Introduction
Lung cancer is the leading cause for
cancer-associated mortality worldwide (1,2). Despite
considerable improvements in diagnosis and chemotherapy, and the
development of molecularly-targeted treatments, the survival rate
for lung cancer remains low; the 5-year survival rate is <20%
(3). Non-small cell lung cancer
(NSCLC) is responsible for >80% of the cases of lung
cancer-associated mortality (4). One
reason for the poor prognosis of lung cancer is metastasis, which
presents a major challenge in the treatment of NSCLC (5). To overcome this challenge, the
elucidation of the molecular mechanism underlying NSCLC metastasis
is required.
MicroRNAs (miRNAs) are small non-coding RNAs that
negatively regulate the translation of mRNA or induce target gene
mRNA degradation (6,7). Accumulating evidence suggests that
miRNAs are associated with a number of cellular biological
processes, including proliferation, apoptosis, drug resistance and
metastasis. Regarding the role of miRNAs in cancer, a number of
potential therapeutic targets were previously identified (8–10). The
mechanisms underlying the effects of miRNAs is a longstanding
research topic, and a number of studies have aimed to identify the
target signaling pathways of cancer-associated miRNAs (11,12).
Proteomics is an emerging field that offers a wide
range of opportunities to investigate the malignancy-associated
molecular alterations at the protein level, and is thus being
increasingly applied in cancer research (13). Isobaric labeling reagents are peptide
tags that produce tandem mass spectrometry (MS/MS)
spectrum-specific fragment ions, used for MS/MS quantification
(14). Isobaric Tag for Relative and
Absolute Quantitation (iTRAQ) labeling technology combined with
nano liquid chromatography-mass spectrometry (NanoLC-MS/MS) has
been employed effectively for biomarker discovery (15). As miRNAs not only induce alterations
of the protein expression level of their target genes, but also
indirectly change the expression of a range of proteins through
interactions with target proteins, cancer research is turning to
proteomics-based strategies to seek the putative targets and
further molecular mechanisms modulated by miRNAs (16–18).
Our previous study (19) demonstrated that miR-148a exerted
metastasis-suppressive effects on NSCLC. However, the molecular
mechanisms underlying the metastasis-suppressive effects of
miR-148a on NSCLC remain uncharacterized. In the present study,
iTRAQ labeling technology and NanoLC-MS/MS were used to analyze the
whole protein profiles of SPC-A-1 lung adenocarcinoma cells
following transfection with the miR-148a inhibitor. The data may
provide a general perspective for the analysis of the
metastasis-modulatory mechanism of miR-148a in NSCLC.
Materials and methods
Cell lines and cell culture
SPC-A-1 human lung adenocarcinoma cells were
obtained from the Cellular Institute of the Chinese Academy of
Science (Shanghai, China). The SPC-A1 cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine
serum (Biowest, Nuaillé, France), 100 U/ml penicillin sodium and
100 mg/ml streptomycin sulfate at 37°C in a humidified atmosphere
containing 5% CO2.
Oligonucleotide transfection
An miR-148a inhibitor (miR20000243) and negative
control (miR02101-1-5) were synthesized by Guangzhou RiboBio Ltd.
(Guangzhou, China). On day 1, 5×104 cells were seeded
into a 6-well plate and were transfected on day 2 with the miRNA
inhibitor or a control oligonucleotide using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's instructions. Cells were collected at 48 h after
transfection for RNA extraction and protein preparation.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
assays
miRNA was extracted from the transfected cells using
a mirVana miRNA Isolation kit (Ambion, Thermo Fisher Scientific,
Inc.). The expression level of mature miR-148a was quantified with
specific primers (Guangzhou RiboBio Ltd.) according to the
manufacturer's instructions. RT-qPCR was performed using miRNA
RT-qPCR Starter kit (Guangzhou RiboBio Ltd.) and normalized to U6
small nuclear RNA. U6 primers were synthesized in Guangzhou RiboBio
Ltd. The RT reaction conditions were as follows: 42°C for 60 min,
70°C for 10 min. The PCR was conducted based on following
conditions: pre-denaturation at 95°C for 10 min, followed by 40
cycles of denaturation at 95°C for 2 sec, annealing at 60°C for 20
sec, and extension at 70°C for 10 sec.
Total RNA was extracted from transfected cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions, and was quantified using a
NanoDrop 2000 spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA). First-strand cDNA was
synthesized with a PrimeScript RT Reagent kit and RT-qPCR was
performed with SYBR Green Premix Ex Taq (both from Takara Bio,
Inc., Otsu, Japan) using β-actin as an endogenous control. The
primers were synthesized by Genewiz, Inc. (South Plainfield, NJ,
USA), and the sequences are presented in the Table I. The RT reaction conditions were as
follows: 37°C for 45 min, 85°C for 5 sec. While the PCR was
conducted based on following conditions: pre-denaturation at 95°C
for 10 min, followed by 40 cycles of denaturation at 95°C for 5
sec, annealing and extension at 60°C for 31 sec.
 | Table I.Primer sequences used in
experiments. |
Table I.
Primer sequences used in
experiments.
| Genes | Forward | Reverse |
|---|
| MYH9 |
GACAGCCAGAGCGTTAGAGG |
AGACCAGTGAGGACGAGCTA |
| CD44 |
CCTCCCTCCGTCTTAGGTCA |
ATTCAAATCGATCTGCGCCA |
| ITGB1 |
CCGCGCGGAAAAGATGAAT |
ATGTCATCTGGAGGGCAACC |
| LAMB3 |
GGGAGACCCCCACATTCAAG |
GCAGGGCAAAACACAAGAGG |
| PHGDH |
CTGGCCAGGCAGATTCCC |
AGAGGCCAGATCTCCTCCAG |
| ACTN4 |
TGACAAGCTGAGGAAGGACG |
ATTATGGCCTTCTCGTCGGG |
| LMNA |
ATCGCTTGGCGGTCTACATC |
TTGGTATTGCGCGCTTTCAG |
| VIM |
GGACCAGCTAACCAACGACA |
AAGGTCAAGACGTGCCAGAG |
| PSMA7 |
GTGTGCGCTTTTGAGAGTCG |
TCTTCCTCGAACACCAACCG |
| MTHFD1 |
TCCAGTAGTAGTGGCCGTGA |
GCTTTGTGTTGAGCTTCGGG |
| GSTM3 |
TGCACAGTTGGAGAGAGCAG |
TGTACACAGGACGGTTTCCG |
| FH |
AGCCGCCCAGAAATTCTACC |
TTTTGGCTTGCCATTCGAGC |
| HSPB1 |
CGCGGAAATACACGCTGC |
CGGATTTTGCAGCTTCTGGG |
| LRPPRC |
TGGCCGGAGGACTACTGAG |
GCAAGGCATGACTACCACCT |
| CPT2 |
AAGAAGCAGCAATGGGCCAG |
AGGGTCCAGGTAGAGCTCAG |
| IDH2 |
TTTGCAACGCCATAGGCTTC |
CTCATCAGGGGTGATGGTGG |
| β-actin |
TTGTTACAGGAAGTCCCTTGCC |
ATGCTATCACCTCCCCTGTGTG |
The relative expression level of miR-148a and
differentially expressed gene were analyzed using the comparative
Cq method (20).
Protein preparation
All protein extraction procedures were performed on
ice. The cell pellets were dissociated in lysis buffer (Pierce;
Thermo Fisher Scientific, Inc.) supplemented with protease and
phosphatase inhibitors, followed by 40 min incubation on ice. The
lysate was centrifuged at 12,000 × g for 15 min at 4°C, and
the supernatants were collected. The protein concentrations were
determined using a bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions, using bovine serum albumin (Roche Diagnostics, Basel,
Switzerland) as the standard.
iTRAQ labeling
The following iTRAQ experiments were performed as
described previously (15), with some
modifications. iTRAQ labeling was achieved using an iTRAQ Reagent
4-Plex kit (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The cell lysates of
SPC-A1 cells transfected with the control or miR-148a inhibitor
were labeled with iTRAQ labeling reagents 116 and 117,
respectively. In brief, 100 µg lysate from each sample was reduced
with tris-(2-carboxyethyl) phosphine, alkylated with methyl
methanethiosulfonate (MMTS) and digested overnight at 37°C using
trypsin (mass spectrometry grade; Promega Corporation, Madison, WI,
USA) at a trypsin to protein ratio of 1:20 (w/w). The iTRAQ labeled
samples were combined and transferred into a 1.5 ml tube, desalted
with Oasis HLB cartridges (Waters Corporation, Milford, MA, USA)
and dried in a vacuum centrifuge (Concentrator Plus; Eppendorf,
Hamburg, Germany) at 45°C for 1 h, 60°C for 1 to 2 h. The iTRAQ
workflow is illustrated in Fig.
1A.
