Fig. 2 and Table IV).
 | Table III.Deleterious and novel mutations
(n=61). |
Table III.
Deleterious and novel mutations
(n=61).
| Gene | Known (%) | Novel (%) | Total |
|---|
| BRCA1 | 21 (65.6) | 8 (27.6) | 29 |
| BRCA2 | 11 (34.4) | 21 (72.4) | 32 |
| Total | 32 (52.5) | 29 (47.5) | 61 |
 | Table IV.BRCA known deleterious
mutation sites (n=32). |
Table IV.
BRCA known deleterious
mutation sites (n=32).
| Gene | Location | Exon | Mutation type | AA change | Probands |
|---|
| BRCA1 |
c.190T>Cb | 4 | M | p.Cys64Arg | HOBC |
|
|
c.212G>Ac | 4 | M | p.Arg71Lys | HBOC |
|
|
c.212+1G>Tb | Intr | S | – | MBC/BCFH |
|
|
c.441+1G>A | Intr | S | – | EO |
|
|
c.1660G>Ta | 10 | N | p.Glu554Ter | BCFH |
|
|
c.1674del | 10 | FS | p.Gly559fs | MBC |
|
|
c.2014A>Tb | 10 | N | p.Lys672Ter | BCFH |
|
|
c.2572C>Td | 10 | N |
| BBC |
|
|
c.3329dup | 10 | FS | p.Gln1111fs | EO |
|
|
c.3400G>T | 10 | N | p.Glu1134Ter | BBC/BCFH |
|
|
c.3472G>T | 10 | N | p.Glu1158Ter | EO |
|
|
c.3607C>T | 10 | FS | p.Arg1203Ter | BCFH |
|
|
c.3626T>G | 10 | N | p.Leu1209Ter | BBD |
|
|
c.3640G>Tb | 10 | N | p.Glu1214Ter | BCFH |
|
|
c.4065_4068del | 10 | FS | p.Asn1355fs | HBOC |
|
|
c.4484+1G>A | Intr | S | – | BCFH |
|
|
c.4801A>Td | 15 | N | p.Lys1601Ter | HBOC |
|
|
c.5251C>T | 19 | N | p.Arg1751Ter | EO |
|
|
c.5278-1G>C | Intr | S | – | BBD |
|
|
c.5431C>T | 22 | N | p.Gln1811Ter | HBOS |
|
|
c.5470_5477deld | 23 | FS | p.Ile1824fs | BBD/BCFH |
| BRCA2 |
c.961C>T | 10 | N | p.Gln321Ter | EO |
|
|
c.1310_1313delb | 10 | FS | p.Lys437Ilefs | BCFH |
|
|
c.1399A>T | 10 | N | p.Lys467Ter | EO |
|
|
c.2806_2809deld | 11 | FS | p.Asp936fs | EO |
|
|
c.3109C>Ta,d | 11 | FS | p.Gln1037Ter | BBD/BCFH |
|
|
c.5682C>Aa | 11 | N | p.Tyr1894Ter | BCFH |
|
|
c.7007G>Ta | 14 | S | p.Arg2336Leu | MBC |
|
|
c.8504C>G | 20 | N | p.Ser2835Ter | BCFH |
|
|
c.8517C>A | 20 | N | p.Tyr2839Ter | BCFH |
|
|
c.9100C>T | 23 | N | p.Gln3034Ter | EO |
|
|
c.9117G>A | 23 | Syn | p.Pro3039= | EO |
 | Table V.Novel variations (n=29). |
Table V.
Novel variations (n=29).
