Introduction
Ovarian cancer is the most lethal gynecological
malignancy in the world (1). Due to
the limited number of specific symptoms, women usually only seek
medical help once the disease is at an advanced stage, with distant
metastases (2). Overall, 90% of
ovarian cancer cases are epithelial ovarian cancer (EOC) (3). Standard therapy for advanced EOC
involves a combination of cytoreductive surgery and platinum-based
chemotherapy, with the combination of paclitaxel and platinum being
the standard adjuvant chemotherapy regimen for EOC (4). Paclitaxel is an important agent for EOC
treatment, and is an effective first-line therapy for advanced
ovarian cancer. However, recurrence still affects the majority of
patients a short period following chemotherapeutic intervention
(5). The main cause for the failure
of chemotherapy is chemoresistance of the tumor tissues, which
adversely affects the prognosis of patients with ovarian cancer.
Furthermore, patients with ovarian carcinoma may have variable
responses to the standard chemotherapeutic regimen, even when they
have the same histologic type. Heterogeneity of the tumor tissue,
one of the primary features of malignancies, is thought to be the
main factor causing this difference (6). With the incidence of paclitaxel
resistance increasing, it is necessary to identify novel, specific
biomarkers that predict chemosensitivity to paclitaxel to improve
outcomes for patients with ovarian cancer (7,8).
The chemosensitivity test is an in vitro,
predictive assay for used to assess cancer cell sensitivity to a
range of chemotherapeutic agents. Adenosine triphosphate-tumor
chemosensitivity assays (ATP-TCA) are sensitive assays that have
been widely used to determine the drug sensitivity of solid tumors
in the past few years (9). ATP-TCA
measures the intracellular ATP levels of drug-exposed cells and
untreated controls to assess tumor cell viability. This method has
notable advantages for guiding the design of chemotherapy protocols
and individualized treatments, and assessing novel chemotherapeutic
drugs. Since its introduction, a number of studies have reported
that ATP-TCA have a high sensitivity and a positive predictive
value, and accurately predict the response to chemotherapy in
ovarian cancer (10,11).
In the present study, ATP-TCA was used to assess the
chemosensitivity of EOC to paclitaxel. Parameters determined by
analyzing the correlation between the inhibition rate and
paclitaxel doses were measured as follows: Inhibitory concentration
(IC)90 and IC50, (90 or 50% growth inhibition
in vitro, respectively), and sensitivity index (SI), which
was calculated by summation of the percentage of tumor growth
inhibition (TGI) at each concentration detected (12). SI >250 was suggested to be the
optimal standard for predicting chemoresistance. Therefore, 250
were selected as the cut-off point for SI in the present study
(13).
Paclitaxel is known to induce cytotoxicity by
triggering apoptosis via regulation of the expression of
apoptosis-associated proteins in the caspase-independent and
caspase-dependent pathways, or by preventing tubulin
depolymerization during the metaphase to anaphase transition of
mitosis (14). However, paclitaxel
resistance limits its use in the long-term management of EOC, and
the molecular mechanisms underlying this resistance remain to be
fully elucidated. Therefore, the identification of specific markers
for ovarian cancer with paclitaxel resistance is a long-term goal
of the medical community. The present study aimed to identify
proteins associated with paclitaxel resistance in ovarian cancer,
in order to investigate the molecular mechanisms underlying
paclitaxel resistance and discover potential novel drug targets for
paclitaxel-resistant ovarian cancer (15).
In the present study, two approaches for
quantitative proteomic analysis were selected for identifying the
differentially expressed proteins between paclitaxel-resistant and
paclitaxel-sensitive groups of ovarian cancer tissues: iTRAQ
analysis and two-dimensional electrophoresis coupled to liquid
chromatography tandem mass spectrometry (LC-MS/MS). iTRAQ is a gel
free mass spectrometry technique, applying isobaric amine specific
tags to compare peptide intensities between samples, then inferring
quantitative values for the corresponding proteins. LC-MS/MS is
based on the differential two-dimensional gel electrophoresis
pattern between protein samples and provides additional biological
information, including molecular weight alterations or isoelectric
point drift, based on which protein functions are implicated
(16). The present study aimed to
identify biomarkers, which were associated with
paclitaxel-resistant ovarian cancer, providing information to aid
our understanding of the underlying molecular mechanisms and to
predict treatment responses to therapeutic agents. The ovarian
cancer-specific proteins identified were further confirmed by
western blot analysis (17).