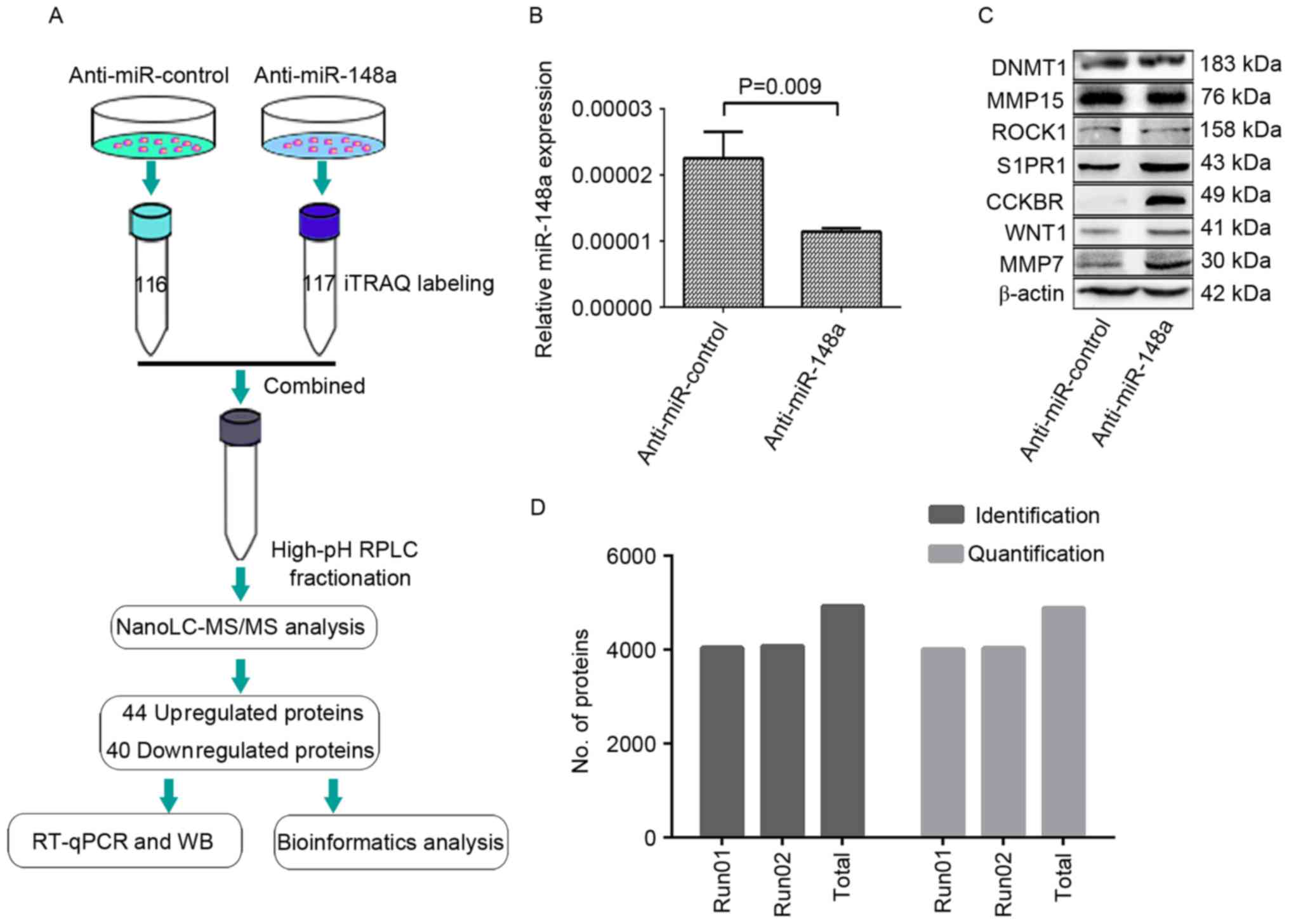 | Figure 1.Quantitative proteomic analysis
experimental workflow and results. (A) SPC-A-1 cells were
transfected with a miR-148a inhibitor or a control oligonucleotide.
At 48 h, proteins of the transfected cells were digested with
trypsin and labeled with the iTRAQ tags 116 and 117. The labeled
peptides were then separated using offline strong cation exchange
LC and analyzed using NanoLC-MS/MS. The experiment was performed in
duplicate, and identified 44 upregulated/40 downregulated proteins,
for a total of 84 differentially expressed proteins, which were
then analyzed with bioinformatics. A number of the differentially
expressed proteins were also verified with WB and RT-qPCR. (B)
RT-qPCR quantification of miR-148a in the SPC-A-1 cells at 48 h
after transfection. (C) Expression levels of miR-148a-associated
proteins as determined by WB in the SPC-A-1 cells at 48 h after
transfection, to demonstrate the downregulation of miR-148a at the
functional level. β-actin served as an internal control. (D) Across
the two MS experiments, a total of 4,934 proteins were identified,
and 4,885 proteins were quantified, with the criteria of an unused
protein score >1.3 and a number of peptides ≥2. The label rate
was 99.0% (4,885/4,934). miR, microRNA; iTRAQ, isobaric tag for
relative and absolute quantitation; LC, liquid chromatography; MS,
mass spectrometry; WB, western blotting; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; DNMT1, DNA
methyltransferase 1; MMP15, matrix metalloproteinase 15; ROCK1, Rho
associated protein kinase 1; S1PR1, sphingosine 1-phosphate
receptor 1; CCKBR, cholecystokinin B receptor; MMP7, matrix
metalloproteinase 7. |
Strong cation exchange (SCX)
The iTRAQ-labeled peptides were fractionated by SCX
chromatography using a 20AD HPLC system (Shimadzu Corporation,
Kyoto, Japan) and a polysulfethyl column (2.1×100 mm, 5 µg, 200 Å;
The Nest Group, Inc., Southborough, MA, USA). The peptide mixture
was dissolved in 80 µl buffer A [10 mM KH2PO4
in 25% acetonitrile (ACN; pH 3.0); Thermo Fisher Scientific, Inc.]
and loaded onto the column. The peptides were separated in a
gradient of 0–80% buffer B (as buffer A, with the addition of 350
mM KCl) at a flow rate of 200 µl/min over 60 min. A total of 20 RP
fractions were collected, desalted using C18 cartridges
(UltraMicroSpin; The Nest Group, Inc.), dried and reconstituted
using 20 µl 0.1% formic acid (FA) for NanoLC-MS/MS analysis.
NanoLC-MS/MS analysis
A NanoLC system (NanoLC-2D Ultra; Eksigent
Technologies, Dublin, CA, USA) combined with a Triple TOF 5600 mass
spectrometer (AB Sciex LLC, Framingham, MA, USA) were utilized for
analysis. The peptides were enriched on a reversed-phase trap
column (ProteoPepII C18 column, 5 µm, 300 Å, 0.15×25 mm; New
Objective IntegraFrit; Scientific Instrument Services, Inc.,
Ringoes, NJ, USA) and eluted onto an analytical column (ProteoPep
C18 column, 5 µm, 300 Å, 0.075×150 mm; New Objective IntegraFrit;
Scientific Instrument Services, Inc.). The NanoLC gradient was
5–35% buffer C (98% ACN, 2% H2O, 0.1% FA) over 120 min
at a flow rate of 300 nl/min. The MS analysis was performed in the
positive-ion mode with a nano ion spray voltage being typically
maintained at 2.3 kV and a scan range of 350 to 1,500 (m/z).
Full-scan MS spectra were acquired from 40 precursors selected for
MS/MS from the 100–1,500 m/z range, utilizing a dynamic exclusion
of 30 sec. The IDA collision energy (CE) parameter script,
selecting up to 40 precursors with charge states of 2+
to 4+, controlled the CE automatically. The tryptic
peptides of β-galactosidase were used to calibrate the mass
spectrometer.
Protein identification and
quantification
Analysis of the proteins was performed using
ProteinPilot 4.1 software (AB Sciex). The search parameters were
specified as follows: i) Sample type, iTRAQ 4-plex
(peptide-labeled); ii) cysteine alkylation, MMTS; iii) digestion,
trypsin; iv) instrument, TripleTOF 5600; v) special factors, none;
vi) species, Homo sapiens; vii) ID Focus, biological
modifications; viii) database, UniProtKB/Swiss-Prot FASTA (as
released in November 2013 with 176,592 human sequences); and ix)
search effort, thorough ID. The peptides for iTRAQ quantitation
were automatically chosen by the Pro Group™ algorithm from the
ProteinPilot software to calculate the reporter peak area and the
false discovery rate (FDR) using a reverse database search
strategy. A qualification criterion of unused confidence score
>1.3 was enforced, corresponding to a peptide confidence level
of 95%. When the iTRAQ ratios were >1.5 or <0.67 between the
SPC-A-1 cells transfected with the control miR and with the
miR-148a inhibitor, the protein expression levels were considered
to be differential.
Western blotting (WB)
A total of 20 µg of protein samples from the
collected cells were loaded onto 8–12% SDS-PAGE gels and
transferred onto nitrocellulose filter membranes (EMD Millipore,
Billerica, MA, USA). Subsequent to blocking (20°C for 1 h) in 5%
non-fat milk in PBS, the membranes were incubated overnight at 4°C
with rabbit primary antibodies against the following: DNA
methyltransferase 1 (DNMT1, dilution 1:500, D160261), matrix
metallopeptidase 15 (MMP15, 1:500 dilution, D120991),
Rho-associated protein kinase 1 (ROCK1, 1:1,000 dilution, D221198),
sphingosine-1-phosphate receptor 1 (S1PR1, 1:500 dilution,
D161195), cholecystokinin B receptor (CCKBR, 1:500 dilution,
D160389), WNT1 (1:200 dilution, D261302), (MMP7, 1:500 dilution,
D120096), actinin α4 (ACTN4, 1:500 dilution, D221929), fumarate
hydratase (FH, 1:500 dilution, D222390), heat shock protein β1
(HSPB1, 1:500 dilution, D163024), lactate dehydrogenase (LDHB,
1:1,000 dilution, D151002), fatty acid synthase (FASN, 1:200
dilution, D190620) and catalase (CAT, 1:500 dilution, D122036) (all
from BBI Life Sciences Corporation, Shanghai, China), vimentin
(VIM, 1:200 dilution, sc-5565) and laminin β3 (LAMB3, 1:200
dilution, sc-20775) (both from Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), phosphoglycerate dehydrogenase (PHGDH; 1:200
dilution, AP2936c; Abgent, San Diego, CA, USA) and isocitrate
dehydrogenase (NADP+) 2 (IDH2, 1:200 dilution, ab131263;
Epitomics, Burlingame, CA, USA).