| Gene | Location | Exon | Mutation type | AA change | Probands |
|---|
| BRCA1 |
c.1934del | 10 | FS | p.Ser645fs | EO |
|
|
c.2957del | 10 | FS | p.Ile986fs | HBOC |
|
|
c.3294del | 10 | FS | p.Leu1098fs | BCFH |
|
|
c.3621del | 10 | FS | p.Lys1207fs | BBD |
|
|
c.3859del | 10 | FS | p.Glu1287fs | BCFH |
|
|
c.4013del | 10 | FS | p.Lys1338fs | EO |
|
|
c.4676-1G>T | Intr | S | – | EO |
|
|
c.5156del | 18 | FS | p.Val1719fs | BCFH |
| BRCA2 | c.31del | 2 | FS | p.Phe11fs | EO |
|
|
c.767_771del | 9 | FS | p.Thr256fs | EO |
|
|
c.988del | 10 | FS | p.Lys330fs | BCFH |
|
|
c.3364del | 11 | FS | p.Gly1122fs | EO |
|
|
c.426-2A>T | Intr | S | – | HBOC |
|
|
c.4410_4413del | 11 | FS | p.Ile1470fs | BCFH |
|
|
c.5480del | 11 | FS | p.Ile1827fs | BCFH |
|
|
c.5495del | 11 | FS | p.Ser1832fs | MBC/BCFH |
|
|
c.5599_5602del | 11 | FS | p.Tre1867fs | EO |
|
|
c.5718_5719del | 11 | FS | p.Asn1906fs | BBC |
|
|
c.5753dela | 11 | FS | p.His1918fs | BCFH |
|
|
c.6288_6289dela | 11 | FS | p.Pro2096fs | BCFH |
|
|
c.6462_6465del | 11 | FS | p.Tyr2154fs | BBC/EO |
|
|
c.6552del | 11 | FS | p.Glu2184fs | BCFH |
|
|
c.6698_6699insTTTT | 11 | FS | p.Ala2233fs | HBOC |
|
|
c.7178_7179del | 14 | FS | p.Met2393fs | BCFH |
|
|
c.8019_8020insAT | 18 | N | p.Lys2673fs | BBD |
|
|
c.8039_8040del | 18 | FS | p.Asp2680fs | EO |
|
| c.8367_8369
TAC>A | 19 | FS | p.Tyr2789Ter | EO |
|
|
c.8400_8402delinsAAAA | 19 | FS | p.Phe2801fs | EO |
|
|
c.9090dupa,b | 23 | FS | p.Thr3030fs | EO |
Thirty-four points (55.7%) were frame shift,
followed by 17 (27.9%) nonsenses, 7 splices (11.5%), 2 pathogenic
missenses (3.3%), and 1 synonymous mutation (1.6%). Extra 10
missenses were found as variants of uncertain significance (VUS)
(Table VI), accounting for 14.1% of
all the variants including 61 deleterious mutation.
 | Table VI.Information of VUS (n=10). |
Table VI.
Information of VUS (n=10).
| Gene | Location | AA change | Proband | Family history |
|---|
| BRCA1 |
c.446A>C | p.E149A | p.(Glu149Ala) | EO | – |
|
|
c.1669A>C | p.T557P | p.(Thr557Pro) | MBC (TNBC) | Uterine cancer |
|
|
c.4580A>T | p.E1527V | p.(Glu1527Val) | BBC | – |
|
|
c.5156T>C | p.V1719A | p.(Val1710Ala) | BBC | – |
|
|
c.5498T>A | p.V1833E | p.(Val1833Glu) | MBC | – |
| BRCA2 |
c.6875A>C | p.E2292A | p.(Glu2292Ala) | BBC (TNBC) | – |
|
|
c.7811T>C | p.L2604P | p.(Leu260Pro) | EO | – |
|
|
c.7967T>C | p.L2656P | p.(Leu2656Pro) | EO | – |
|
|
c.8162T>C | p.L2721P | p.(Leu2721Pro) | BBC | one BC sister |
|
|
c.9374T>C | p.L3125P | p.(Leu3125Pro) | BBC (TNBCs) | – |
Eleven different mutations in 10 families were
related to hereditary breast and ovarian cancer syndrome (HBOC),
