Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally (1). Non-small-cell lung cancer (NSCLC) is the
most common type of lung cancer, accounting for ~85% of all lung
cancers, and >40% of NSCLC cases are metastatic (stage IV) at
the point of diagnosis (2,3). Lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC) are the major pathological subtypes
of NSCLC. NSCLC treatment has improved in the past decade, with
more effective screening, diagnosis, and systemic treatments
contributing to increased survival rates (3). Lung cancer cells frequently participate
in cancer-associated pathways, including growth signaling,
programmed cell death, angiogenesis, cell invasion, and metastasis
(4). Transformation from a normal
cell to a malignant lung cancer cell phenotype is considered to
occur in a multistep manner through a series of genetic and
epigenetic alterations; ultimately evolving into invasive cancer by
clonal expansion (5). Dysfunctional
noncoding RNAs, including microRNAs (miRNAs/miRs), have been
reported to be critical in carcinogenesis (6). MiRNAs are small, non-protein-coding
RNAs, 19–24 nucleotides in length, which negatively modulate gene
expression by suppressing mRNA translation or accelerating mRNA
degradation (7–9). A single miRNA may regulate >100
target genes by targeting their 3′-untranslated regions. Therefore,
miRNAs typically only fine-tune mRNA expression, suggesting that
co-expressed miRNAs may function simultaneously to co-modulate a
signaling pathway to achieve a biological function (10). Numerous dysregulated miRNAs have been
reported in lung cancer cells, including let-7 and miR-17, −20a,
−29, −107, −126, −138, and −185 (11–17).
Dysfunctional expression of these miRNAs serves a critical function
in the co-modulation of cell cycle progression, angiogenesis,
apoptosis, adhesion and motility of lung cancer cells. In addition
to miRNA-mRNA interaction networks, miR-#-5p and −3p arm selection
preference is a complex mechanism that regulates the biological
functions of miRNAs. Although the 5p and 3p arms of miRNA are
generated from an identical pre-miRNA structure during the
maturation process, the expression of certain miRNAs is altered in
different tissues, developmental stages and species, as well as
during cancer progression (18–27).
However, the underlying mechanisms and functions of miRNA arm
selection preference in lung cancer cells remain unclear. In the
present study, multiple miRNA candidates with significantly altered
arm selection preference in lung cancer were identified. Among
these, miR-324 was selected for further examination in the present
study. Our previous study revealed that miR-324-5p and −3p serve
tumor suppressive functions in breast cancer and colon cancer
(28). However, the function of
miR-324-5p and −3p remains unclear in lung cancer. The present
study is the first to report that miR-324-5p and −3p serve
oncogenic functions in lung cancer cells. The results suggested
that miR-324 may be a novel potential therapeutic target. These
results may provide novel insights into lung cancer therapy and
direct future research.
Materials and methods
Identification of miRNA arm selection
preference using The Cancer Genome Atlas (TCGA)
A total of 891 small RNA expression profiles of LUAD
(46 normal and 458 tumor tissues) and LUSC (45 normal and 342 tumor
tissues) were downloaded from TCGA (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm).
A total of 253 miRNAs were selected for further examination of
their arm selection preference, with the simultaneous expression of
the 5p and 3p arms (expression of miR-#-5p or miR-#-3p >1
transcripts per million) in LUAD and LUSC. The 5p:3p arm expression
ratio was assessed in LUAD, LUSC, and adjacent normal tissues. A
fold change of >1.5 or <0.75 and P<0.05 was considered to
indicate a significant difference in the arm selection
preference.
Cell lines
A human lung cancer cell line, A549, was obtained
from the American Type Culture Collection (ATCC, Manassas, VA,
USA). The cells were maintained in Dulbecco's modified Eagle's
medium (Biological Industries, Cromwell, CT, USA), supplemented
with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) and penicillin-streptomycin (penicillin,
100 U/ml and streptomycin, 100 µg/ml; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) in a humidified atmosphere containing 5%
CO2 at 37°C.
Ectopic expression of miR-324-5p and
−3p following transfection with mimics
The A549 cells were seeded in a 25T flask at a
density of 1×106 cells/ml and were transfected with 10
nM miRNA-324-5p mimics; sense, 5′-CGCAUCCCCUAGGGCAUUGGUGU-3′ and
antisense, 5′-ACCAAUGCCCUAGGGGAUGCGUU-3′, miR-324-3p mimics; sense,
5′-ACUGCCCCAGGUGCUGCUGG-3′ and antisense,
5′-AGCAGCACCUGGGGCAGUUU-3′ or a scramble control; sense,
5′-GCGACGAUCUGCCUAAGAUdTdT-3′ and antisense,
5′-AUCUUAGGCAGAUCGUCGCdTdT-3′ (GenDiscovery Biotechnology, Inc.,
New Taipei, Taiwan), using Lipofectamine RNAiMAX reagent, according
to the manufacturer's instructions (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). After 24 h of transfection,
total RNA was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), following the manufacturer's
instructions. Briefly, tissue samples were homogenized in 1 ml
TRIzol reagent and mixed with 0.2 ml chloroform for protein
extraction, and RNA was precipitated by adding 0.5 ml isopropanol.
The concentration, purity, and total RNA content were determined
using a NanoDrop 1000 spectrophotometer (Nanodrop Technologies;
Thermo Fisher Scientific, Inc.). The transfection efficiency was
examined through stem-loop reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis.