HRP-conjugated anti-rabbit IgG (1:5,000 dilution,
A0545; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used as
the secondary antibody in which membranes were incubated at 20°C
for 2 h. A total of 3 washes in PBS-Tween were performed following
each antibody incubation. SuperSignal West Femto Maximum
Sensitivity substrate (Thermo Fisher Scientific, Inc.) was used for
the visualization of the proteins, and β-actin was used as a
loading control (1:30,000 dilution, A3854; Sigma-Aldrich, Merck
KGaA).
Statistical analysis
The differentially expressed proteins were input
into the Database for Annotation, Visualization and Integrated
Discovery (DAVID; http://david.abcc.ncifcrf.gov/) and the Search Tool
for the Retrieval of Interacting genes/proteins (STRING; http://string.embl.de). The DAVID search tool was used
for the Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis of the differentially expressed
proteins, with a threshold of P<0.05. STRING was used to predict
protein-protein interactions with a weight score threshold of
≤0.4.
Statistical analysis
t-tests performed with SPSS 19.0 software (IBM
Corp., Armonk, NY, USA) were used to analyze differences between
the protein profiles, the level of miR-148a expression and the mRNA
expression of the differentially expressed proteins between the
cells treated with a miR-148a inhibitor and the control. P<0.05
was considered to indicate a statistically significant
difference.
Results
Quantitative proteomic analysis of
miR-148a-regulated downstream proteins
To identify downstream proteins regulated by
miR-148a, global protein expression changes in the expression
profile of SPC-A-1 cells transfected with a miR-148a inhibitor
compared with SPC-A-1 cells transfected with a control
oligonucleotide were identified using the iTRAQ-labeling proteomic
approach.
The knockdown of miR-148a in SPC-A-1 cells was
detected using RT-qPCR (Fig. 1B) and
validated at a functional level by performing western blots for 7
well-established targets of miR-148a: DNMT1, MMP15, ROCK1, S1PR1,
CCKBR, WNT-1 and MMP7, which were selected according to previous
studies (21–26). No evident change was observed for
DNMT1, MMP15 or ROCK1, whereas S1PR1, CCKBR, WNT1 and MMP7
expression levels were distinctly upregulated in SPC-A1 cells
treated with the miR-148a inhibitor compared with SPC-A-1 cells
treated with the control (Fig.
1C).
With the criteria of unused protein score >1.3
and number of peptides ≥2, two iTRAQ experiments identified 4,048
and 4,083 proteins, as annotated with ProteinPilot software (global
FDR, <1%), and 4,014 and 4,039 proteins were quantified.
Cumulatively, 4,934 proteins were identified and 4,885 proteins
were quantified (Fig. 1D); the label
rate was 99.0% (4,885/4,934). A total of 44 upregulated and 40
downregulated proteins were identified in the SPC-A-1 cells treated
with the miR-148a inhibitor compared with the control cells
(P<0.05; Table II).
 | Table II.Differentially expressed proteins in
microRNA-148a inhibitor-transfected cells compared with control
cells. |
Table II.
Differentially expressed proteins in
microRNA-148a inhibitor-transfected cells compared with control
cells.
| A, Upregulated
differentially expressed proteins |
|---|
|
|---|
| Accession | Protein | Unused score | Coverage,
%a | Peptide 95%
CLa | iTRAQ
ratea |
P-valuea |
|---|
| sp|Q09666 | AHNAK | 457.84±0.62 | 66.71±1.69 | 352.5±12.02 | 4.57±0.18 | <0.0001 |
| sp|P02545 | LMNA | 91.37±2.67 | 66.04±3.30 | 75.5±3.54 | 9.83±0.77 | <0.0001 |
| tr|F5GZS6 | SLC3A2 | 54.32±0.18 | 45.74±0.23 | 37.5±0.71 | 4.45±0.06 | <0.0001 |
| sp|O43707 | ACTN4 | 130.18±3.61 | 71.84±4.11 | 167.5±7.78 | 2.54±0.80 | <0.0001 |
| sp|P35579 | MYH9 | 222.67±10.28 | 59.18±2.38 | 319.5±9.19 | 1.65±0.05 | <0.0001 |
| tr|E7EPC6 | CD44 | 20.93±1.51 | 15.41±0.38 | 14.5±0.71 | 5.85±0.19 | <0.0001 |
| sp|P16615-5 | ATP2A2 | 48.62±5.43 | 30.59±3.83 | 32.5±0.71 | 2.10±0.18 | <0.0001 |
| sp|O43175 | PHGDH | 42.32±4.59 | 56.01±6.50 | 43.5±4.94 | 4.43±1.80 | 0.0004 |
| sp|Q07065 | CKAP4 | 37.42±3.34 | 37.13±0.12 | 23.5±2.12 | 1.84±0.07 | 0.0084 |
| sp|P05556 | ITGB1 | 34.97±1.27 | 25.50±0.27 | 22.5±3.54 | 2.35±0.33 | 0.0002 |
| sp|Q9Y6N5 | SQRDL | 33.74±1.28 | 44.56±4.24 | 20.0±1.41 | 2.12±0.18 | 0.0007 |
| sp|Q13501 | SQSTM1 | 31.33±1.22 | 63.41±0.00 | 25.5±2.12 | 4.04±0.08 | 0.0002 |
| sp|P07355-2 | ANXA2 | 75.77±0.13 | 72.13±1.39 | 135.5±9.19 | 1.86±0.04 | 0.0012 |
| sp|P21980 | TGM2 | 46.15±2.05 | 45.30±6.38 | 30.0±2.83 | 2.12±0.28 | 0.0002 |
| tr|H0Y323 | CAPN2 | 49.09±2.84 | 48.01±5.43 | 34.5±3.54 | 2.39±1.14 | 0.0004 |
| tr|F5H0Q5 | AHSG | 8.43±0.08 | 6.93±0.00 | 22.0±0.00 | 4.06±0.11 | 0.0014 |
| sp|Q13751 | LAMB3 | 14.62±2.70 | 11.48±1.39 | 8.5±0.71 | 3.12±0.45 | 0.0018 |
| sp|Q96AG4 | LRRC59 | 24.42±0.61 | 48.21±0.00 | 15.0±1.41 | 2.76±0.29 | 0.0006 |
| sp|P37802 | TAGLN2 | 48.05±5.01 | 75.88±0.71 | 76.5±0.71 | 4.01±1.16 | 0.0014 |
| sp|O15231 | ZNF185 | 6.63±0.86 | 10.45±1.44 | 3.5±0.71 | 3.60±0.05 | 0.0145 |
| sp|P08670 | VIM | 78.14±2.44 | 69.52±0.30 | 128.5±2.12 | 6.78±0.66 | 0.0009 |
| sp|P07203 | GPX1 | 27.36±0.85 | 84.24±0.00 | 17.5±0.71 | 4.32±0.48 | 0.0013 |
| sp|P46060 | RANGAP1 | 44.64±3.22 | 53.24±0.12 | 28.5±0.71 | 1.66±0.02 | 0.0079 |
| sp|O00159-2 | MYO1C | 51.34±4.12 | 34.29±0.48 | 30.5±4.95 | 3.26±1.68 | 0.0058 |