Materials and methods
Ethics statement
The study protocol received approval from the Ethics
Committee of the Beijing Shijitan Hospital, Capital Medical
University (Beijing, China). Written, signed informed consent was
obtained from all patients and their family members prior to
surgery. All procedures were carried out in agreement with the Code
of Ethics of the World Medical Association (Declaration of
Helsinki, 1964; as revised in 2004).
Tumor samples
A total of 54 fresh specimens were obtained from
patients with EOC who underwent staging surgery at the Beijing
Shijitan Hospital, Beijing University People's Hospital (Beijing,
China), People's Liberation Army General Hospital (Beijing, China)
and Beijing Obstetrics and Gynecology Hospital (Beijing, China),
between March 2013 and December 2014. Routine histopathology was
conducted on formalin-fixed and paraffin-embedded samples, which
were obtained from the same tissues, by at least two experienced
gynecological pathologists (the Beijing Shijitan Hospital) in order
to determine the malignancy and the stage of the tumor samples.
Each fresh collected sample was divided into two fractions: One was
prepared for ATP-TCA, and the other was stored at −80°C for
subsequent tests. The ATP-TCA was conducted as a routine procedure
immediately following surgery using residual primary tumor samples
which were not required for histopathology. The sensitivity of
viable ovarian cancer cells harvested from malignant tissues to
paclitaxel (Corden Pharma Latina S.P.A., Sermoneta Italy) was then
detected as follows.
In vitro ATP-TCA
An ATP-TCA kit, containing serum-free complete assay
medium, digestive enzyme and luciferin-luciferase reagent (Huzhou
Haichuang Biotech Co., Ltd., Huzhou, China) was used for the
assessment of chemosensitivity. The ATP-TCA was performed as
previously described (12,18). Briefly, samples (1–2 cm3)
were harvested from solid tumors during surgical resection and cut
into smaller fragments (1 mm3). The fragments were then
incubated with 5–10 ml sterile digestive enzyme reagent for 2–3 h
at 37°C in a 5% CO2 incubator, and dissociated to form a
single cell suspension. Once the concentration of the cell
suspension was adjusted to 2–4×105/ml, 100 µl cell
suspension was seeded into a 96-well polypropylene microplate.
Cells were incubated with 5% CO2 at 37°C for 5 days, and
treated with five different doses (12.5, 25, 50, 100 and 200%) of
the test drug concentration (TDC) derived from the plasma peak
concentrations, which were in turn determined by pharmacokinetic
and clinical information (19). The
standard 100% TDC value of paclitaxel was 13.8 g/ml. The assay was
performed in duplicate wells, with positive and negative controls.
For each dose, two controls were included in each plate: A drug
free control comprised of media only (M0) and a maximum inhibitor
(MI) control which kills all cells present. At the end of the 5
day-incubation, the cells were lysed with 50 µl ATP extraction
reagent, and 50 µl luciferin-luciferase reagent was added to each
well. A luminometer (Orion II; Berthold Technologies GmbH & Co.
KG, Bad Wildbad, Germany) was used to assess the level of ATP
present, and an inhibition curve was plotted.
iTRAQ combined with LC-MS/MS
According to the results of the ATP-TCA, tumor
specimens were divided into three main types: Sensitive, weakly
sensitive and resistant. In order to screen the altered proteins
associated with paclitaxel resistance more effectively, sensitive
specimens (S group, n=8) and resistant specimens (R group, n=8)
were selected for iTRAQ analysis. Frozen tissues were homogenized
and sonicated (20 kHz) using 0.5% sodium dodecyl sulfate (SDS) with
a cell disperser, followed by centrifugation at 20,000 × g for 30
min at 4°C to eliminate the cell debris. Following this, the
supernatant was collected, and the Bradford assay was used to
determine protein concentration. Next, 100 µg protein per condition
were treated with dithiothreitol (10 mM) and iodoacetamide (55 mM)
for reduction and alkylation. Following this, the proteins were
digested with trypsin (Promega Corporation, Madison, WI, USA), and
the resultant peptides mixture was further labeled using chemicals
from the iTRAQ reagent kit (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocols. The samples were marked with iTRAQ tags as follows:
iTRAQ115 for the S group and iTRAQ116 for the R group.