Stem-loop RT-qPCR
Total RNA (1 µg) was reverse-transcribed in a
stem-loop RT reaction using RT primers and SuperScript III reverse
transcriptase, according to the manufacturer's instructions
(Invitrogen; Thermo Fisher Scientific, Inc.). The RT reaction,
included 1 µg RNA, 1X RT buffer (200 mM Tris-HCl, 500 mM KCl; pH
8.4), 2.5 mM dNTP (Invitrogen; Thermo Fisher Scientific, Inc.) and
0.5 mM stem-loop miR-324-5p or −3p-RT primer (Gemomics BioSci and
Tech, New Taipei, Taiwan). The reaction was performed under the
following incubation conditions: 30 min at 16°C, followed by 50
cycles of 20°C for 30 sec, 42°C for 30 sec and 50°C for 1 sec. The
enzyme was subsequently inactivated by incubation at 85°C for 5
min. qPCR was performed using an miRNA-specific forward primer and
universal reverse primer, and the reaction mixture was incubated at
94°C for 10 min, followed by 40 cycles of 94°C for 15 sec and 60°C
for 32 sec. Gene expression was detected using a SYBR Green I assay
(Applied Biosystems; Thermo Fisher Scientific, Inc.), and miRNA
expression was normalized to that of U6. The primer sequences used
to examine miRNAs were as follows: miR-324-5p-RT primer:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACACCAAT-3′; miR-324-5p
gene-specific forward (GSF), 5′-CGGCGGCGCATCCCCTAGGGCAT-3′;
miR-324-3p-RT, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCAGCAGC-3′;
miR-324-3p-GSF, 5′-CGGCGGACTGCCCCAGGTGC-3′; universal reverse,
5′-CTGGTGTCGTGGAGTCGGCAATTC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation and colony
formation assay
For the clonogenic assay, a gradient number of A549
cells, 1,000, 2,000 and 4,000, were seeded in 6-well plates and
transfected with 10 nM miR-324-5p mimics, miR-324-3p mimics or a
scramble control as aforementioned. The cells were incubated in a
CO2 incubator at 37°C for 2 weeks until colony
formation. The cells were then fixed with 4% formaldehyde
(Sigma-Aldrich; Merck KGaA) for 10 min at room temperature and
stained with crystal violet solution (0.05% crystal violet, 1%
formaldehyde and 1% methanol; all from Sigma-Aldrich; Merck KGaA)
for 20 min at room temperature. Then, the images of a
representative colony formation were examined using a light
microscope (magnification, ×100) (CKX41; Olympus Corporation,
Tokyo, Japan). Finally, 1 ml of 10% acetic acid (Sigma-Aldrich;
Merck KGaA) was added to each well to dissolve the crystal violet.
The absorbance of individual wells was determined at 595 nm by
using Multiskan FC (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). For the cell proliferation assay, 1×103 A549 cells
transfected with 10 nM miR-324-5p mimics, miR-324-3p mimics, or a
scramble control were seeded in a 96-well plate. Proliferation was
determined at 0, 1, 2, 3, and 4 days using the CellTiter-Glo One
Solution assay, according to the manufacturer's instructions
(Promega Corporation, Madison, WI, USA). All experiments were
independently repeated 3 times.
Cell invasion assays
A549 cell invasion was analyzed using Transwell
assays. Briefly, 4.5×105 cells were suspended in a
serum-deprived DMEM (Biological Industries), supplemented with
penicillin-streptomycin (penicillin, 100 U/ml and streptomycin, 100
µg/ml; Sigma-Aldrich; Merck KGaA) and seeded on the upper chamber
of the Transwell plates (Falcon; Corning Inc., Corning, NY, USA)
with a Matrigel coating, and DMEM (Biological Industries),
supplemented with 10% fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) and
penicillin-streptomycin (penicillin, 100 U/ml and streptomycin, 100
µg/ml; Sigma-Aldrich; Merck KGaA) was added to the lower chambers
for the invasion assay. The chambers were incubated in a
CO2 incubator at 37°C for 12 or 24 h. The remaining
cells in the upper chamber were then removed using cotton swabs,
and the cells under the surface of the Transwell plates were fixed
in 10% formaldehyde solution for 10 min at room temperature. The
cells were stained with crystal violet solution (0.05% crystal
violet, 1% Formaldehyde and 1% methanol) (Sigma-Aldrich; Merck
KGaA) for 20 min at room temperature, and the number of lung cancer
cells in 3 fields of view was counted through phase-contrast
microscopy. All experiments were repeated 3 times.
Identification of putative target
genes of miR-324 with bioinformatics
A total of 82 putative target genes of miR-324-5p
were identified using the TargetScan tool (release no. 7.0)
(29). The expression levels of these
putative target genes in LUAD and LUSC were obtained from TCGA. The
correlations between putative target gene and miR-324-5p expression
in LUAD or LUSC were examined using Spearman's correlation
analysis.
Statistical analysis
All statistical analyses were carried out using the
SPSS 22.0 statistical software package (IBM Corp., Armonk, NY,
USA). Changes in the arm selection preference of miRNAs in lung
cancer cells from TCGA were analyzed using unpaired Student's
t-tests. Correlations between the miR-324-5p:-3p ratio in LUAD,
LUSC and normal tissues were determined using Pearson's correlation
coefficient analysis, with R2 as indicated. Spearman's
correlation analysis was applied to examine the correlations
between miR-324-5p and its target gene expression in lung cancer.