| sp|P04083 | ANXA1 | 52.37±6.22 | 68.64±1.02 | 91.5±9.19 | 1.84±0.17 | 0.0033 |
| sp|Q92597 | NDRG1 | 20.49±0.41 | 37.82±0.00 | 20.0±1.41 | 3.36±1.03 | 0.0019 |
| tr|B4E0H8 | ITGA3 | 23.64±2.69 | 16.20±2.59 | 16.5±3.54 | 2.09±0.54 | 0.0087 |
| tr|E7ESU5 | ALB | 33.95±4.02 | 41.66±1.03 | 72.5±6.36 | 2.26±0.13 | 0.0023 |
| tr|D6RAK8 | GC | 9.94±2.45 | 17.95±1.29 | 10.0±1.41 | 4.52±0.82 | 0.0036 |
| tr|Q5T985 | ITIH2 | 11.62±0.23 | 9.68±0.68 | 9.0±0.00 | 3.99±2.19 | 0.0080 |
| sp|P26639 | TARS | 77.47±5.34 | 57.74±1.27 | 68.5±2.12 | 2.26±0.32 | 0.0040 |
| sp|P49589-3 | CARS | 38.44±3.73 | 32.61±4.94 | 18.0±2.90 | 2.12±0.10 | 0.0176 |
| tr|F5H7K4 | NCEH1 | 17.57±0.62 | 26.90±0.79 | 10.5±0.71 | 2.25±0.61 | 0.0241 |
| sp|P02788 | LTF | 8.71±1.27 | 9.86±0.00 | 12.5±0.71 | 7.29±0.62 | 0.0188 |
| sp|Q92621 | NUP205 | 33.73±1.67 | 12.23±2.81 | 20.5±3.54 | 2.33±0.29 | 0.0318 |
| sp|Q9NZM1-6 | MYOF | 140.84±3.56 | 47.85±2.60 | 87.0±4.24 | 2.24±0.16 | 0.0062 |
| sp|Q9Y2T3-3 | GDA | 39.77±1.44 | 59.45±0.00 | 38.0±1.41 | 3.18±0.40 | 0.0062 |
| sp|P29317 | EPHA2 | 15.51±5.52 | 11.99±2.03 | 10.0±1.41 | 3.46±0.34 | 0.0083 |
| sp|P80723 | BASP1 | 25.19±1.68 | 72.03±4.05 | 27.0±1.41 | 7.59±2.89 | 0.0101 |
| sp|P48681 | NES | 23.27±1.01 | 10.73±0.87 | 12.5±0.71 | 2.86±0.11 | 0.0240 |
| sp|P31947 | SFN | 20.68±0.27 | 67.34±1.71 | 37.0±1041 | 2.37±0.65 | 0.0190 |
| sp|P16144-4 | ITGB4 | 41.89±1.24 | 18.83±0.69 | 25.5±0.71 | 1.81±0.01 | 0.0364 |
| sp|Q01995 | TAGLN | 6.56±2.14 | 25.13±3.88 | 5.0±1.41 | 2.31±0.35 | 0.0373 |
| sp|Q6P9B6 | KIAA1609 | 8.96±1.87 | 16.01±0.62 | 6.0±1.41 | 3.02±0.20 | 0.0326 |
|
| B, Downregulated
differentially expressed proteins |
|
|
Accession | Protein | Unused
scorea | Coverage,
%a | Peptide 95%
CLa | iTRAQ
ratea |
P-valuea |
|
| sp|A0MZ66 | KIAA1598 | 50.71±0.09 | 51.74±0.78 | 31.0±2.12 | 0.58±0.04 | 0.0011 |
| sp|O00232 | PSMD12 | 35.44±1.51 | 47.03±2.02 | 24.0±2.83 | 0.47±0.04 | 0.0029 |
| sp|O14818 | PSMA7 | 29.16±1.11 | 62.70±0.28 | 23.0±2.83 | 0.47±0.09 | 0.0237 |
| sp|O60749 | SNX2 | 20.85±4.76 | 29.57±4.22 | 16.0±2.12 | 0.51±0.03 | 0.0015 |
| sp|O75533 | SF3B1 | 69±0.34 | 36.5±2.60 | 40.0±1.41 | 0.41±0.00 | 0.0014 |
| sp|O95347 | SMC–2 | 56.22±3.49 | 30.91±2.01 | 37.0±4.95 | 0.58±0.04 | 0.0068 |
| sp|O95573 | ACSL3 | 35.36±1.34 | 36.04±0.10 | 21.0±2.83 | 0.58±0.07 | 0.0096 |
| sp|O95861 | BPNT1 | 23.27±0.62 | 56.82±0.92 | 15.0±0.71 | 0.49±0.20 | 0.0206 |
| sp|P00390 | GSR | 17.07±0.64 | 32.47±3.11 | 11.0±0.00 | 0.45±0.16 | 0.0018 |
| sp|P00505 | GOT2 | 53.15±1.19 | 66.62±0.16 | 39.0±4.24 | 0.53±0.02 | 0.0172 |
| sp|P04040 | CAT | 34.55±2.03 | 48.29±2.81 | 22.0±0.71 | 0.49±0.01 | 0.0015 |
| sp|P04075 | ALDOA | 97.5±2.79 | 87.23±1.36 | 212.0±7.07 | 0.50±0.02 | 0.0011 |
| sp|P04792 | HSPB1 | 41.05±1.21 | 79.51±8.28 | 53.0±1.41 | 0.23±0.23 | 0.0034 |
| sp|P05455 | SSB | 41.26±0.87 | 51.10±1.21 | 26.0±1.41 | 0.49±0.03 | 0.0034 |
| sp|P06744 | GPI | 51.08±0.08 | 55.19±1.52 | 65.0±4.24 | 0.41±0.10 | 0.0078 |
| sp|P07195 | LDHB | 33.47±2.41 | 65.72±4.48 | 73.0±1.41 | 0.32±0.12 | 0.0161 |
| sp|P07339 | CTSD | 33.9±0.68 | 50.85±4.98 | 36.0±2.12 | 0.54±0.04 | 0.0010 |
| sp|P07942 | LAMB1 | 42.6±3.96 | 18.47±1.27 | 27.0±3.54 | 0.54±0.06 | 0.0215 |
| sp|P07954 | FH | 38.99±0.31 | 50.09±0.42 | 37.0±2.12 | 0.58±0.05 | 0.0065 |
| sp|P09972 | ALDOC | 32.73±1.24 | 70.05±2.33 | 96.0±0.00 | 0.21±0.01 | 0.0020 |
| sp|P11586 | MTHFD1 | 76.09±4.05 | 50.16±4.99 | 53.0±3.54 | 0.60±0.04 | 0.0055 |
| sp|P12004 | PCNA | 27.15±3.05 | 58.62±2.16 | 37.0±3.54 | 0.61±0.04 | 0.0032 |
| sp|P13611-2 | VCAN | 25.2±1.73 | 8.053±0.53 | 17.0±0.71 | 0.37±0.12 | 0.0065 |
| sp|P13797 | PLS3 | 74.13±3.57 | 67.69±1.01 | 69.0±4.95 | 0.24±0.07 | 0.0037 |
| sp|P21266 | GSTM3 | 33.26±1.87 | 72.22±3.46 | 21.0±1.41 | 0.38±0.04 | 0.0050 |
| sp|P23786 | CPT2 | 27.57±5.16 | 33.06±5.06 | 17.0±3.54 | 0.61±0.04 | 0.0274 |
| sp|P25789 | PSMA4 | 27.86±0.86 | 65.13±1.63 | 32.0±4.95 | 0.58±0.00 | 0.0133 |
| sp|P27824 | CANX | 57.96±1.73 | 62.92±3.71 | 53.0±0.71 | 0.31±0.10 | 0.0016 |
| sp|P37268 | FDFT1 | 17.91±0.20 | 32.85±2.71 | 12.0±2.12 | 0.30±0.10 | 0.0052 |
| sp|P40937 | RFC5 | 14.34±1.11 | 27.06±3.75 | 12.0±2.12 | 0.48±0.02 | 0.0278 |
| sp|P42166 | TMPO | 48.96±6.58 | 57.28±2.55 | 35.0±3.54 | 0.28±0.04 | 0.0004 |
| sp|P42704 | LRPPRC | 129.11±3.57 | 58.21±3.70 | 95.0±12.02 | 0.44±0.09 | 0.0003 |
| sp|P48449 | LSS | 28.14±3.34 | 26.57±0.10 | 16.0±0.71 | 0.43±0.07 | 0.0314 |
| sp|P48735 | IDH2 | 36.91±0.72 | 44.02±4.38 | 21.0±0.71 | 0.49±0.07 | 0.0010 |
| sp|P49321 | NASP | 50.23±3.21 | 55.08±2.87 | 38.0±2.12 | 0.29±0.00 | 0.0175 |
| sp|P49327 | FASN | 200.88±9.07 | 56.81±0.37 | 167.0±3.54 | 0.21±0.01 | <0.0001 |
| sp|P49419 | ALDH7A1 | 40.15±4.71 | 48.24±0 | 32.0±3.54 | 0.49±0.00 | 0.0238 |
| sp|P51659 | HSD17B4 | 48.92±2.98 | 50.87±0.86 | 36.0±4.95 | 0.47±0.06 | 0.0345 |
| sp|P52209 | PGD | 60±8.37 | 66.97±9.81 | 54.0±4.24 | 0.48±0.11 | 0.0226 |
| sp|P52943 | CRIP2 | 16.69±0.41 | 44.47±2.38 | 12.0±1.41 | 0.58±0.11 | 0.0371 |
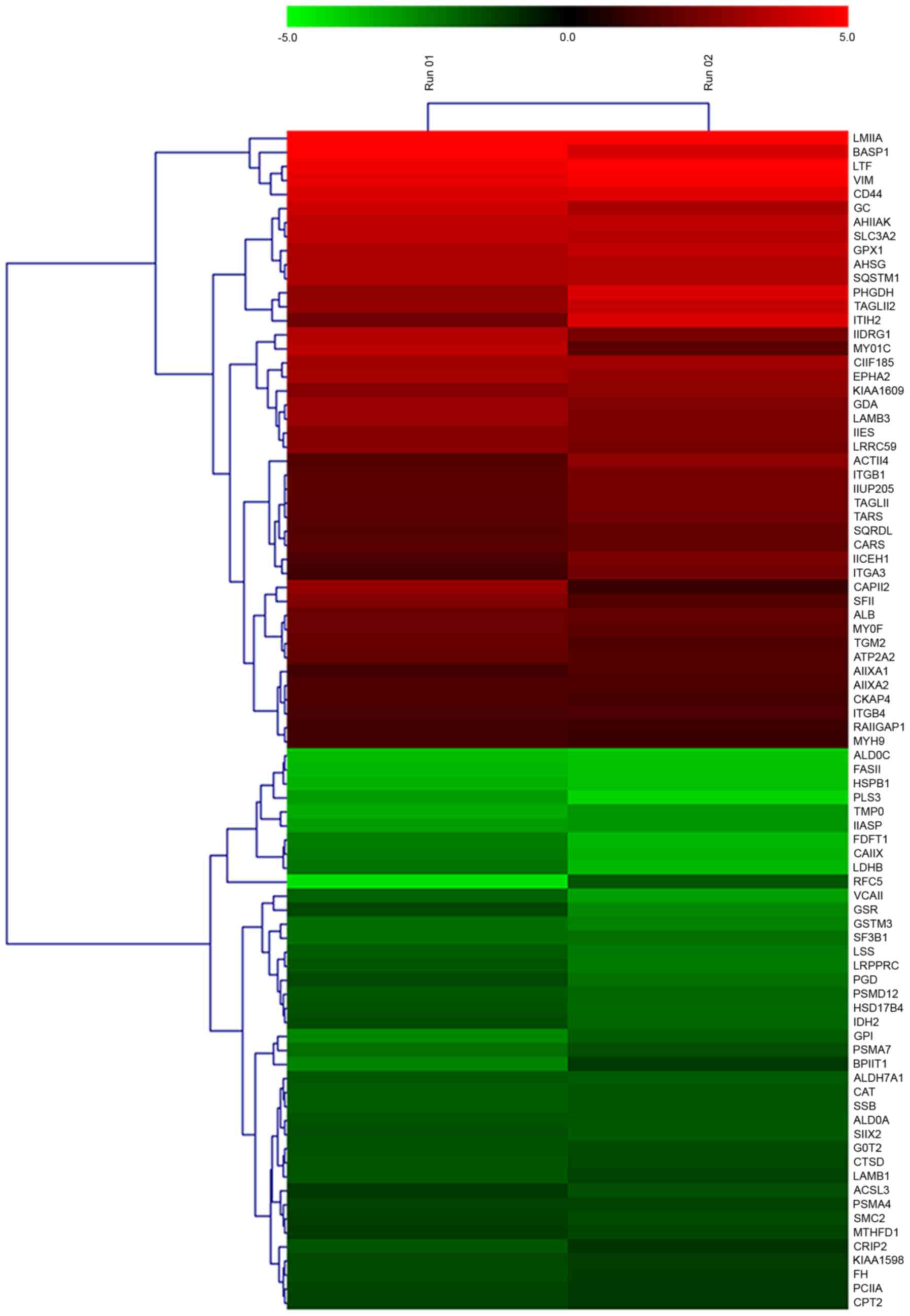
Cluster analysis of miR-148a-regulated
downstream proteins
A heat map was generated for the 84 differentially
expressed proteins subsequent to miR-148a inhibitor transfection
(Fig. 2). The identified
differentially expressed proteins may have been directly or
indirectly regulated by miR-148a.