four in BRCA2 were novel (Table
VII). Moreover, none of the identified mutations were shared
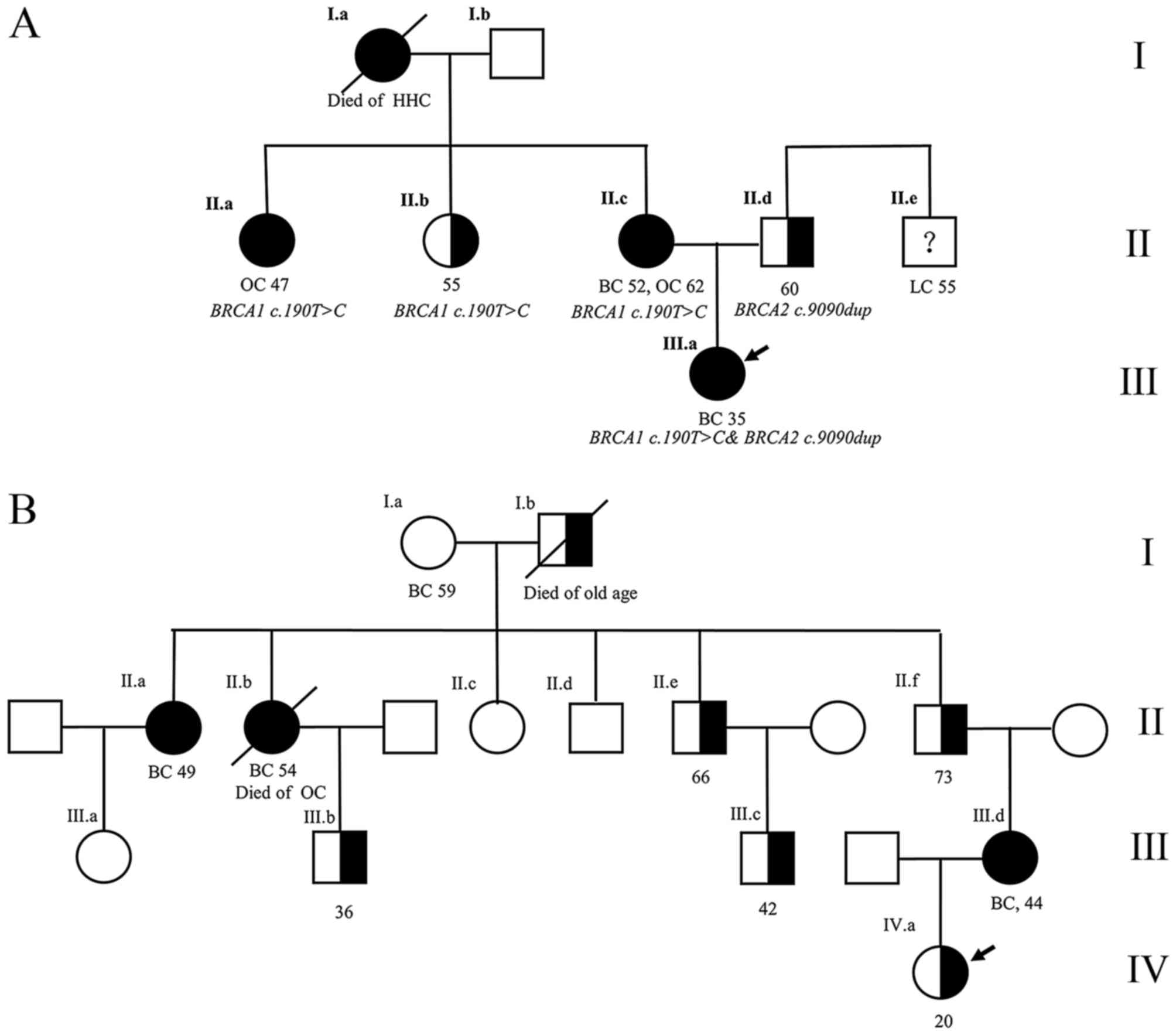
between the families. BRCA1 c.190T>C&BRCA2 c.9090dup
were found in one same family, and were carried by a proband from
maternal and paternal line respectively. The proband's BC was
inherited from maternal HBOC (Fig.
3A). In BRCA1 c.5431C>T hereditary family, all the
middle-age females had BC/OC, but all the male carriers of four
general relatives were healthy (Fig.
3B).
 | Table VII.Familial mutations information
(n=11). |
Table VII.
Familial mutations information
(n=11).
| Gene | Location | Family history
features |
Chinesea |
|---|
| BRCA1 |
c.190T>Cb | HBOC of maternal
hereditary | N |
|
|
c.1660G>T | BC sisters | Y |
|
|
c.2014A>T | BCs of maternal
history | N |
|
|
c.212+G>T | BCs of maternal
history | Y |
|
|
c.3640G>T | BC sisters | N |
|
|
c.5431C>T | HBOC of paternal
hereditary | N |
| BRCA2 |
c.988del | BC sisters | N (Novel) |
|
|
c.1310_1313del | BCs maternal
history | N |
|
|
c.5753del | BCs maternal
history | N (Novel) |
|
|
c.6288_6289del | BCs maternal
history | N (Novel) |
|
|
c.9090dupb | Paternal line | N (Novel) |
Clinical analysis
410 precise tumor node metastasis TNM results were
obtained. No significant difference was found among wild type
BRCA (BRCA−), BRCA1 and
BRCA2 mutation patients (Table
VIII).
 | Table VIII.TNM in three BRCA groups
(n=410). |
Table VIII.
TNM in three BRCA groups
(n=410).
|
| TNM (%) |
|
|---|
|
|
|
|
|---|
| Gene | I (n=126) | II (n=199) | III (n=67) | IV (n=18) | P-trend |
|---|
|
BRCA− | 99
(28.9)a | 171 (49.9) | 57 (16.6) | 16 (4.7) | 0.33 |
| BRCA1 | 17 (48.6) | 14 (40) | 3 (8.6) | 1 (2.9) |
|
| BRCA2 | 10 (31.3) | 14 (43.8) | 7 (21.9) | 1 (3.1) |
|
Univariate analysis demonstrated higher expression
of ER negative (ER−) and PR negative (PR−) in
BRCA1 group compared to BRCA− group (72.6%
vs. 35.1%, P<0.0001; 82.4% vs. 40.6%, P<0.0001). Conversely,
higher expression of ER positive (ER+) and PR positive
(PR+) were found in BRCA2 group compered to
BRCA− group patients (93.5% vs. 64.9%, P=0.001;
87.1% vs. 59.4%, P=0.002). Moreover, BRCA1 and BRCA2
groups were both more frequently HER2 negative (HER2−)
compared to BRCA− group (97.1, 96.8% vs. 74.3%,
P=0.003 and 0.005 respectively). Based on the multivariate
analysis, PR− and HER2− were the independent
risk factors for BRCA1 mutation, HER2− alone for
BRCA2 (Tables IX and X). Then, compared with
BRCA−, BRCA1 mutation tended to be TNBC
(68.6% vs. 24.8%, P<0.0001), while BRCA2 mutation had
higher proportion of HRs positive (HRs+) BC (93.5% vs.