Data concerning miR-324-5p and −3p expression in paired LUAD or
LUSC tissues were analyzed using paired Student's t-tests. Cell
proliferation and invasion were assessed in triplicate, and
histograms represent the mean ± standard deviation. The data of
cell growth and invasion were analyzed using one-way ANOVA.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of changes in candidate
miRNA arm selection preference in lung cancer using TCGA
Our group has previously demonstrated that the arm
selection preference of certain miRNAs is altered significantly in
breast cancer tissues compared with adjacent normal tissues
(27). However, the arm selection
preference of miRNAs in lung cancer remains to be clarified. To
identify candidate miRNAs with an altered arm preference in lung
cancer, small RNA profiles of 458 LUAD and 46 adjacent normal
tissues, and 342 LUSC and 45 adjacent normal tissues, were
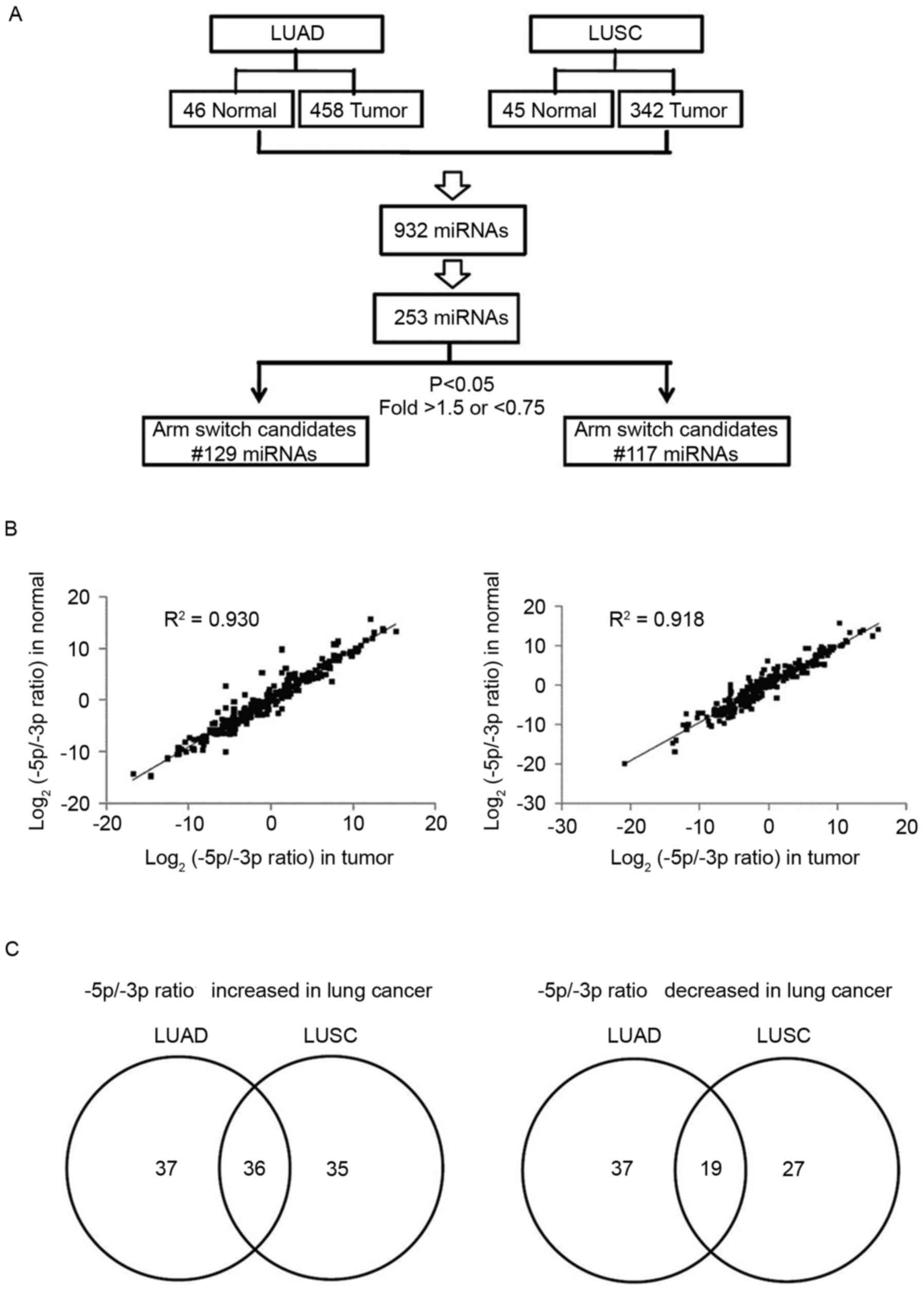
downloaded from TCGA. As presented in Fig. 1A, 932 pre-miRNAs were detected in lung
cancer (transcripts/million >1). Among these, 253 pre-miRNAs
simultaneously produced miR-#-5p and miR-#-3p. The 5p:3p ratio was
further analyzed in identical pre-miRNAs, and the 5p:3p ratio of
the majority of miRNAs was revealed to be consistent between lung
cancer and adjacent normal tissues (LUAD: R2=0.9304 and
LUSC: R2=0.9188; Fig. 1B).
Only a few miRNAs had significantly altered arm preference in LUAD
or LUSC compared with adjacent normal tissues (fold change >1.5
or <0.75; P<0.05). In LUAD, the 5p:3p ratio of 73 miRNAs was
significantly increased, and the 5p:3p ratio of 56 miRNAs was
significantly decreased. In LUSC, the 5p:3p ratio of 71 miRNAs was
significantly increased, and the 5p:3p ratio of 46 miRNAs was
significantly decreased. Alterations in the arm selection
preference of common miRNA candidates were further identified using
a Venn diagram tool (version 2.1) (30). The results revealed that the arm
selection preference of 36 miRNAs was upregulated and that of 19
miRNAs was downregulated in lung cancer (Fig. 1C; Table
I).
 | Table I.Candidate miRNAs indicate changes in
arm selection preferences in lung cancer tissues compared with
normal tissues. |
Table I.
Candidate miRNAs indicate changes in
arm selection preferences in lung cancer tissues compared with
normal tissues.