Next, the iTRAQ-labeled peptides were pooled and
fractionated by strong cation exchange (SCX) chromatography on a
SCX column (5 µm, 100A; Phenomenex, Torrance, CA, USA) with a
linear gradient from 0% B to 100% B in 90 min at a flow rate of 1
ml/min (solution A: 10 mM KH2PO4, pH 3.0, 25%
acetonitrile; solution B: 2 M KCl, 10 mM
KH2PO4, pH 3.0, 25% acetonitrile). According
to the chromatography results, the collected fractions were
recombined into 16 fractions and then freeze-dried (−10°C).
Following this, each freeze-dried fraction from the SCX column was
re-dissolved in 100 µl 0.1% formic acid aqueous solution, and then
desalted using a strata-X C18 column (Phenomenex). The sample was
then extracted and analyzed using nano-LC-MS/MS with a
quadrupole-Orbitrap mass spectrometer (Q-Exactive; Thermo Fisher
Scientific, Inc.) as previously described (20).
Western blot analysis
Based on the proteomic results, two proteins of
interest, plexin domain containing 2 (Plxdc2) and cytokeratin 7
(CK7), were expressed at higher levels in paclitaxel-resistant
tissues than paclitaxel-sensitive tissues. Western blot analysis
was used to examine the expression of CK7 and Plxdc2 in EOC tissues
with different chemosensitivities (sensitive, weakly sensitive and
resistant). The protein selections were based on a high fold change
(FC) and high significance (Plxdc2, P<0.05; FC=1.539; CK7,
P<0.05; FC=1.724). The extracted proteins (20 µg) were separated
by 12% SDS-PAGE and transferred onto nitrocellulose membranes.
Following blocking with 5% non-fat milk in Tris-buffered saline
with 0.1% Tween-20 at room temperature for 1 h, the membranes were
probed with the following primary antibodies: Rabbit anti-human
polyclonal Plxdc2 (dilution, 1:5,000; cat. no. NBP1-76858; Novus
Biologicals, LLC, Littleton, CO, USA) and rabbit anti-human
polyclonal CK7 (dilution, 1:10,000; cat. no. ab154334; Abcam,
Cambridge, MA, USA) at 4°C overnight. Following washing with
Tris-buffered saline with Tween-20 three times, the membranes were
incubated with a horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin G antibody (dilution, 1:5,000; cat. no. ab97051;
Abcam, Cambridge, MA, USA) at room temperature for 1–2 h. Membranes
were washed as aforementioned and analyzed using a two-color
infrared imaging system (Odyssey; Li-COR Biosciences, Lincoln, NE,
USA). The gray level of each band was calculated using image
processing ImageJ software (version 1.48; National Institutes of
Health, Bethesda, MA, USA). Densitometric analysis of the bands was
conducted three times and normalized to GAPDH (dilution, 1:5,000;
cat. no. ab9485; Abcam, Cambridge, MA, USA).
Data analysis
ATP-TCA
ATP-TCA data were exported to a Microsoft Excel 2010
spreadsheet (Microsoft Corporation, Redmond, WA, USA), and the
parameters SI (SI=500 - sum of % TGI at 200, 100, 50, 25 and 12.5%
TDC), IC90 and IC50 were compared. The three
types of in vitro sensitivity were defined below: Sensitive
(S), IC50 <25% TDC and IC90 ≤100% TDC;
weakly sensitive (WS), IC50 <25% TDC and
IC90 ≤100% TDC or SI ≤250; and resistant (R), SI
>250. Quality control for each assay was conducted as follows:
Two measurements of every drug-treated sample were used for
controlling the variability of individual ATP values. Samples with
coefficient of variation (CV) >0.15 were rejected and retested.