Verification of iTRAQ MS results using
RT-qPCR and western blotting
Among the globally dysregulated group of
miR-148a-regulated proteins, a number of the proteins, including
IDH2 (27), PHGDH (28), VIM (29), SLC3A2 (30), ACTN4 (31), MYH9 (32), ITGB1 (33), LAMB3 (34), were previously associated with
migration in cancer cells. To verify the iTRAQ results, the mRNA
levels of eight of the upregulated proteins (including MYH9, CD44,
ITGB1, LAMB3, PHGDH, ACTN4, LMNA and VIM) and eight of the
downregulated proteins (including PSMA7, MTHFD1, GSTM3, FH, HSPB1,
LRPPRC, CPT2 and IDH2) were determined by RT-qPCR. As illustrated
in Fig. 3A, the expression of the 16
genes was consistent with the MS analysis. Similar results were
also demonstrated for the protein levels of LAMB3, VIM, PHGDH,
ACTN4, FH, HSPB1, IDH2, LDHB, FASN and CAT, as revealed by WB
analysis (Fig. 3B).
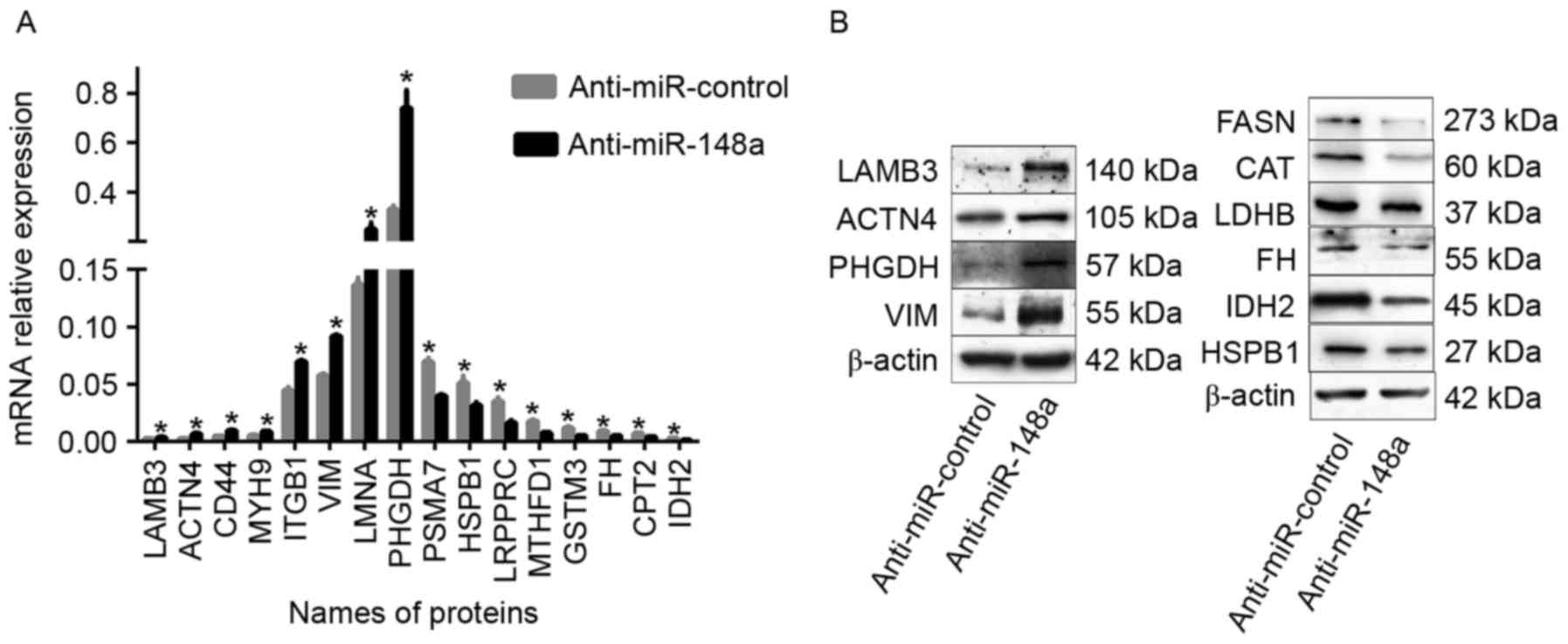 | Figure 3.Identified differentially expressed
proteins verification by RT-qPCR and western blot analysis. (A)
MicroRNA levels of 8 upregulated proteins (MYH9, CD44, ITGB1,
LAMB3, PHGDH, ACTN4, LMNA and VIM) and 8 downregulated proteins
(PSMA7, MTHFD1, GSTM3, FH, HSPB1, LRPPRC, CPT2 and IDH2) were
determined by RT-qPCR following the transfection of SPC-A-1 cells
with an miR-148a inhibitor or control oligonucleotide. β-actin
served as an internal control. (B) Protein levels of LAMB3, VIM,
PHGDH, ACTN4, FH, HSPB1, IDH2, LDHB, FASN and CAT were determined
by western blot analysis following the transfection of SPC-A-1
cells with a miR-148a inhibitor or control oligonucleotide. β-actin
served as an internal control. The data were representative of
three independent experiments. *P<0.05. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; MYH9, myosin
heavy chain 9; CD44, cluster of differentiation 44; ITGB1, integrin
β1; LAMB3, laminin subunit β3; PHGDH, phosphoglycerate
dehydrogenase; ACTN4, α-actinin-4; LMNA, lamin A/C; VIM, vimentin;
PSMA7, proteasome subunit α7; MTHFD1, methylenetetrahydrofolate
dehydrogenase; GSTM3, glutathione S-transferase M3; FH, fumarate
hydratase; HSPB1, heat shock protein β1; LRPPRC, leucine rich
pentatricopeptide repeat; CPT2, carnitine palmitoyltransferase 2;
IDH2, isocitrate dehydrogenase 2; LDHB, lactate dehydrogenase B;
FASN, fatty acid synthase; CAT, catalase. |
Functional enrichment of
miR-148a-regulated proteins
As demonstrated in Table
III, the most significantly enriched KEGG pathways for the
differentially expressed proteins were ‘focal adhesion’,
‘arrhythmogenic right ventricular cardiomyopathy (ARVC)’,
‘ECM-receptor interaction’, ‘glutathione metabolism’ and ‘small
cell lung cancer’. The proteins associated with each pathway are
displayed in Table III.
 | Table III.Significantly enriched Kyoto
Encyclopedia of Genes and Genomes pathways associated with the
identified differentially expressed proteins. |
Table III.
Significantly enriched Kyoto
Encyclopedia of Genes and Genomes pathways associated with the
identified differentially expressed proteins.