| A, Upregulated
miRNAs |
|---|
|
|---|
|
| 5p/3p ratio fold
change (tumor/normal) |
|---|
|
|
|
|---|
| miRNAs | Lung squamous cell
carcinoma (LUSC) | Lung adenocarcinoma
(LUAD) |
|---|
| hsa-mir-9-1 | 9.2 | 10.9 |
| hsa-mir-9-2 | 7.0 | 14.6 |
| hsa-mir-17 | 2.7 | 1.9 |
| hsa-mir-27b | 1.8 | 1.6 |
| hsa-mir-29a | 2.5 | 4.5 |
| hsa-mir-30c-1 | 2.5 | 4.7 |
| hsa-mir-30c-2 | 6.3 | 8.7 |
| hsa-mir-30e | 1.9 | 2.1 |
| hsa-mir-33a | 3.0 | 9.0 |
| hsa-mir-33b | 2.0 | 14.4 |
| hsa-mir-96 | 19.8 | 69.5 |
| hsa-mir-101-1 | 1.6 | 2.2 |
| hsa-mir-125b-2 | 2.0 | 4.4 |
| hsa-mir-127 | 2.2 | 4.5 |
| hsa-mir-130a | 1.9 | 1.9 |
| hsa-mir-136 | 3.3 | 2.2 |
| hsa-mir-140 | 2.1 | 1.9 |
| hsa-mir-143 | 3.7 | 3.2 |
| hsa-mir-149 | 3.2 | 3.5 |
| hsa-mir-182 | 11.2 | 38.8 |
| hsa-mir-205 | 8.7 | 11.3 |
| hsa-mir-218-1 | 1.8 | 2.1 |
| hsa-mir-224 | 4.3 | 2.4 |
| hsa-mir-324 | 1.5 | 2.2 |
| hsa-mir-337 | 2.0 | 4.9 |
| hsa-mir-362 | 2.0 | 2.9 |
| hsa-mir-425 | 2.0 | 1.8 |
| hsa-mir-502 | 2.1 | 1.6 |
| hsa-mir-556 | 3.0 | 5.4 |
| hsa-mir-616 | 1.7 | 5.2 |
| hsa-mir-671 | 4.3 | 3.4 |
| hsa-mir-877 | 5.0 | 3.0 |
| hsa-mir-2277 | 1.9 | 11.1 |
| hsa-mir-3065 | 2.2 | 1.7 |
| hsa-mir-3607 | 1.8 | 23.6 |
| hsa-mir-3679 | 6.7 | 8.6 |
|
| B, Downregulated
miRNAs |
|
|
| 5p/3p ratio fold
change (tumor/normal) |
|
|
|
| miRNAs | Lung squamous
cell carcinoma (LUSC) | Lung squamous
cell carcinoma (LUSC) |
|
| hsa-let-7a-3 | 0.6 | 0.6 |
| hsa-let-7g | 0.5 | 0.1 |
| hsa-mir-20a | 0.5 | 0.7 |
| hsa-mir-26a | 0.5 | 0.7 |
| hsa-mir-27a | 0.3 | 0.4 |
| hsa-mir-29b | 0.5 | 0.1 |
| hsa-mir-138 | 0.3 | 0.5 |
| hsa-mir-142 | 0.6 | 0.3 |
| hsa-mir-335 | 0.7 | 0.5 |
| hsa-mir-374b | 0.7 | 0.6 |
| hsa-mir-423 | 0.6 | 0.3 |
| hsa-mir-455 | 0.2 | 0.4 |
| hsa-mir-497 | 0.3 | 0.7 |
| hsa-mir-509 | 0.6 | 0.3 |
| hsa-mir-532 | 0.5 | 0.5 |
| hsa-mir-589 | 0.4 | 0.6 |
| hsa-mir-629 | 0.3 | 0.4 |
| hsa-mir-744 | 0.3 | 0.2 |
| hsa-mir-769 | 0.6 | 0.4 |
miR-324-5p and −3p serve distinctive
functions in lung cancer