75.2%, P=0.002) (Table XI).
 | Table IX.HRs and HER2 comparisons between
BRCA1 and BRCA− groups (n=376a). |
Table IX.
HRs and HER2 comparisons between
BRCA1 and BRCA− groups (n=376a).
| Molecular
markers | BRCA1
(%) |
BRCA−b (%) | Univariate
P-value | Multivariate
P-value | OR (95%CI) |
|---|
| ER |
|
|
|
|
|
|
<1% | 24 (70.6) | 120 (35.1) | <0.0001 | 0.73 | 1.2 (0.4–3.3) |
|
≥1% | 10 (29.4) | 222 (64.9) | – |
|
|
| PR |
|
|
|
|
|
|
<1% | 28 (82.4) | 139 (40.6) | <0.0001 | 0.003 | 6.3 (1.9–20.6) |
|
≥1% | 6 (17.6) | 203 (59.4) | – |
|
|
| HER2 |
|
|
|
|
|
|
− | 33 (97.1) | 254 (74.3) | 0.003 | 0.01 | 12.7
(1.7–95.6) |
|
+ | 1 (2.9) | 88 (25.7) | – |
|
|
 | Table X.HRs and HER2 comparisons between
BRCA2 and BRCA- groups (n=373a). |
Table X.
HRs and HER2 comparisons between
BRCA2 and BRCA- groups (n=373a).
| Molecular
markers | BRCA2
(%) |
BRCA−b (%) | Univariate
P-value | Multivariate
P-value | OR (95%CI) |
|---|
| ER |
|
|
|
|
|
|
<1% | 2 (6.5) | 120 (35.1) | 0.001 | 0.09 | 0.2 (0.4–1.3) |
|
≥1% | 29 (93.5) | 222 (64.9) | – |
|
|
| PR |
|
|
|
|
|
|
<1% | 4 (12.9) | 139 (40.6) | 0.002 | 0.41 | 0.6 (0.2–2.2) |
|
≥1% | 27 (87.1) | 203 (59.4) | – |
|
|
| HER-2 |
|
|
|
|
|
|
− | 30 (96.8) | 254 (74.3) | 0.005 | 0.03 | 9.4 (1.3–71.0) |
|
+ | 1 (3.2) | 88 (25.7) |
|
|
|
 | Table XI.Molecular types comparison in three
BRCA groups (n=409a). |
Table XI.
Molecular types comparison in three
BRCA groups (n=409a).
| Gene | TNBC (%) | HR+b (%) | P-value |
|---|
|
BRCA-c | 85 (24.8) | 258 (75.2) | – |
| BRCA1 | 24 (68.6) | 11 (31.4) | <0.0001 |
| BRCA2 | 2 (6.5) | 29 (93.5) | 0.02 |
| Total | 111 | 298 |
|
Discussion
The results from the present study, which is to our
knowledge, the largest screening study ever performed in China,
reveal that the total BRCA mutation rate is 17.4% for breast
cancer patients at risk of hereditary BRCA mutation across
China with no observed geographical differences.
One of the main components of the present study has
to do with 21.7% of mutation rate which lies in BC/OC family
history subgroup. This finding is in line with a Korean study
conducted across 36-centers (22.3%) (17). The mutation rate appears generally
lower compared to Western countries (23~35.3%) (18–22), but
higher compared to Peking or Shanghai regions (10.5~18.2%)
(23–25). Lower prevalence of BRCA
mutation is in line with comparisons of BC incidence with the
Western countries. It clearly suggests essential distinction in
BRCA mutation between Asian and Euromerican people that goes
well beyond different study design biases. Furthermore, thus far
observed domestic inconsistencies may be caused by the limitations
related to areas and criteria. The present study covers most of the
Chinese regions, i.e. areas with huge concentrations of Chinese
populations. Moreover, the present study does not impose the BC
onset age limitation for family cancer history, which allows for
wider screening rang. However, some families with late-onset
hereditary BC/OC that are really in need of BRCA testing may
be excluded from testing due to young cutoff diagnose age
established by BRCA testing guideline. Most of all, our
results are representative of the real data on hereditary risk for
breast cancer patients in China.