| Pathway | Count | P-value | Associated
genes |
|---|
| Arrhythmogenic
right ventricular cardiomyopathy | 6 | 0.0007 | ATP2A2, ACTN4,
LMNA, ITGB4, ITGA3, ITGB1 |
| ECM-receptor
interaction | 6 | 0.0012 | LAMB3, CD44, ITGB4,
ITGA3, LAMB1, ITGB1 |
| Glutathione
metabolism | 5 | 0.0013 | GSR, GPX1, GSTM3,
PGD, IDH2 |
| Pentose phosphate
pathway | 4 | 0.0017 | ALDOA, GPI, ALDOC,
PGD |
|
Glycolysis/Gluconeogenesis | 5 | 0.0025 | ALDOA, GPI, LDHB,
ALDH7A1, ALDOC |
| Hypertrophic
cardiomyopathy (HCM) | 5 | 0.0087 | ATP2A2, LMNA,
ITGB4, ITGA3, ITGB1 |
| Dilated
cardiomyopathy | 5 | 0.0114 | ATP2A2, LMNA,
ITGB4, ITGA3, ITGB1 |
| Focal adhesion | 7 | 0.0120 | LAMB3, ACTN4,
ITGB4, ITGA3, LAMB1, CAPN2, ITGB1 |
| Small cell lung
cancer | 4 | 0.0465 | LAMB3, ITGA3,
LAMB1, ITGB1 |
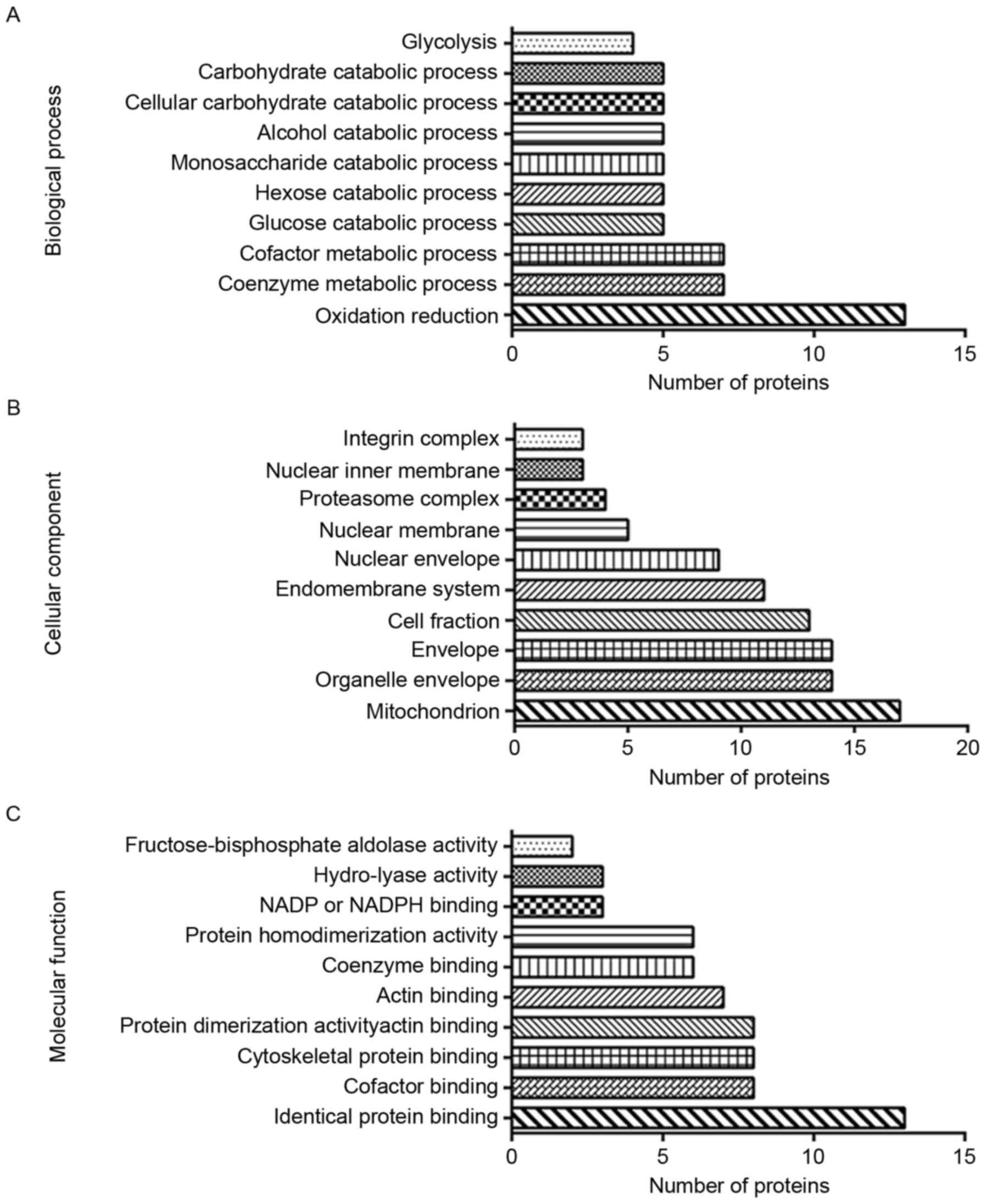
GO annotation included the biological process,
cellular component and molecular function GO categories. In
Biological Process, the top 3 were ‘oxidation reduction’, ‘coenzyme
metabolic process’ and ‘cofactor metabolic process’. In cellular
component, differentially expressed proteins were most likely to be
associated with ‘mitochondrion’, ‘organelle envelope’, ‘envelope’
and ‘cell fraction’. Molecular Function analysis indicated that the
proteins were chiefly involved in ‘identical protein binding’,
‘cofactor binding’, ‘cytoskeletal protein binding’ and ‘actin
binding’. The results of GO annotation are illustrated in Fig. 4.
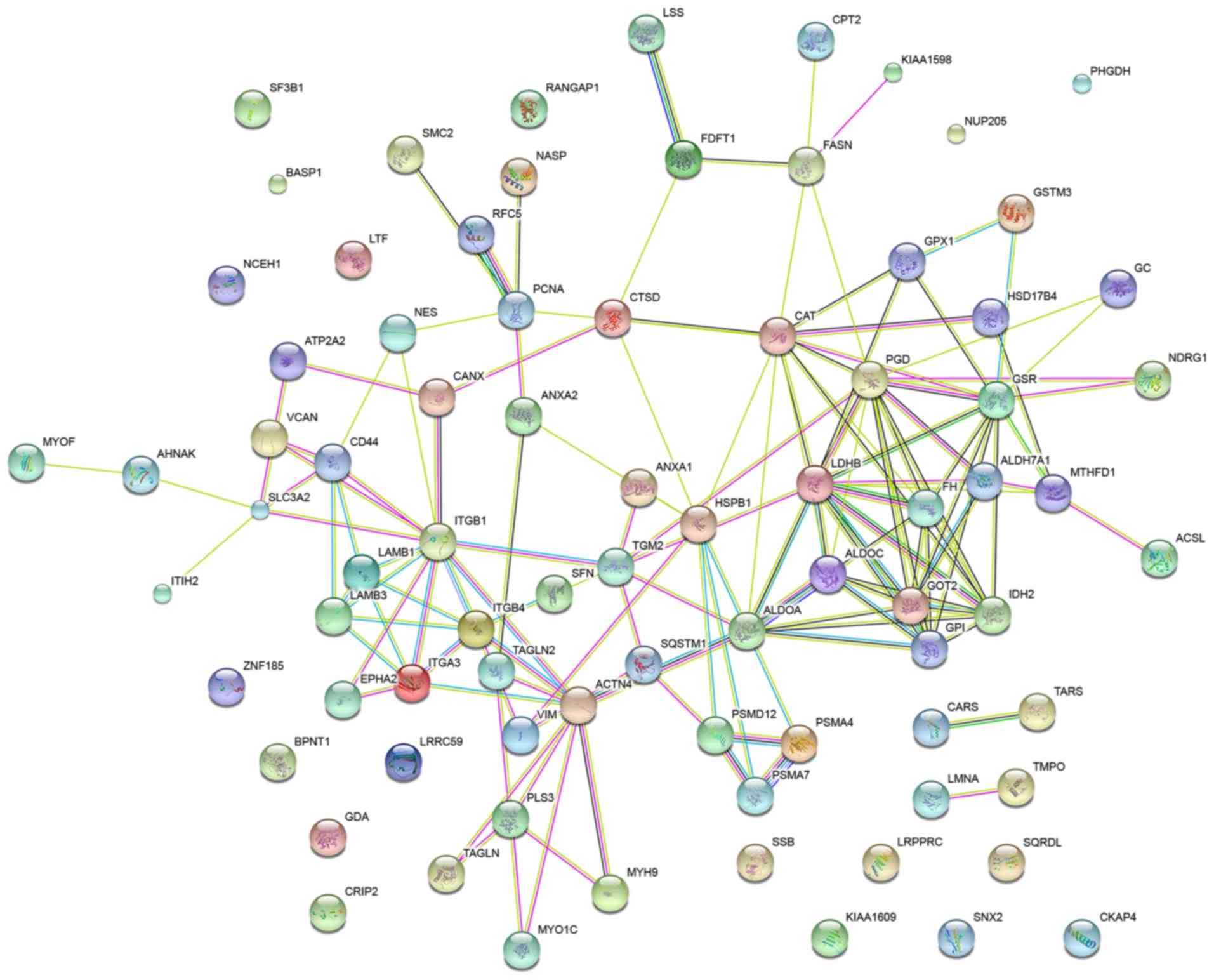
Protein-protein interaction
network
To elucidate the protein-protein interactions of the
globally differentially expressed proteins in the SPC-A-1 cells
treated with a miR-148a inhibitor, a STRING database search was
performed. The database identified interactions for 81 proteins at
the medium confidence level (STRING score, 0.4). The network is
displayed in Fig. 5.
GO analysis in STRING revealed that the GO terms
most significantly associated with the differentially expressed
proteins in the network were ‘small molecule metabolic process’,
‘epithelium development’, ‘carboxylic acid metabolic process’,
‘oxoacid metabolic process’ and ‘organonitrogen compound metabolic
process’. GSR, IDH2, ALDH7A1, GOT2, PMSA4 and PMSA7 were associated
with ‘small molecule metabolic process’, VIM, LAMB3, CAT, TAGLN and
TAGLN2 with ‘epithelium development’ and CARS, TARS, GPI, ALDOA and
PGD with ‘carboxylic acid metabolic process’.
Discussion
It has been identified that miR-148a serves
important functions in various types of cancer (35,36). Our
previous study demonstrated that miR-148a exerted
metastasis-suppressive effects in NSCLC, suppressing NSCLC invasion
and metastasis in vitro and in vivo (19). These results have been corroborated by
other studies (21,37), indicating that miR-148a may provide a
promising therapeutic target against NSCLC. However, the molecular
mechanisms underlying the metastasis-suppressive effects of
miR-148a on NSCLC remain poorly understood. An increasing number of
researchers employ proteomic strategies to seek downstream putative
targets and molecular mechanisms modulated by miRNAs (16–18). In
the present study, iTRAQ technology combined with NanoLC-MS/MS was
used to analyze the global protein expression profiles of SPC-A-1
cells treated with the miR-148a inhibitor and control, and
therefore, to explore the molecular mechanisms modulated by
miR-148a. A total of 84 differentially expressed proteins were
identified, of which 44 proteins were upregulated and 40 were
downregulated. A number of these miR-148a-regulated proteins may be
associated with cancer migration.
The protein expression levels of four upregulated
proteins (LAMB3, VIM, PHGDH and ACTN4) and six downregulated
proteins (FH, HSPB1, IDH2, LDHB, FASN and CAT) were examined by WB
analysis; all results were consistent with the MS results. LAMB3 is
a member of the laminin family of large glycoproteins, present in
various types of basement membrane (BM) (38). LAMB3 is associated with the metastasis
of a number of types of tumor (39–41).