Our previous study revealed that miR-324-5p and
miR-324-3p play distinct biological functions in breast cancer
(28). However, the role of
miR-324-5p and miR-324-3p was still unknown in lung cancer. Among
the miRNA candidates with a switched arm preference, miR-324 was
selected for further examination in the present study. According to
the miRbase database, the pre-mir-324 gene is located in
chr17:7223297-7223379, and an identical pre-mir-324 simultaneously
produces miR-324-5p and −3p in humans (31–35).
MiR-324-5p and −3p were significantly overexpressed in LUAD and
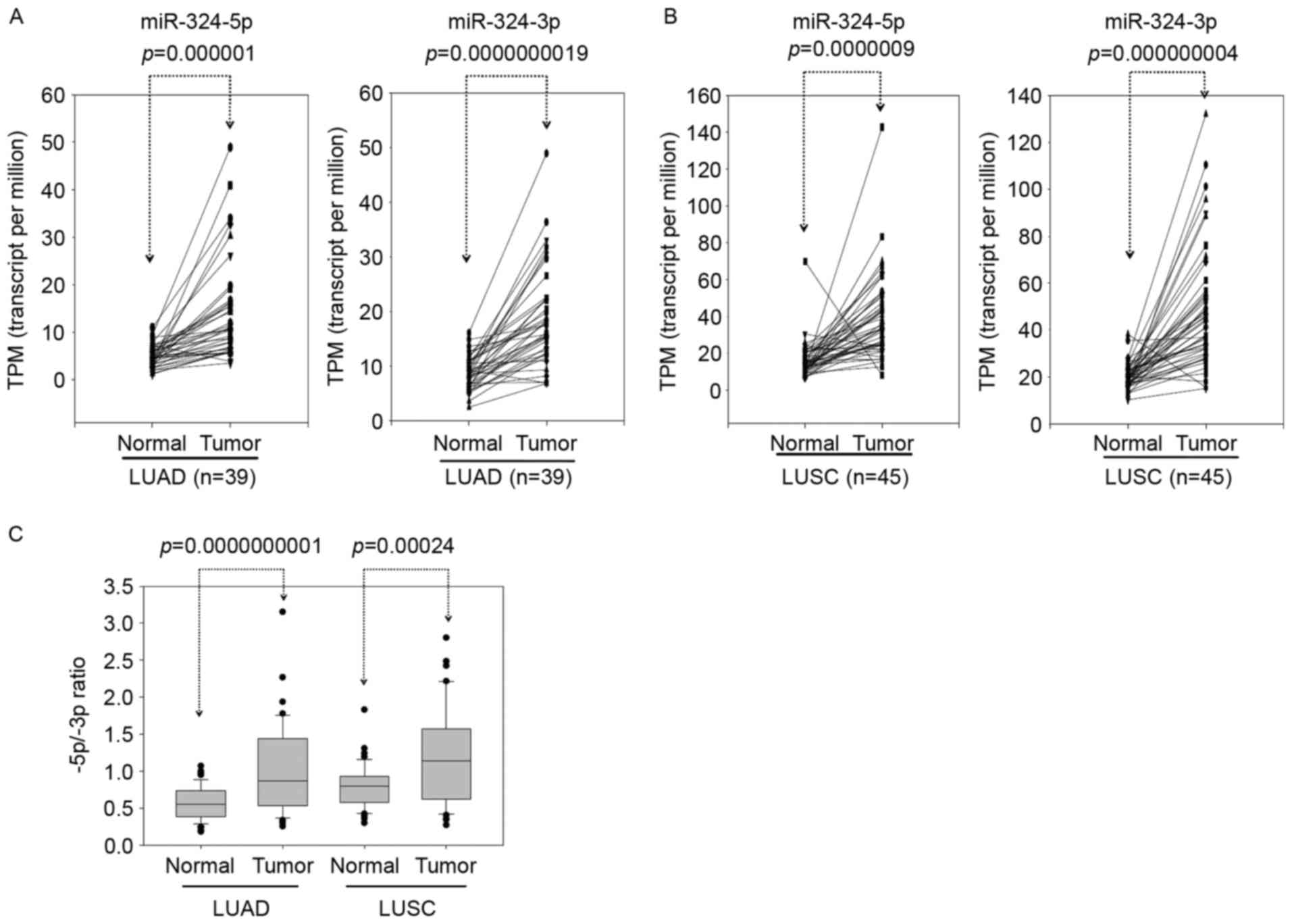
LUSC compared with adjacent normal tissues (Fig. 2A and B). The expression of miR-324-5p
was increased compared with miR-324-3p in lung cancer (Fig. 2C). As presented in Fig. 2C, the 5p:3p ratio in miR-324 was
significantly increased (~2.2-fold) in LUAD compared with adjacent
normal tissues. Furthermore, the 5p:3p ratio in miR-324 was
significantly increased (~1.5-fold) in LUSC compared with adjacent
normal tissues. Therefore, the 5p arm selection of miR-324 was more
preferred in tumors compared with adjacent normal tissues. To
investigate the biological functions of the individual arms of
miR-324, miR-324-5p and miR-324-3p mimics were transfected into the
A549 cells. Following transfection, the expression of miR-324-5p
and −3p significantly increased in the transfected cells compared
with the scramble controls (data not shown). Subsequently, cell
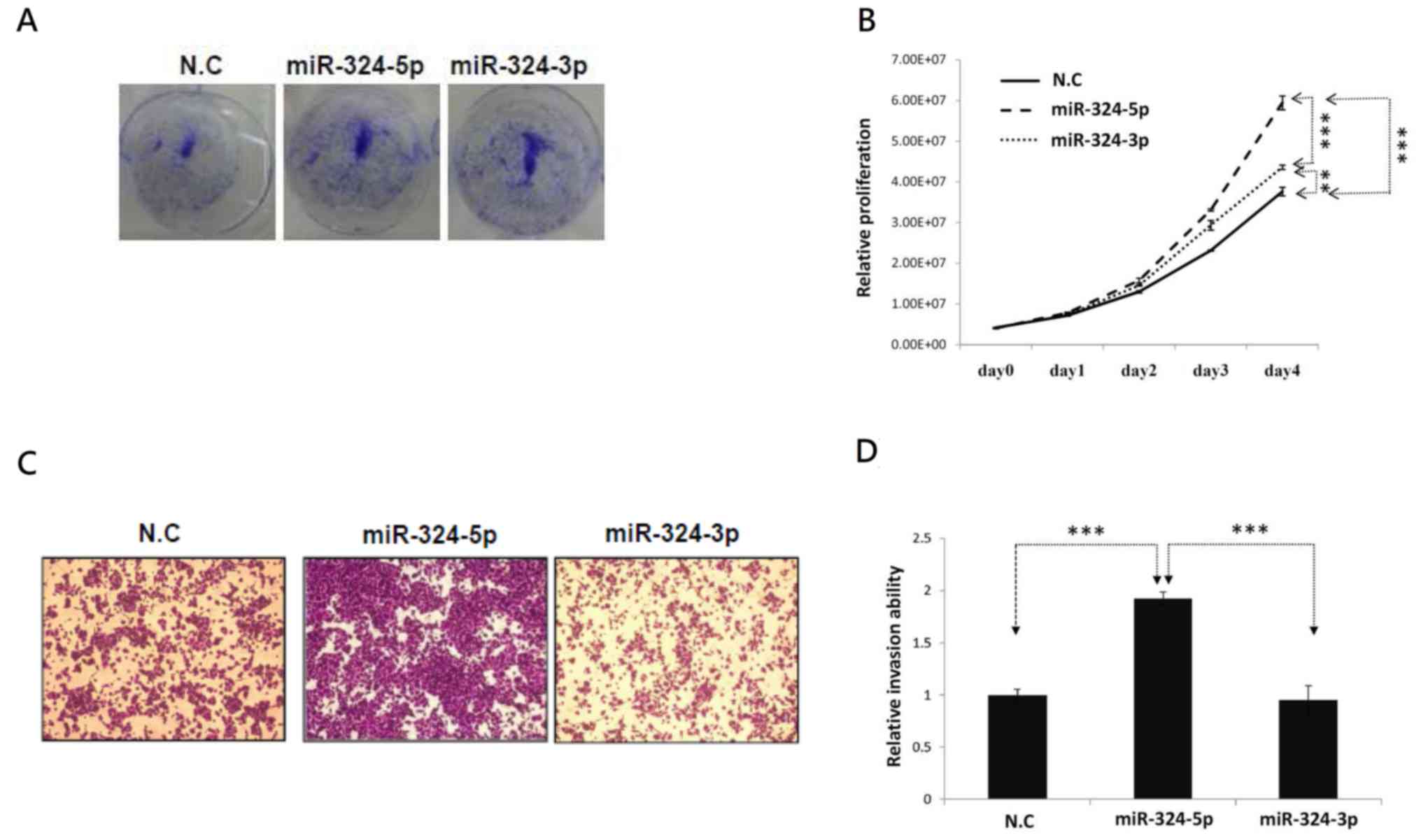
proliferation and invasion were examined. As presented in Fig. 3A and B, ectopic miR-324-5p and −3p
increased the proliferation of A549 cells. However, only miR-324-5p
significantly increased cell invasion in lung cancer cells
(Fig. 3C and D). These results
revealed that miR-324-5p and −3p were generated from an identical
pre-mir-324; however, the arm selection preference of miR-324
differed significantly between lung cancer and normal tissues.