The sporadic early-onset (≤35 years) BC mutation
rates (9.2%) are nearly twice higher compared to those obtained by
the recent studies from China (5%, ≤40 years) (24) and from Western countries (5.9%, <36
years) (26), but lower compared to
other 5 risk factor groups in this study. The observed discrepancy
may come from different sample sizes and study populations.
Moreover, different age limitations for early-onset suggest that
cutoff age may impact the mutation rates in younger patients.
Currently, the age of 45 is set as upper limitation of young BC for
BRCA testing according to Chinese BC Treatment Guideline
(27). Nevertheless, increasing trend
of breast cancer incidence in younger patients in China (6) may imply more patients with sporadic
early-onset from the whole population. Ten years interval, between
the ages 35 and 45, may actually double the difference on
sensitivity and specificity for BRCA screening in Chinese
patients with breast cancer. Additionally, besides onset age,
family history, bilateral BC, with OC and similar should also be
considered in the early-onset BC to improve BRCA test
indications (24). Consequently, we
suggest re-evaluation of the early-onset age for BRCA test
to increase sensitivity of BRCA mutation screening and to
fit current BC epidemiology in China.
BRCA mutation sites in our study suggest
special features of BRCA mutation in China. Four sites are
hereditary, and nearly half are novel. Also, 14.1% VUS were
slightly higher than Asian data (13.6%) obtained from a study
conducted by Hall et al. Interestingly, their data were
already more than 2.5 times higher compared to European VUS
(28). Then, BRCA1
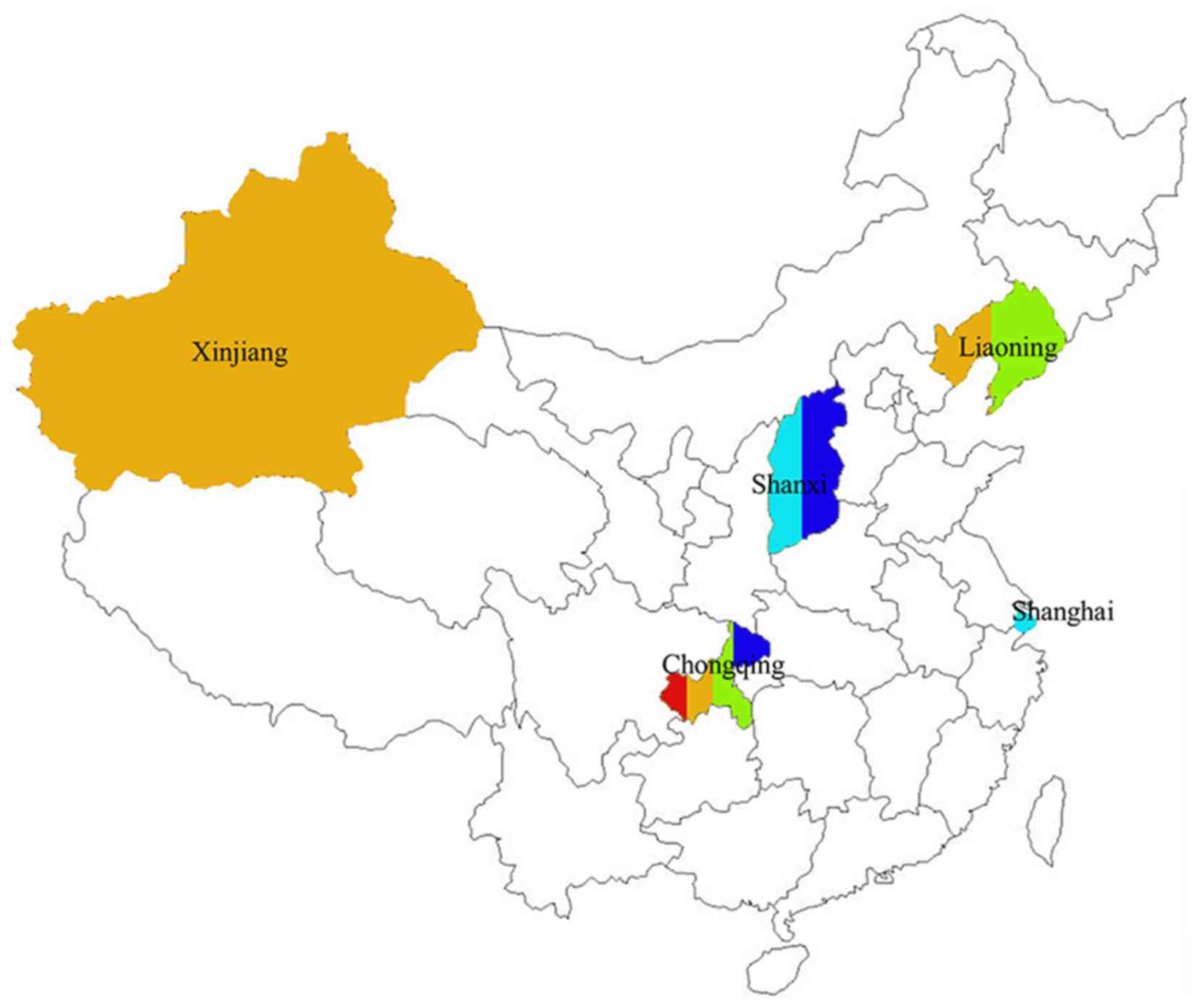
c.2572C>T reappeared twice in two unrelated patients from
Chongqing and Shanxi Provinces, respectively. Up to now, it has
been found in unrelated patients from five different provinces in
China (29). Nevertheless, it has not
been frequently reported in international studies, meaning it might
be one of founder mutation candidates among Chinese populations.