For the present study, the mean CV was 0.048 (range,
0.023–0.114).
iTRAQ assay
LC-MS/MS analysis of iTRAQ-labeled peptides was
performed using Mascot (version 2.3.0) and Proteome Discoverer
Version 1.3 software (Thermo Fisher Scientific, Inc.) and
identification of the proteins was conducted by utilizing the raw
MS data (21). For quantitative iTRAQ
analysis, the peptide was automatically selected by Protein
Discoverer with the Pro Group™ algorithm, and the error
factor, P-value and the reporter peak area were calculated. If the
iTRAQ ratio (sensitive tissues/resistant tissues) was <0.83 or
>1.2 (P<0.05), the protein was considered to be
differentially expressed (22). Next,
Gene Ontology enrichment analysis was conducted to analyze
functions of the differentially expressed proteins using
Bioconductor 3.0 software (https://www.bioconductor.org), and biological process,
molecular function and cellular component were included. For
significant enrichment of the protein sets, a false discovery rate
of <0.05 was considered as a threshold (23–25).
Statistical analysis
All results are expressed as the mean ± standard
deviation. Statistical analysis between groups was performed using
SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA), and comparisons
were made using an unpaired Student's t-test, χ2 test
and one-way analysis of variance (ANOVA). Fishers least significant
difference test was performed on ANOVA data in order to determine
statistical significance. P<0.05 was considered to indicate a
statistically significant difference.
Results
In vitro ATP-TCA
The patients were aged between 20–76 years, with a
median age of 51 years. The tumor characteristics of the samples
are listed in Table I. Notable
heterogeneity in chemosensitivity was observed among the tumor
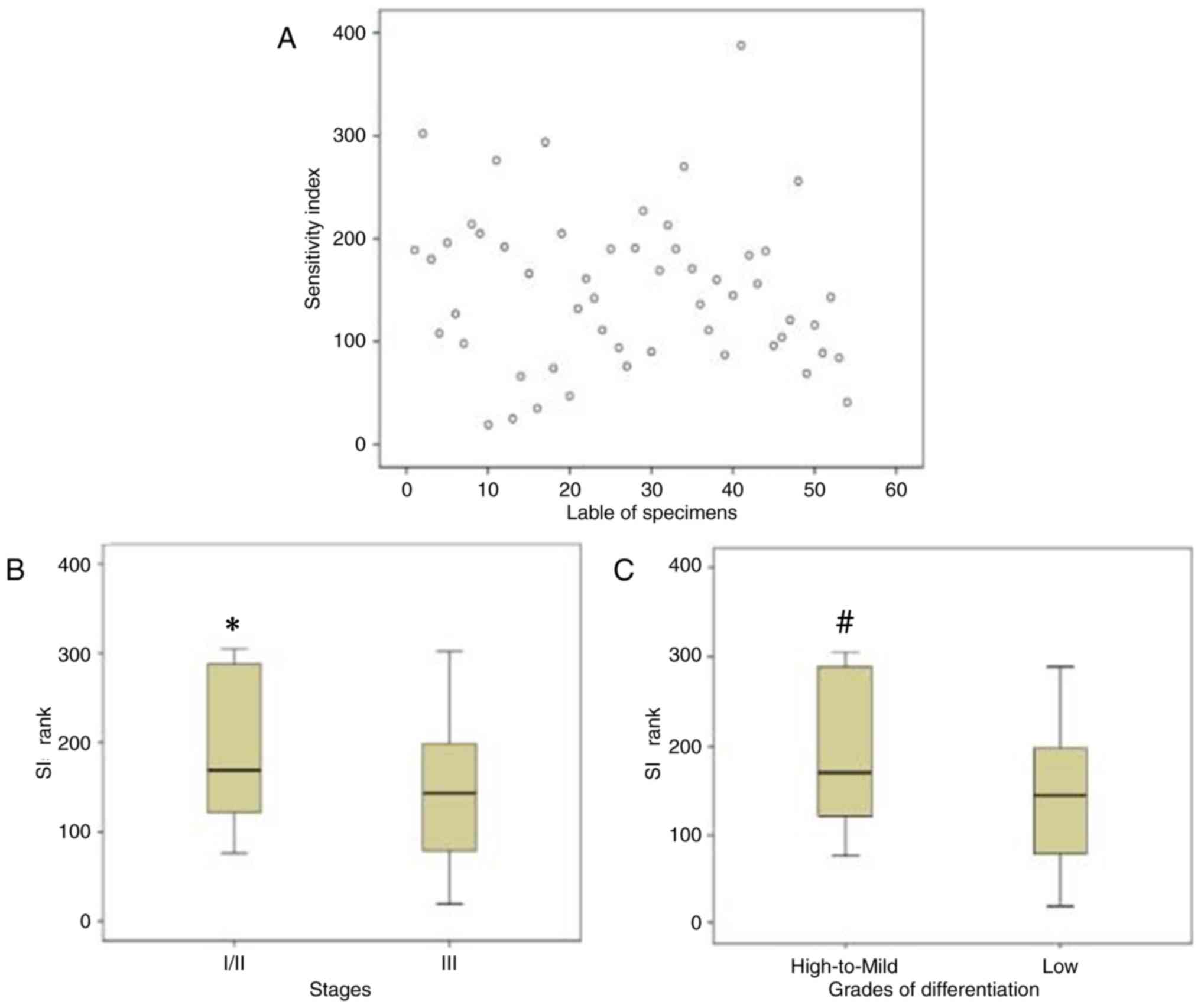
samples examined (Fig. 1A). There was
a significant association between clinical indicators of the tumor
samples and the ATP-TCA results. The associations between the stage
or differentiation grade of the tumor samples and the ATP-TCA