Different arms of miR-324 may serve distinct biological functions
in the modulation of lung cancer cell invasion.
Identification of putative target
genes of miR-324-5p in lung cancer
The data revealed that miR-324-5p significantly
promoted lung cancer cell invasion. Therefore, putative target
genes of miR-324-5p were identified using a bioinformatics
approach. A total of 83 putative target genes of miR-324-5p were
identified using TargetScan prediction tools. The expression levels
of the target genes should be negatively correlated with miR-324-5p
expression in lung cancer. Among these identified target genes, 11
gene expression levels were negatively correlated with miR-324-5p
expression in LUAD and LUSC (Fig.
4A). As the expression levels of miR-324-5p were increased in
lung cancer, those of the target genes should be decreased in lung
cancer. The expression levels of the predicted candidates were
further analyzed using TCGA, which revealed that glutathione
peroxidase 3 (GPX3), regulator of calcineurin 1
(RCAN1) and mannosyl (β-1,4-) glycoprotein
β-1,4-N-acetylglucosaminyltransferase (MGAT3)
expression levels were significantly decreased (fold change
<0.5) in LUAD and LUSC compared with adjacent normal tissues
(Fig. 4B and C). Previous studies
have reported that GPX3, RCAN1 and MGAT3 expression
is significantly downregulated and serves a tumor-suppressive
function in human cancer, including prostate cancer, lung cancer,
ovarian cancer and lymphoma (36–39).
According to the aforementioned data, miR-324-5p may promote lung
cancer cell proliferation and invasion by suppressing GPX3,
RCAN1 and MGAT3 expression in lung cancer. However, the
underlying mechanisms require further clarification in the
future.
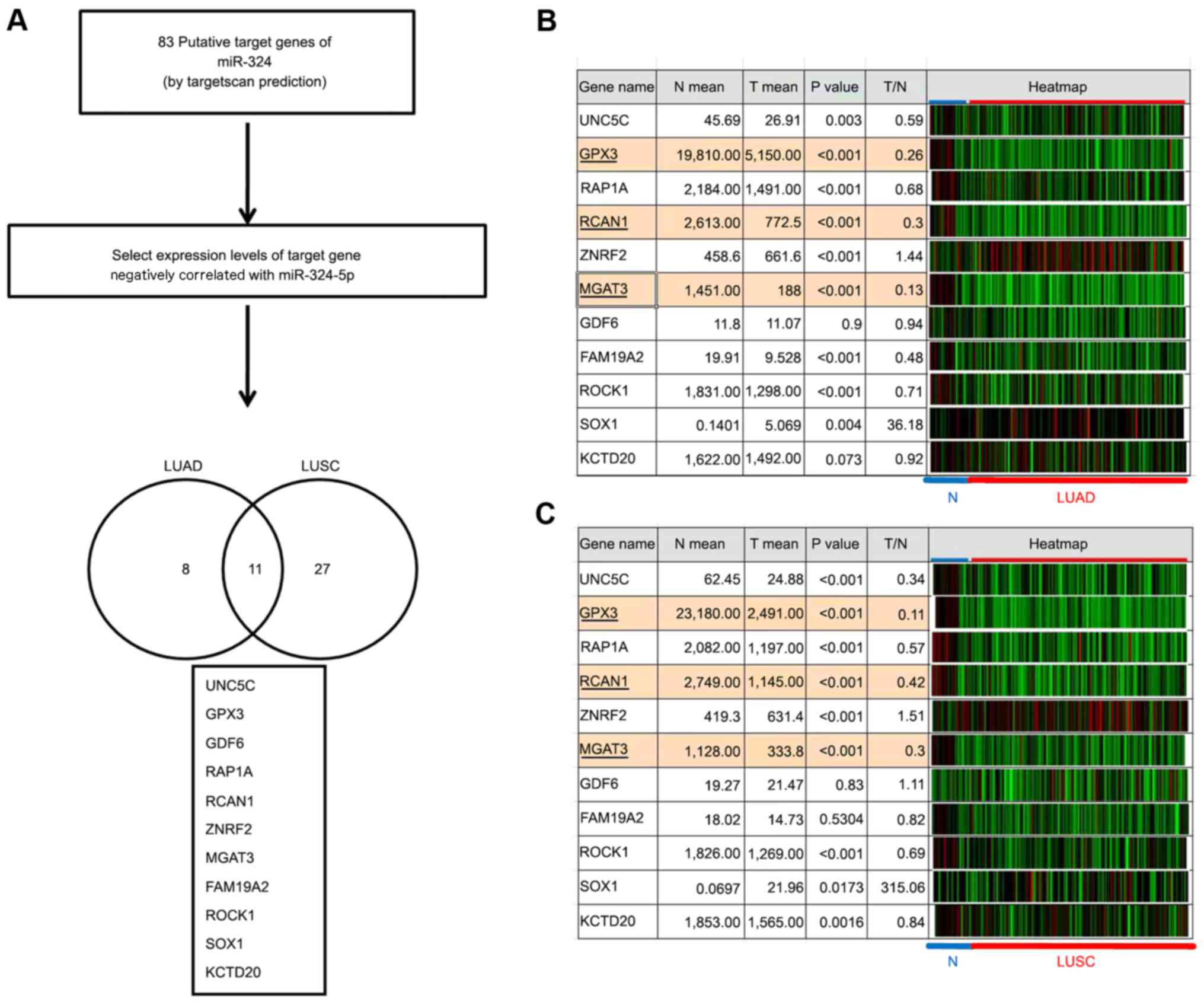 | Figure 4.Identification of putative target
genes of miR-324-5p using a bioinformatics approach. (A) Workflow
for identifying putative target genes of miR-324-5p in lung cancer.