BRCA1 c.5470_5477del is another highly repeated site found
three times in our study. Internationally, the site has been
previously identified in Korean sample (30), while nationally it has been reported
in Shanghai (31), Beijing (32), Zhejiang, and Liaoning (33). Thus far, it has been found in 12
unrelated carriers from six different areas of China, including
HBOC patients (33.3%) (32), sporadic
BC patients (33.3%) (31,33) and early-onset BC (33) or BBCpatients (33.3%) (the present
study). Also, three of the patients (from Zhejiang, Shanghai, and
Liaoning) share the same haplotype (33). To our knowledge, it has only once been
reported in native Asian patient (34), therefore, it is strongly associated
with Asian founder mutation. However, Kwong et al have
recognized BRCA2 c.3109C>T as a founder mutation in
southern Chinese population in Hong Kong (35,36). 81.5%
of probands in the study were immigrants from Guangdong. It was
repeated in two unrelated patients from Chongqing, southwest of
China, but it did not appear in our population sample from southern
China. Further large scale unselected population epidemic research
is necessary to clarify this.
Two confirmed HBOC families have special features.
The proband who carried two mutations is the youngest patient in
her family, thus it is not possible to ignore the effects of
paternal inherited BRCA2 c.9090dup (novel) on the proband
given her uncle has lung cancer (Fig.
3A). BRCA1 c.5431C>T pathogenicity shows a gender
trend: It appears harmful for women, and silent for men (Fig. 3B).
All the information on the mutation sites reported
above suggest quite different BRCA mutation spectrum and
features that exit in Chinese BC patients compared with well-known
white populations.
Similar to other studies, we came to the conclusion
that BRCA1 mutation is concerned with TNBC. However,
BRCA2 mutations tend to be HRs+ BC, which is
inconsistent with results from other studies (37,38). The
different results show heterogeneous of BRCA2 mutant BC
beyond different study populations and regions (39,40). The
present study suggests that BRCA2 mutant BC may responds
better to endocrine therapy due to high proportion of
HRs+ tumor with the same background of hereditary risk
of BC compared with BRCA− and BRCA1 mutant
BC.
In summary, this study provides a general
BRCA mutation profile in China, which enhances the
prevalence of BRCA mutations in non-white populations. The
BRCA screening provides a distinguishing BRCA
mutation profile in China, which compared to the West reveals lower
mutation prevalence, and special mutation spectrum. Cutoff ages for
diagnosis of early-onset and BC/OC family history should be
re-evaluated based on population screening data to improve
BRCA test indications. BRCA2 mutation suggests the
best response to endocrine therapy among BRCA mutant and
BRCA− BCs in this selected hereditary risk
population. However, further studies are necessary to confirm
precise BRCA mutation situation in China.
Acknowledgements
The authors would like to thank Zefei Jiang,
Mengmeng Zhang, Shaohua Wei, Yuanyuan Wang, Dawei Yun, Huiquan
Jiang, Yang Li, Yimeng Hu, Yong Yang and other medical staff for
their involvement in the study design, BRCA lab test, information
collection, data analysis and manuscript writing. The present study
was supported by the China Health Promotion Foundation.
References
|
1
|
Fan L, Zheng Y, Yu KD, Liu GY, Wu J, Lu
JS, Shen KW, Shen ZZ and Shao ZM: Breast cancer in a transitional
society over 18 years: Trends and present status in Shanghai,
China. Breast Cancer Res Treat. 117:409–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hemminki K, Mousavi SM, Sundquist J and
Brandt A: Does the breast cancer age at diagnosis differ by
ethnicity? A study on immigrants to Sweden. Oncologist. 16:146–154.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng S, Bai JQ, Li J, Fan JH, Pang Y,
Song QK, Huang R, Yang HJ, Xu F, Lu N and Qiao YL: The pathologic
characteristics of breast cancer in China and its shift during
1999–2008: A national-wide multicenter cross-sectional image over
10 years. Int J Cancer. 131:2622–2631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song QK, Wang XL, Zhou XN, Yang HB, Li YC,
Wu JP, Ren J and Lyerly HK: Breast cancer challenges and screening
in China: Lessons from current registry data and population
screening studies. Oncologist. 20:773–779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Si W, Li Y, Han Y, Zhang F, Wang Y, Li Y,
Linghu RX, Zhang X and Yang J: Epidemiological and
clinicopathological trends of breast cancer in Chinese patients
during 1993 to 2013: A retrospective study. Medicine (Baltimore).