results were assessed using χ2 tests. It was
demonstrated that specimens with high to mild differentiation or an
early stage (I/II) had lower chemosensitivity to paclitaxel when
compared with low-differentiated or advanced stage (III) specimens,
respectively (Table II).
Furthermore, the SIs of different tumor stages and differentiation
grades were also significantly different (Fig. 1B and C).
 | Table I.Characteristics of tumor samples
(n=54). |
Table I.
Characteristics of tumor samples
(n=54).
|
Characteristics | N | (%) |
|---|
| Histology |
|
|
|
Serous | 41 | 75.9 |
|
Mucinous | 2 | 3.7 |
| Clear
cell | 5 | 9.3 |
|
Endometrioid | 4 | 7.4 |
|
Transitional cell | 2 | 3.7 |
| FIGO stage |
|
|
| I | 8 | 14.8 |
| II | 7 | 13.0 |
|
III | 39 | 72.2 |
| Grade of
differentiation |
|
|
|
High | 5 | 9.3 |
|
Mild | 7 | 13.0 |
|
Low | 42 | 77.8 |
| Primary | 48 | 88.9 |
| Recurrent | 6 | 11.1 |
 | Table II.Associations between the adenosine
triphosphate-tumor chemosensitivity assay results for paclitaxel
resistance and the stage or grade of differentiation of tumor
samples. |
Table II.
Associations between the adenosine
triphosphate-tumor chemosensitivity assay results for paclitaxel
resistance and the stage or grade of differentiation of tumor
samples.
|
| FIGO stage |
Differentiation |
|---|
|
|
|
|
|---|
| Sensitivity to
paclitaxel | I/II | III | P-value | High-mild | Low | P-value |
|---|
| S+WS | 8 | 34 | 0.021 | 5 | 37 | 0.003 |
| R | 7 | 5 |
| 7 | 5 |
|
iTRAQ assay
Proteins from paclitaxel-sensitive tissues and
paclitaxel-resistant tissues were quantified by LC-MS/MS and iTRAQ
analysis. In the present study, a total of 496 significantly
differentially-expressed proteins were identified between
paclitaxel-sensitive and paclitaxel-resistant tissues. The
threshold of the iTRAQ ratio (sensitive tissue/resistant tissue)
was <0.83 or >1.2, which implied lower or higher expression
of proteins in sensitive tissues compared with resistant tissues.
Among them, 233 proteins were upregulated in the
paclitaxel-resistant tissues and 263 proteins were downregulated.