A total of 83 target genes of miR-324-5p were identified using the
TargetScan tool. The correlations between target genes and
miR-324-5p were examined in LUAD and LUSC via TCG A analysis. The
Venn diagrams present the 11 putative target genes with expression
levels negatively correlated with miR-324-5p expression levels in
LUAD and LUSC. The expression levels of potential targets were
analysed in (B) LUAD (26 normal and 227 tumor tissues) and (C) LUSC
(24 normal and 234 tumor tissues) from TCGA. miR, microRNA; TCGA,
The Cancer Genome Atlas; UNC5C, unc-5 netrin receptor C; GPX3,
glutathione peroxidase 3; RAP1A, RAP1A, member of RAS oncogene
family; RCAN1, regulator of calcineurin 1; ZNRF2, zinc and ring
finger 2; MGAT3, mannosyl (β-1,4-) glycoprotein
β-1,4-N-acetylglucosaminyltransferase; GDF6, growth differentiation
factor 6; FAM19A2, family with sequence similarity 19 member A2,
C-C motif chemokine like; ROCK1, Rho associated coiled-coil
containing protein kinase 1; SOX1, SRY-box 1; KCTD20, potassium
channel tetramerization domain containing 20. |
Discussion
An identical pre-miRNA structure consists of two
arms (5p and 3p) and is selected by the RNA-induced silencing
complex to generate two mature miRNAs in the cell (8,9,40). Typically, the arm is selected on the
basis of hydrogen bond theory. However, a number of previous
studies have reported that the arm selection preference of certain
miRNAs may vary in different types of cell or during carcinogenesis
(20,21,27,41). In
humans, significant changes have been reported in the arm selection
preference of certain miRNAs between normal and tumor tissues,
including gastric cancer, hepatocellular carcinoma, and breast
cancer (20,21,27,41). Our
group identified significant changes in the arm selection
preference of 17 candidate miRNAs in breast cancer compared with
adjacent normal tissues by comprehensively analyzing the Sequence
Read Archive database (27). Among
these, miR-324 arm selection was frequently altered in human
cancer, including breast cancer, colon cancer, lung cancer and
bladder cancer (28). The arm
selection preference of miR-324 is a complex mechanism that
modulates its biological function. Through TCGA analysis,
miR-324-5p and miR-324-3p expression was frequently observed to be
abnormal in human cancer (28).
Microarray data have demonstrated that miR-324-5p expression is
upregulated in lung cancer, melanoma, hepatocellular carcinoma and
acute myeloid leukemia but downregulated in medulloblastoma
(42–46). These results consistently indicate the
significant upregulation of miR-324-5p in lung cancer.
miR-324-5p and miR-324-3p sequences differ
considerably, suggesting that miR-324-5p and −3p serve distinct
functions through targeting different protein-coding genes in
different types of cancer. Tsai et al (27) reported that miR-324-5p and −3p
suppressed cell proliferation and invasion in breast cancer, but
only miR-324-5p inhibited cell proliferation and motility in colon
cancer (28). An et al
(39) reported that miR-324-5p
suppressed the invasion of hepatocellular carcinoma cells by
silencing ETS proto-oncogene 1, transcription factor and Sp1
transcription factor expression. The ectopic expression of
miR-324-5p inhibited glioma cell proliferation by suppressing GLI
family zinc finger 1 expression (47). Until now, previous studies have
primarily focused on the function of miR-324-5p in human types of
cancer, while the function of the passenger arm, miR-324-3p,
remains unclear. Li et al (41) reported that miR-324-3p suppressed the
growth of nasopharyngeal carcinoma cells and promoted apoptosis by
targeting Smad family member 7. Tsai et al (27) reported that miR-324-3p silenced the
growth and motility of breast and colon cancer cells. In the
present study, the results revealed that miR-324-5p and −3p
promoted the proliferation of lung cancer cells, but only
miR-324-5p accelerated the invasion of lung cancer cells. The
present study is the first to report variation in the arm selection
preference of miRNA-#-5p and miR-#-3p in lung cancer compared with
healthy tissue. The results provide a novel insight into the arm
selection preference of miRNAs, suggesting that this may be a
mechanism that modulates their function and further complicates
their regulatory network in human types of cancer.
Acknowledgements
The present study was supported by grants from
Kaohsiung Veterans General Hospital (grant nos. VGHKS-105-135,
VGHKS-106-153 and VGHKS-106-006).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Socinski MA, Crowell R, Hensing TE, Langer
CJ, Lilenbaum R, Sandler AB and Morris D: American College of Chest
Physicians: Treatment of non-small cell lung cancer, stage IV: ACCP
evidence-based clinical practice guidelines (2nd edition). Chest.
132 Suppl 3:277S–289S. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nowell PC: The clonal evolution of tumor
cell populations. Science. 194:23–28. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu HH, Lin WC and Tsai KW: Advances in
molecular biomarkers for gastric cancer: miRNAs as emerging novel
cancer markers. Expert Rev Mol Med. 16:e12014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Trabucchi M, Briata P, Filipowicz W,
Rosenfeld MG, Ramos A and Gherzi R: How to control miRNA
maturation? RNA Biol. 6:536–540. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Newman MA and Hammond SM: Emerging
paradigms of regulated microRNA processing. Genes Dev.