Concordantly, our previous study (34) demonstrated that the protein expression
level of LAMB3 was higher in NSCLC compared with non-cancerous
adjacent tissues and that LAMB3 expression was associated with
lymphatic metastasis. LAMB3 may be a suitable therapeutic target in
NSCLC. Vimentin is responsible for maintaining cell shape and
integrity of the cytoplasm, and stabilizing cytoskeletal
interactions. It is a vital mesenchymal marker that participates in
the endothelial-mesenchymal transition (EndMT) and tumor metastasis
(42). The upregulation of vimentin
in NSCLC was detected in a previous study (43). PHGDH is an enzyme that catalyzes the
NAD+-dependent conversion of 3-phosphoglycerate to
phosphohydroxypyruvate. The conversion is the first step in the
de novo serine synthesis pathway (44). In patients with gastric cancer, high
PHGDH protein expression is associated with a poor prognosis
(45). In addition, PHGDH expression
may promote tumor initiation and metastasis in breast cancer
(28). However, the functions of
PHGDH in NSCLC are not clear. ACTN4 participates in the formation
of the filopodia and lamellipodia, which are important for cell
motility, by regulating the flexibility of actin filaments
(46). It has been reported that
ACTN4 may promote the metastatic potential of lung cancer (31).
With regard to the six verified downregulated
proteins, IDH2 is a mitochondrial NADP-dependent isocitrate
dehydrogenase that functions variably in different types of cancer.
It has been reported that IDH2 inhibited the invasion of
hepatocellular carcinoma cells via the regulation of MMP9, and
therefore acted as a tumor suppressor (47). By contrast, in another study the
overexpression of IDH2 promoted cell growth in colon cancer
(48). It has been suggested that FH
suppresses the tumorigenesis, development and invasion of various
types of cancer (49). FH mRNA
expression and protein expression have been observed to be
significantly lower in lung cancer cells and tissue samples
(50). However, the molecular
mechanisms for the tumor suppressive functions of FH are
uncharacterized. HSPB1 has been reported as a multifunctional
molecule; for example, the expression level of HSPB1 is higher in
nasopharyngeal carcinoma compared with the adjacent non-tumor
tissues (51). HSPB1 has also been
demonstrated to suppress pulmonary fibrosis and lung tumorigenesis
through inhibiting the EndMT. However, the association of HSPB1
deficiency with lung metastasis is unknown (52). The EndMT is characterized by the loss
of endothelial marker expression and the acquisition of a
mesenchymal or fibroblastic phenotype, including the production of
fibroblastic protein-1, type I collagen and smooth muscle actin,
resulting in cells that have invasive and migratory potential
(53,54). Cancer-associated fibroblasts (CAFs)
have been indicated to promote tumor cell proliferation by inducing
changes in the tumor microenvironment (55). The EndMT is an important source of
CAFs in pancreatic carcinoma (56)
and enhances the invasiveness of infected endothelial cells,
contributing to malignant progression (57). A previous study suggested the EndMT
may be necessary for metastatic extravasation in brain endothelial
cells (58). TGFβ-associated signals
have been linked to the EndMT in cancer (59); however, the mechanisms of EndMT in
cancer are incompletely characterized and require further
investigation.
Lactate dehydrogenase B (LDHB) has been reported as
a suppressor of glycolysis and pancreatic cancer progression
(60). However, another study
indicated that the high expression of LDHB was crucial for
osteosarcoma cell growth, proliferation, migration and invasion,
and that it predicted a poor prognosis in patients with
osteosarcoma (61). A previous study
indicated that fatty acid synthase promoted the proliferation of
breast cancer cells, and was a primary target of miR-15a and
miR-16-1 in breast cancer (62). An
association between high LDHB expression and reduced survival time
has been suggested for patients with NSCLC (63). Catalase is a key antioxidant enzyme
that protects against oxidative stress; it has been reported to be
a tumor suppressor that inhibits the migration and invasion of lung
cancer cells (64), and it may
therefore serve as a therapeutic target for lung cancer.
There has been increasing research regarding the
epigenetic modifications in the etiology of human diseases,
including various types of cancer. DNA methylation of CpG islands
is established and maintained by DNA methyltransferases (DNMTs),
including DNMT1, DNMT3A and DNMT3B (65). There may be an association between the
expression of DNMT1 and miR-148a; the overexpression of DNMT1 may
induce the hypermethylation of the miR-148a promoter, and DNMT1 may
be a direct target for inhibition by miR-148a. This regulation loop
has been reported in breast and gastric cancer (22,66).
However, to the best of our knowledge, there are no studies
regarding the association between miR-148a and DNMT1 in lung
cancer, and although DNMT was detected in the present study, there
was no alteration to its expression subsequent to the inhibition of
miR-148a. The lack of difference in DNMT1 protein expression in the
present study may be a discrepancy caused by tumor heterogeneity.
Despite the progress in understanding the molecular mechanisms of
miR-148a and its function in different types of cancer, the topic
remains ambiguous, and further investigation is required.
In the present study, differentially expressed
proteins were identified using a proteomics strategy in SPC-A-1
cells after transfection with the miR-148a inhibitor; the
differentially expressed proteins were then analyzed using
bioinformatics tools. The results may provide a deeper insight into
the molecular mechanisms underlying the metastasis-suppressive
effect of miR-148a on NSCLC. The targets of miR-148a may directly
or indirectly interact with tumor-associated proteins to affect the
metastasis of NSCLC via the pathways identified in the
bioinformatics analysis. The present study also supports the
possibility that miR-148a may be a potential therapeutic target
against NSCLC.
Acknowledgements
The present study was supported by the Shanghai
Natural Science Foundation (grant no. 12ZR1428900) and the State
Key Laboratory of Oncogenes and Related Genes Research Fund (grant
no. 91-15-09).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Fedewa SA, Miller KD,
Goding-Sauer A, Pinheiro PS, Martinez-Tyson D and Jemal A: Cancer
statistics for Hispanics/Latinos, 2015. CA Cancer J Clin.
65:457–480. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mizuno K, Mataki H, Seki N, Kumamoto T,
Kamikawaji K and Inoue H: MicroRNAs in non-small cell lung cancer
and idiopathic pulmonary fibrosis. J Hum Genet. 62:57–65. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu
J, Wang W, Pfeffer LM, Shao ZM, Wu ZH and Wu J: MiRNA-621
sensitizes breast cancer to chemotherapy by suppressing FBXO11 and
enhancing p53 activity. Oncogene. 35:448–458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen B, Yu S, Zhang Y, Yuan Y, Li X, Zhong
J and Feng J: miR-590-5p regulates gastric cancer cell growth and
chemosensitivity through RECK and the AKT/ERK pathway. Onco Targets
Ther. 9:6009–6019. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Wu H, Li W, Yin L, Guo S, Xu X,
Ouyang Y, Zhao Z, Liu S, Tian Y, et al: Downregulated miR-506
expression facilitates pancreatic cancer progression and
chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene.
35:5501–5514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ai J, Huang H, Lv X, Tang Z, Chen M, Chen
T, Duan W, Sun H, Li Q, Tan R, et al: FLNA and PGK1 are two
potential markers for progression in hepatocellular carcinoma. Cell
Physiol Biochem. 27:207–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mueller LN, Brusniak MY, Mani DR and
Aebersold R: An assessment of software solutions for the analysis
of mass spectrometry based quantitative proteomics data. J Proteome
Res. 7:51–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin HC, Zhang FL, Geng Q, Yu T, Cui YQ,
Liu XH, Li J, Yan MX, Liu L, He XH, et al: Quantitative proteomic
analysis identifies CPNE3 as a novel metastasis-promoting gene in
NSCLC. J Proteome Res. 12:3423–3433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaller M, Liffers ST, Oeljeklaus S,
Kuhlmann K, Röh S, Hoffmann R, Warscheid B and Hermeking H:
Genome-wide characterization of miR-34a induced changes in protein
and mRNA expression by a combined pulsed SILAC and microarray
analysis. Mol Cell Proteomics. 10:M111.0104622011.doi:
10.1074/mcp.M111.010462. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baek D, Villen J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Litholdo CG Jr, Parker BL, Eamens AL,
Larsen MR, Cordwell SJ and Waterhouse PM: Proteomic identification
of putative microRNA394 target genes in arabidopsis thaliana
identifies major latex protein family members critical for normal
development. Mol Cell Proteomics. 15:2033–2047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Yu T, Cao J, Liu L, Liu Y, Kong HW,
Zhu MX, Lin HC, Chu DD, Yao M and Yan MX: MicroRNA-148a suppresses
invasion and metastasis of human non-small-cell lung cancer. Cell
Physiol Biochem. 37:1847–1856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Joshi P, Jeon YJ, Lagana A, Middleton J,
Secchiero P, Garofalo M and Croce CM: MicroRNA-148a reduces
tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc
Natl Acad Sci USA. 112:8650–8655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu A, Xia J, Zuo J, Jin S, Zhou H, Yao L,
Huang H and Han Z: MicroRNA-148a is silenced by hypermethylation
and interacts with DNA methyltransferase 1 in gastric cancer. Med
Oncol. 29:2701–2709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang SL and Liu L: MicroRNA-148a inhibits
hepatocellular carcinoma cell invasion by targeting
sphingosine-1-phosphate receptor 1. Exp Ther Med. 9:579–584. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang R, Li M, Zang W, Chen X, Wang Y, Li
P, Du Y, Zhao G and Li L: miR-148a regulates the growth and
apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour
Biol. 35:837–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Q, He M, Ma MT, Wu HZ, Yu ZJ, Guan
S, Jiang LY, Wang Y, Zheng DD, Jin F and Wei MJ: MicroRNA-148a
inhibits breast cancer migration and invasion by directly targeting
WNT-1. Oncol Rep. 35:1425–1432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sakamoto N, Naito Y, Oue N, Sentani K,
Uraoka N, Oo Zarni H, Yanagihara K, Aoyagi K, Sasaki H and Yasui W:
MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and
indicates tumor invasiveness and poor prognosis. Cancer Sci.