94:e8202015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lynch HT, Snyder C and Lynch J: Hereditary
breast cancer: Practical pursuit for clinical translation. Ann Surg
Oncol. 19:1723–1731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shuen AY and Foulkes WD: Inherited
mutations in breast cancer genes-risk and response. J Mammary Gland
Biol Neoplasia. 16:3–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kurian AW, Gong GD, John EM, Miron A,
Felberg A, Phipps AI, West DW and Whittemore AS: Performance of
prediction models for BRCA mutation carriage in three racial/ethnic
groups: Findings from the Northern California Breast Cancer Family
Registry. Cancer Epidemiol Biomarkers Prev. 18:1084–1091. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurian AW, Gong GD, Chun NM, Mills MA,
Staton AD, Kingham KE, Crawford BB, Lee R, Chan S, Donlon SS, et
al: Performance of BRCA1/2 mutation prediction models in Asian
Americans. J Clin Oncol. 26:4752–4758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vogel KJ, Atchley DP, Erlichman J, Broglio
KR, Ready KJ, Valero V, Amos CI, Hortobagyi GN, Lu KH and Arun B:
BRCA1 and BRCA2 genetic testing in Hispanic patients: Mutation
prevalence and evaluation of the BRCAPRO risk assessment model. J
Clin Oncol. 25:4635–4641. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim YC, Zhao L, Zhang H, Huang Y, Cui J,
Xiao F, Downs B and Wang SM: Prevalence and spectrum of BRCA
germline variants in mainland Chinese familial breast and ovarian
cancer patients. Oncotarget. 7:9600–9612. 2016.PubMed/NCBI
|
|
15
|
den Dunnen JT and Antonarakis SE: Mutation
nomenclature extensions and suggestions to describe complex
mutations: A discussion. Hum Mutat. 15:7–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang E, Seong MW, Park SK, Lee JW, Lee J,
Kim LS, Lee JE, Kim SY, Jeong J, Han SA, et al: The prevalence and
spectrum of BRCA1 and BRCA2 mutations in Korean population: Recent
update of the Korean hereditary breast cancer (KOHBRA) study.
Breast Cancer Res Treat. 151:157–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seong MW, Cho SI, Kim KH, Chung IY, Kang
E, Lee JW, Park SK, Lee MH, Choi DH, Yom CK, et al: A
multi-institutional study of the prevalence of BRCA1 and BRCA2
large genomic rearrangements in familial breast cancer patients.
BMC Cancer. 14:6452014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weitzel JN, Clague J, Martir-Negron A,
Ogaz R, Herzog J, Ricker C, Jungbluth C, Cina C, Duncan P, Unzeitig
G, et al: Prevalence and type of BRCA mutations in Hispanics
undergoing genetic cancer risk assessment in the southwestern
United States: A report from the Clinical Cancer Genetics Community
Research Network. J Clin Oncol. 31:210–216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blay P, Santamaria I, Pitiot AS, Luque M,
Alvarado MG, Lastra A, Fernández Y, Paredes A, Freije JM and Balbin
M: Mutational analysis of BRCA1 and BRCA2 in hereditary breast and
ovarian cancer families from Asturias (Northern Spain). BMC Cancer.
13:2432013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Konstantopoulou I, Tsitlaidou M, Fostira
F, Pertesi M, Stavropoulou AV, Triantafyllidou O, Tsotra E,
Tsiftsoglou AP, Tsionou C, Droufakou S, et al: High prevalence of
BRCA1 founder mutations in Greek breast/ovarian families. Clin
Genet. 85:36–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Riahi A, Kharrat M, Ghourabi ME, Khomsi F,
Gamoudi A, Lariani I, May AE, Rahal K and Chaabouni-Bouhamed H:
Mutation spectrum and prevalence of BRCA1 and BRCA2 genes in
patients with familial and early-onset breast/ovarian cancer from
Tunisia. Clin Genet. 87:155–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Pei R, Pang Z, Ouyang T, Li J,
Wang T, Fan Z, Fan T, Lin B and Xie Y: Prevalence and
characterization of BRCA1 and BRCA2 germline mutations in Chinese
women with familial breast cancer. Breast Cancer Res Treat.
132:421–428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Sun J, Chen J, Yao L, Ouyang T,
Li J, Wang T, Fan Z, Fan T, Lin B and Xie Y: Comprehensive analysis
of BRCA1 and BRCA2 germline mutations in a large cohort of 5931
Chinese women with breast cancer. Breast Cancer Res Treat.