Certain proteins with important biological functions are listed in
Table III. In order to investigate
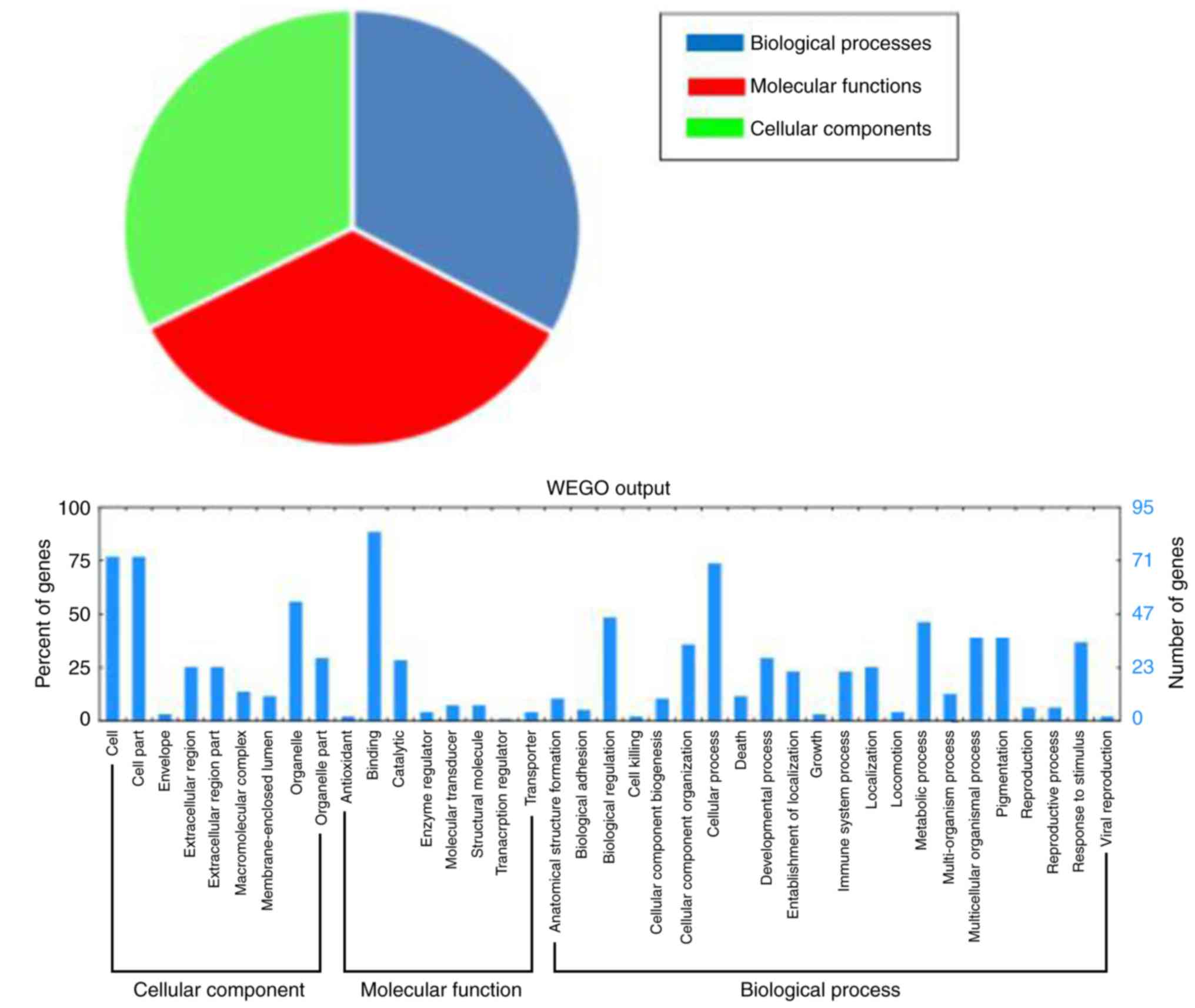
the functions of the differentially expressed proteins, Gene
Ontology enrichment analysis was performed to analyze the functions
of those proteins. A total of 96 differentially expressed proteins
were divided into three categories: ‘Molecular functions’ (92.7%),
‘cellular components’ (87.5%), and ‘biological processes’ (88.5%;
Fig. 2).
 | Table III.Differentially expressed proteins in
S tissues compared with R tissues. |
Table III.