105:236–243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yi WR, Li ZH, Qi BW, Ernest ME, Hu X and
Yu AX: Downregulation of IDH2 exacerbates the malignant progression
of osteosarcoma cells via increased NF-κB and MMP-9 activation.
Oncol Rep. 35:2277–2285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Samanta D, Park Y, Andrabi SA, Shelton LM,
Gilkes DM and Semenza GL: PHGDH expression is required for
mitochondrial redox homeostasis, breast cancer stem cell
maintenance and lung metastasis. Cancer Res. 76:4430–4442. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu QS, Rosenblatt K, Huang KL, Lahat G,
Brobey R, Bolshakov S, Nguyen T, Ding Z, Belousov R, Bill K, et al:
Vimentin is a novel AKT1 target mediating motility and invasion.
Oncogene. 30:457–470. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen CL, Chung T, Wu CC, Ng KF, Yu JS,
Tsai CH, Chang YS, Liang Y, Tsui KH and Chen YT: Comparative tissue
proteomics of microdissected specimens reveals novel candidate
biomarkers of bladder cancer. Mol Cell Proteomics. 14:2466–2478.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao Y, Li G, Sun L, He Y, Li X, Sun Z,
Wang J, Jiang Y and Shi J: ACTN4 and the pathways associated with
cell motility and adhesion contribute to the process of lung cancer
metastasis to the brain. BMC Cancer. 15:2772015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou W, Fan MY, Wei YX, Huang S, Chen JY
and Liu P: The expression of MYH9 in osteosarcoma and its effect on
the migration and invasion abilities of tumor cell. Asian Pac J
Trop Med. 9:597–600. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qin Q, Wei F, Zhang J and Li B: miR-134
suppresses the migration and invasion of non-small cell lung cancer
by targeting ITGB1. Oncol Rep. 37:823–830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang XM, Li J, Yan MX, Liu L, Jia DS, Geng
Q, Lin HC, He XH, Li JJ and Yao M: Integrative analyses identify
osteopontin, LAMB3 and ITGB1 as critical pro-metastatic genes for
lung cancer. PLoS One. 8:e557142013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang H, Wang Y, Xu T, Li C, Wu J, He Q,
Wang G, Ding C, Liu K, Tang H and Ji F: Increased expression of
microRNA-148a in osteosarcoma promotes cancer cell growth by
targeting PTEN. Oncol Lett. 12:3208–3214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lombard AP, Mooso BA, Libertini SJ, Lim
RM, Nakagawa RM, Vidallo KD, Costanzo NC, Ghosh PM and Mudryj M:
miR-148a dependent apoptosis of bladder cancer cells is mediated in
part by the epigenetic modifier DNMT1. Mol Carcinog. 55:757–767.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li J, Song Y, Wang Y, Luo J and Yu W:
MicroRNA-148a suppresses epithelial-to-mesenchymal transition by
targeting ROCK1 in non-small cell lung cancer cells. Mol Cell
Biochem. 380:277–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Simon-Assmann P, Orend G, Mammadova-Bach
E, Spenlé C and Lefebvre O: Role of laminins in physiological and
pathological angiogenesis. Int J Dev Biol. 55:455–465. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tanis T, Cincin ZB, Gokcen-Rohlig B,
Bireller ES, Ulusan M, Tanyel CR and Cakmakoglu B: The role of
components of the extracellular matrix and inflammation on oral
squamous cell carcinoma metastasis. Arch Oral Biol. 59:1155–1163.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamamoto N, Kinoshita T, Nohata N, Itesako
T, Yoshino H, Enokida H, Nakagawa M, Shozu M and Seki N: Tumor
suppressive microRNA-218 inhibits cancer cell migration and
invasion by targeting focal adhesion pathways in cervical squamous
cell carcinoma. Int J Oncol. 42:1523–1532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kinoshita T, Hanazawa T, Nohata N, Kikkawa
N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto
Y and Seki N: Tumor suppressive microRNA-218 inhibits cancer cell
migration and invasion through targeting laminin-332 in head and
neck squamous cell carcinoma. Oncotarget. 3:1386–1400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tadokoro A, Kanaji N, Liu D, Yokomise H,
Haba R, Ishii T, Takagi T, Watanabe N, Kita N, Kadowaki N and
Bandoh S: Vimentin regulates invasiveness and is a poor prognostic
marker in non-small cell lung cancer. Anticancer Res. 36:1545–1551.
2016.PubMed/NCBI
|
|
44
|
Unterlass JE, Basle A, Blackburn TJ,
Tucker J, Cano C, Noble ME and Curtin NJ: Validating and enabling
phosphoglycerate dehydrogenase (PHGDH) as a target for
fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget. 13139–13153. 2018.PubMed/NCBI
|
|
45
|
Xian Y, Zhang S, Wang X, Qin J, Wang W and
Wu H: Phosphoglycerate dehydrogenase is a novel predictor for poor
prognosis in gastric cancer. Onco Targets Ther. 9:5553–5560. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shao H, Wang JH, Pollak MR and Wells A:
α-actinin-4 is essential for maintaining the spreading, motility
and contractility of fibroblasts. PLoS One. 5:e139212010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tian GY, Zang SF, Wang L, Luo Y, Shi JP
and Lou GQ: Isocitrate dehydrogenase 2 suppresses the invasion of
hepatocellular carcinoma cells via matrix metalloproteinase 9. Cell
Physiol Biochem. 37:2405–2414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lv Q, Xing S, Li Z, Li J, Gong P, Xu X,
Chang L, Jin X, Gao F, Li W, et al: Altered expression levels of
IDH2 are involved in the development of colon cancer. Exp Ther Med.
4:801–806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tomlinson IP, Alam NA, Rowan AJ, Barclay
E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et
al: Germline mutations in FH predispose to dominantly inherited
uterine fibroids, skin leiomyomata and papillary renal cell cancer.
Nat Genet. 30:406–410. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ming Z, Jiang M, Li W, Fan N, Deng W,
Zhong Y, Zhang Y, Zhang Q and Yang S: Bioinformatics analysis and
expression study of fumarate hydratase in lung cancer. Thorac
Cancer. 5:543–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cai XZ, Zeng WQ, Xiang Y, Liu Y, Zhang HM,
Li H, She S, Yang M, Xia K and Peng SF: iTRAQ-based quantitative
proteomic analysis of nasopharyngeal carcinoma. J Cell Biochem.
116:1431–1441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Choi SH, Nam JK, Kim BY, Jang J, Jin YB,
Lee HJ, Park S, Ji YH, Cho J and Lee YJ: HSPB1 inhibits the
endothelial-to-mesenchymal transition to suppress pulmonary
fibrosis and lung tumorigenesis. Cancer Res. 76:1019–1030. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Potenta S, Zeisberg E and Kalluri R: The
role of endothelial-to-mesenchymal transition in cancer
progression. Br J Cancer. 99:1375–1379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin F, Wang N and Zhang TC: The role of
end othelial-mesenchymal transition in development and pathological
process. IUBMB Life. 64:717–723. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Augsten M: Cancer-associated fibroblasts
as another polarized cell type of the tumor microenvironment. Front
Oncol. 4:622014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zeisberg EM, Potenta S, Xie L, Zeisberg M
and Kalluri R: Discovery of endothelial-to-mesenchymal transition
as a source for carcinoma-associated fibroblasts. Cancer Res.
67:10123–10128. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gasperini P, Espigol-Frigole G, McCormick
PJ, Salvucci O, Maric D, Uldrick TS, Polizzotto MN, Yarchoan R and
Tosato G: Kaposi sarcoma herpesvirus promotes
endothelial-to-mesenchymal transition through notch-dependent
signaling. Cancer Res. 72:1157–1169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Krizbai IA, Gasparics Á, Nagyőszi P,
Fazakas C, Molnár J, Wilhelm I, Bencs R, Rosivall L and Sebe A:
Endothelial-mesenchymal transition of brain endothelial cells:
Possible role during metastatic extravasation. PLoS One.
10:e01238452015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
van Meeteren LA and ten Dijke P:
Regulation of endothelial cell plasticity by TGF-β. Cell Tissue
Res. 347:177–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cui J, Quan M, Jiang W, Hu H, Jiao F, Li
N, Jin Z and Wang L, Wang Y and Wang L: Suppressed expression of
LDHB promotes pancreatic cancer progression via inducing glycolytic
phenotype. Med Oncol. 32:1432015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li C, Chen Y, Bai P, Wang J, Liu Z, Wang T
and Cai Q: LDHB may be a significant predictor of poor prognosis in
osteosarcoma. Am J Transl Res. 8:4831–4843. 2016.PubMed/NCBI
|
|
62
|
Wang J, Zhang X, Shi J, Cao P, Wan M,
Zhang Q, Wang Y, Kridel SJ, Liu W, Xu J, et al: Fatty acid synthase
is a primary target of miR-15a and miR-16-1 in breast cancer.
Oncotarget. 7:78566–78576. 2016.PubMed/NCBI
|
|
63
|
Cerne D, Zitnik IP and Sok M: Increased
fatty acid synthase activity in non-small cell lung cancer tissue
is a weaker predictor of shorter patient survival than increased
lipoprotein lipase activity. Arch Med Res. 41:405–409. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tsai JY, Lee MJ, Chang Dah-Tsyr M and
Huang H: The effect of catalase on migration and invasion of lung
cancer cells by regulating the activities of cathepsin S, L and K.
Exp Cell Res. 323:28–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang W and Xu J: DNA methyltransferases
and their roles in tumorigenesis. Biomark Res. 5:12017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y,
Qi YT, Xu Q, Li W, Lu B, et al: A regulatory circuit of
miR-148a/152 and DNMT1 in modulating cell transformation and
tumorangiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 5:3–13.
2013. View Article : Google Scholar : PubMed/NCBI
|