158:455–462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang X, Wu J, Lu J, Liu G, Di G, Chen C,
Hou Y, Sun M, Yang W, Xu X, et al: Identification of a
comprehensive spectrum of genetic factors for hereditary breast
cancer in a Chinese population by next-generation sequencing. PLoS
One. 10:e01255712015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peto J, Collins N, Barfoot R, Seal S,
Warren W, Rahman N, Easton DF, Evans C, Deacon J and Stratton MR:
Prevalence of BRCA1 and BRCA2 gene mutations in patients with
early-onset breast cancer. J Natl Cancer Inst. 91:943–949. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Anonymity: Guidelines and specifications
for the diagnosis and treatment of breast cancer in Chinese Cancer
Society (Version 2015). China Oncology. 25:692–754. 2015.
|
|
28
|
Hall MJ, Reid JE, Burbidge LA, Pruss D,
Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA and Noll
WW: BRCA1 and BRCA2 mutations in women of different ethnicities
undergoing testing for hereditary breast-ovarian cancer. Cancer.
115:2222–2233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li WF, Hu Z, Liu XY, Zhang B, Cao MZ, Wang
YS, Zhao L, Liu YB, Yuan WT, Shen ZZ, et al: BRCA1 germ line
mutations in Chinese early-onset breast cancer patients. Zhonghua
Yi Xue Yi Chuan Xue Za Zhi. 24:499–504. 2007.(In Chinese).
PubMed/NCBI
|
|
30
|
Choi DH, Lee MH, Bale AE, Carter D and
Haffty BG: Incidence of BRCA1 and BRCA2 mutations in young Korean
breast cancer patients. J Clin Oncol. 22:1638–1645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suter NM, Ray RM, Hu YW, Lin MG, Porter P,
Gao DL, Zaucha RE, Iwasaki LM, Sabacan LP, Langlois MC, et al:
BRCA1 and BRCA2 mutations in women from Shanghai China. Cancer
Epidemiol Biomarkers Prev. 13:181–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li N, Zhang X, Cai Y, Xu X, Zhang L, Pan
KF, Wu LY and Wang MR: BRCA1 germline mutations in Chinese patients
with hereditary breast and ovarian cancer. Int J Gynecol Cancer. 16
Supp 1:S172–S178. 2006. View Article : Google Scholar
|
|
33
|
Hu Z, Li WF, Liu XY, Zhang B, Cao MZ, Wang
YS, Zhao L, Song CG, Lu JS, Wu J, et al: 5589del8: The recurrent
mutation of BRCA1 gene in Chinese breast cancer patients. Zhonghua
Yi Xue Yi Chuan Xue Za Zhi. 24:378–381. 2007.(In Chinese).
PubMed/NCBI
|
|
34
|
Anonymity: ClinVar, . NCBI. https://www.ncbi.nlm.nih.gov/clinvar/variation/55591/#summary-evidence12–Jan.
2016
|
|
35
|
Kwong A, Wong LP, Wong HN, Law FB, Ng EK,
Tang YH, Chan WK, Ho LS, Kwan KH, Poon M, et al: A BRCA2 founder
mutation and seven novel deleterious BRCA mutations in southern
Chinese women with breast and ovarian cancer. Breast Cancer Res
Treat. 117:683–686. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kwong A, Ng EK, Wong CL, Law FB, Au T,
Wong HN, Kurian AW, West DW, Ford JM and Ma ES: Identification of
BRCA1/2 founder mutations in Southern Chinese breast cancer
patients using gene sequencing and high resolution DNA melting
analysis. PLoS One. 7:e439942012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Adem C, Soderberg CL, Hafner K, Reynolds
C, Slezak JM, Sinclair CS, Sellers TA, Schaid DJ, Couch F, Hartmann
LC and Jenkins RB: ERBB2, TBX2, RPS6KB1, and MYC alterations in
breast tissues of BRCA1 and BRCA2 mutation carriers. Genes
Chromosomes Cancer. 41:1–11. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Eerola H, Heikkilä P, Tamminen A,
Aittomäki K, Blomqvist C and Nevanlinna H: Histopathological
features of breast tumours in BRCA1, BRCA2 and mutation-negative
breast cancer families. Breast Cancer Res. 7:R93–R100. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shao J, Yang J, Wang JN, Qiao L, Fan W,
Gao QL and Feng YJ: Effect of BRCA2 mutation on familial breast
cancer survival: A systematic review and meta-analysis. J Huazhong
Univ Sci Technolog Med Sci. 35:629–634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhong Q, Peng HL, Zhao X, Zhang L and
Hwang WT: Effects of BRCA1- and BRCA2-related mutations on ovarian
and breast cancer survival: A meta-analysis. Clin Cancer Res.
21:211–220. 2015. View Article : Google Scholar : PubMed/NCBI
|