Differentially expressed proteins in
S tissues compared with R tissues.
| Serial no. | Protein | Fold-change for
S/R |
|---|
| Upregulated in R
tissues |
|
|
|
P02765 |
α-2-HS-glycoprotein | 0.346 |
|
Q9BW30 | Tubulin
polymerization-promoting protein family member 3 | 0.359 |
|
Q92954 | Proteoglycan 4 | 0.463 |
|
P00734 | Prothrombin | 0.467 |
|
P07602 | Proactivator
polypeptide | 0.485 |
|
P35080 | Profilin-2 | 0.517 |
|
Q9UNP9 | Peptidyl-prolyl
cis-trans isomerase E | 0.524 |
|
Q14508 | WAP four-disulfide
core domain protein 2 | 0.525 |
|
Q6ZU11 | Uncharacterized
protein C9orf142 | 0.539 |
|
Q9H6Y7 | E3
ubiquitin-protein ligase RNF167 | 0.547 |
|
P09758 | Tumor-associated
calcium signal transducer 2 | 0.552 |
|
P84157 |
Matrix-remodeling-associated protein
7 | 0.565 |
|
Q9H4G0 | Band 4.1-like
protein 1 | 0.567 |
|
P08729 | Cytokeratin 7, type
II cytoskeletal 7 | 0.580 |
|
P42330 | Aldo-keto reductase
family 1 member C3 | 0.587 |
|
O75882 | Attractin | 0.592 |
|
Q969E4 | Transcription
elongation factor A protein-like 3 | 0.595 |
|
Q9Y240 | C-type lectin
domain family 11, member A | 0.604 |
|
P05783 | Cytokeratin 7, type
I cytoskeletal 18 | 0.623 |
|
P81605 | Dermcidin | 0.644 |
|
P09455 | Retinol-binding
protein 1 | 0.649 |
|
Q6UX71 | Plexin
domain-containing protein 2 | 0.650 |
|
O43175 |
D-3-phosphoglycerate dehydrogenase | 0.651 |
|
P55809 |
Succinyl-CoA:3-ketoacid-coenzyme A
transferase 1, mitochondrial | 0.653 |
|
Q7L2H7 | Eukaryotic
translation initiation factor 3 subunit M | 0.688 |
|
Q12805 | EGF-containing
fibulin-like extracellular matrix protein 1 | 0.689 |
|
Q8TEQ8 | GPI ethanolamine
phosphate transferase 3 | 0.691 |
|
Q9C0H2 | Protein tweety
homolog 3 | 0.695 |
|
P00751 | Complement factor
B | 0.698 |
|
Q14676 | Mediator of DNA
damage checkpoint protein 1 | 0.701 |
|
Q9BUH6 | Uncharacterized
protein C9orf142 | 0.702 |
|
Q9BX66 | Sorbin and SH3
domain-containing protein 1 | 0.702 |