24:1086–1092. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen WS, Chen TW, Yang TH, Hu LY, Pan HW,
Leung CM, Li SC, Ho MR, Shu CW, Liu PF, et al: Co-modulated
behavior and effects of differentially expressed miRNA in
colorectal cancer. BMC Genomics. 14 Suppl 5:S122013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsubara H, Takeuchi T, Nishikawa E,
Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M,
Nimura Y, et al: Apoptosis induction by antisense oligonucleotides
against miR-17-5p and miR-20a in lung cancers overexpressing
miR-17-92. Oncogene. 26:6099–6105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: MiR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: miR-107 and MiR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao L, Zhou H, Li XP, Chen J, Fang C, Mao
CX, Cui JJ, Zhang W, Zhou HH, Yin JY and Liu ZQ: MicroRNA-138 acts
as a tumor suppressor in non small cell lung cancer via targeting
YAP1. Oncotarget. 7:40038–40046. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Leung CM, Li SC, Chen TW, Ho MR, Hu LY,
Liu WS, Wu TT, Hsu PC, Chang HT and Tsai KW: Comprehensive microRNA
profiling of prostate cancer cells after ionizing radiation
treatment. Oncol Rep. 31:1067–1078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheng WC, Chung IF, Huang TS, Chang ST,
Sun HJ, Tsai CF, Liang ML, Wong TT and Wang HW: YM500: A small RNA
sequencing (smRNA-seq) database for microRNA research. Nucleic
Acids Res. 41:(Database Issue). D285–D294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li SC, Liao YL, Ho MR, Tsai KW, Lai CH and
Lin WC: miRNA arm selection and isomiR distribution in gastric
cancer. BMC Genomics. 13 Suppl 1:S132012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang HT, Li SC, Ho MR, Pan HW, Ger LP, Hu
LY, Yu SY, Li WH and Tsai KW: Comprehensive analysis of microRNAs
in breast cancer. BMC Genomics. 13 Suppl 7:S182012.PubMed/NCBI
|
|
22
|
Li SC, Liao YL, Chan WC, Ho MR, Tsai KW,
Hu LY, Lai CH, Hsu CN and Lin WC: Interrogation of rabbit miRNAs
and their isomiRs. Genomics. 98:453–459. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Griffiths-Jones S, Hui JH, Marco A and
Ronshaugen M: MicroRNA evolution by arm switching. EMBO Rep.
12:172–177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cloonan N, Wani S, Xu Q, Gu J, Lea K,
Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al:
MicroRNAs and their isomiRs function cooperatively to target common
biological pathways. Genome Biol. 12:R1262011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marco A, Hui JH, Ronshaugen M and
Griffiths-Jones S: Functional shifts in insect microRNA evolution.
Genome Biol Evol. 2:686–696. 2010.PubMed/NCBI
|
|
26
|
Guo L, Li H, Liang T, Lu J, Yang Q, Ge Q
and Lu Z: Consistent isomiR expression patterns and 3′ addition
events in miRNA gene clusters and families implicate functional and
evolutionary relationships. Mol Biol Rep. 39:6699–6706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsai KW, Leung CM, Lo YH, Chen TW, Chan
WC, Yu SY, Tu YT, Lam HC, Li SC, Ger LP, et al: Arm selection
preference of MicroRNA-193a varies in breast cancer. Sci Rep.
6:281762016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuo WT, Yu SY, Li SC, Lam HC, Chang HT,
Chen WS, Yeh CY, Hung SF, Liu TC, Wu T, et al: MicroRNA-324 in
Human cancer: miR-324-5p and miR-324-3p have distinct biological
functions in human cancer. Anticancer Res. 36:5189–5196. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oliveros JC: Venny. An interactive tool
for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html2007–2015.
|
|
31
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:(Database issue). D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:(Database issue).
D140–D144. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:(Database issue). D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:(Database issue). D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Griffiths-Jones S: The microRNA registry.
Nucleic Acids Res. 32:(Database issue). D109–D111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu C, Zheng L, Wang H, Ran X, Liu H and
Sun X: The RCAN1 inhibits NF-κB and suppresses lymphoma growth in
mice. Cell Death Dis. 6:e19292015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kohler RS, Anugraham M, López MN, Xiao C,
Schoetzau A, Hettich T, Schlotterbeck G, Fedier A, Jacob F and
Heinzelmann-Schwarz V: Epigenetic activation of MGAT3 and
corresponding bisecting GlcNAc shortens the survival of cancer
patients. Oncotarget. 7:51674–51686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chang SN, Lee JM, Oh H and Park JH:
Glutathione peroxidase 3 inhibits prostate tumorigenesis in TRAMP
mice. Prostate. 76:1387–1398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
An BC, Jung NK, Park CY, Oh IJ, Choi YD,
Park JI and Lee SW: Epigenetic and glucocorticoid receptor-mediated
regulation of glutathione peroxidase 3 in lung cancer cells. Mol
Cells. 39:631–638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Slezak-Prochazka I, Durmus S, Kroesen BJ
and van den Berg A: MicroRNAs, macrocontrol: Regulation of miRNA
processing. Rna. 16:1087–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li SC, Tsai KW, Pan HW, Jeng YM, Ho MR and
Li WH: MicroRNA 3′ end nucleotide modification patterns and arm
selection preference in liver tissues. BMC Syst Biol. 6 Suppl
2:S142012. View Article : Google Scholar
|
|
42
|
Dixon-McIver A, East P, Mein CA, Cazier
JB, Molloy G, Chaplin T, Lister Andrew T, Young BD and Debernardi
S: Distinctive patterns of microRNA expression associated with
karyotype in acute myeloid leukaemia. PLoS One. 3:e21412008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schultz J, Lorenz P, Gross G, Ibrahim S
and Kunz M: MicroRNA let-7b targets important cell cycle molecules
in malignant melanoma cells and interferes with
anchorage-independent growth. Cell Res. 18:549–557. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang
Y, Tantoso E, Li KB, Ooi LL, Tan P and Lee CG: Profiling microRNA
expression in hepatocellular carcinoma reveals microRNA-224
up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific
target. J Biol Chem. 283:13205–13215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Crawford M, Batte K, Yu L, Wu X, Nuovo GJ,
Marsh CB, Otterson GA and Nana-Sinkam SP: MicroRNA 133B targets
pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem
Biophys Res Commun. 388:483–489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu HS, Zong HL, Shang M, Ming X, Zhao JP,
Ma C and Cao L: MiR-324-5p inhibits proliferation of glioma by
target regulation of GLI1. Eur Rev Med Pharmacol Sci. 18:828–832.
2014.PubMed/NCBI
|