|
P02786 | Transferrin
receptor protein 1 | 0.706 |
|
P01861 | Ig γ-4 chain C
region | 0.706 |
|
O15305 | Phosphomannomutase
2 | 0.707 |
|
O43752 | Syntaxin-6 | 0.731 |
|
Q86SX6 |
Glutaredoxin-related protein 5 | 0.732 |
|
Q8NFV4 | Abhydrolase
domain-containing protein 11 | 0.736 |
|
Q14696 | LDLR chaperone
MESD | 0.736 |
|
P17931 | Galectin-3 | 0.739 |
|
Q8WWF6 | DnaJ homolog
subfamily B member 3 | 0.741 |
| Downregulated in R
tissues |
|
|
|
Q15063 | Periostin | 2.041 |
|
P49913 | Cathelicidin
antimicrobial peptide | 2.064 |
|
P41218 | Myeloid cell
nuclear differentiation antigen | 2.111 |
|
P01814 | Ig heavy chain V–II
region OU | 2.145 |
|
Q9HCF4 | Protein ALO17 | 2.231 |
|
P59665 | Neutrophil defensin
1 | 2.232 |
|
P05164 |
Myeloperoxidase | 2.246 |
|
P20962 | Parathymosin | 2.283 |
|
P61626 | Lysozyme C | 2.284 |
| A8MW06 | Thymosin β-4-like
protein 3 | 2.329 |
| Q9NP78 | ATP-binding
cassette sub-family B member 9 | 2.337 |
| P02671 | Fibrinogen α
chain | 2.554 |
| P08311 | Cathepsin G | 2.763 |
| P02675 | Fibrinogen β
chain | 2.784 |
Verification by western blot
analysis
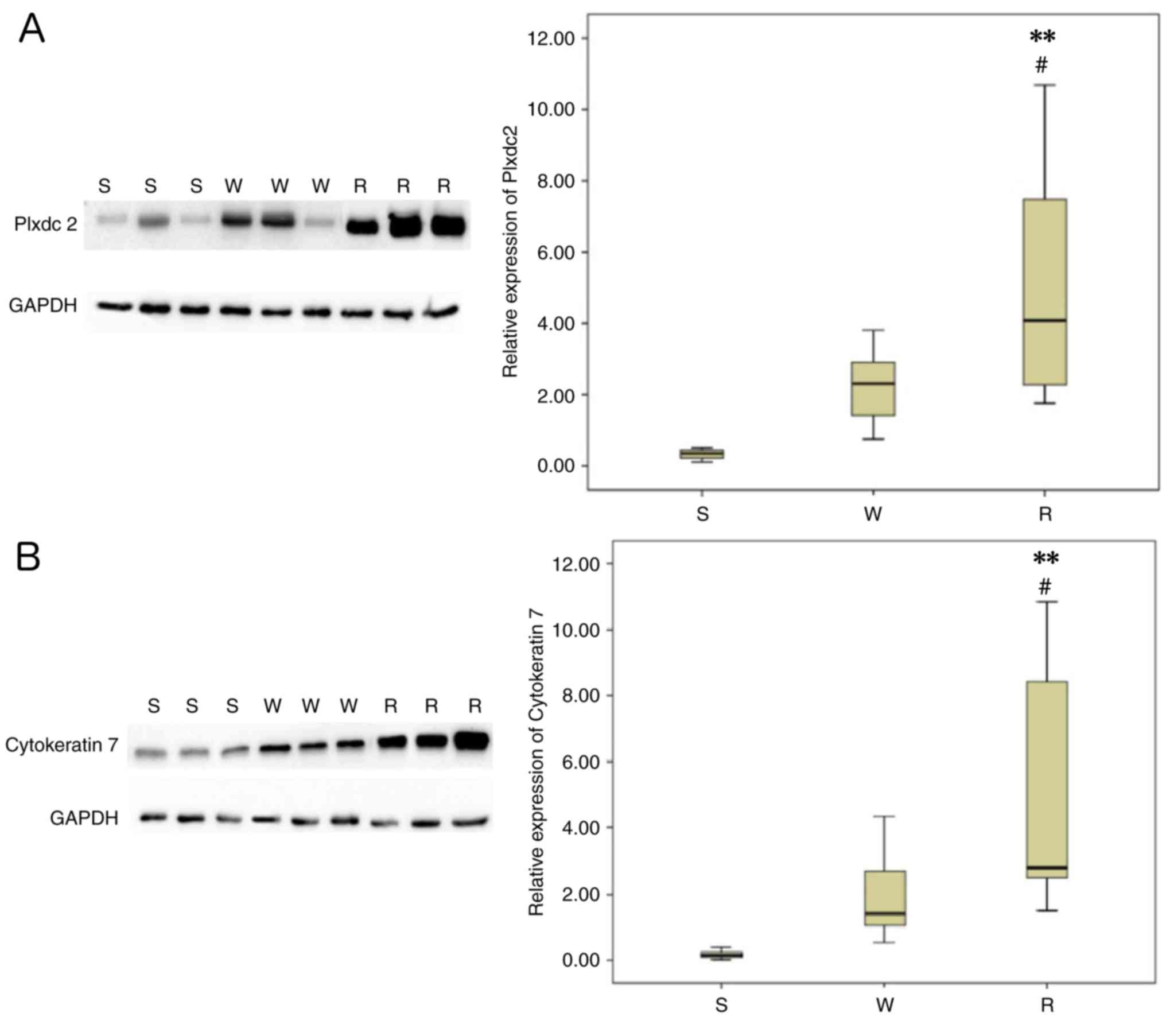
To validate the expression of the two selected
proteins (Plxdc2 and CK7) identified by iTRAQ in EOC tissues with
different chemosensitivities (sensitive, weakly sensitive and
resistant), western blotting was performed and normalized
densitometry data from the western blotting were used for the
determination of relative expression values. Commercially available
antibodies were used for probing the proteins, which were extracted
from eight individuals with each type of tissues. The results were
in concordance with those of the iTRAQ: the protein expression
levels of Plxdc2 and CK7 were significantly increased in the
paclitaxel-resistant tissues compared with the other two types of
tissues (Fig. 3A and B).
Discussion
Ovarian cancer is the most lethal gynecologic
malignancy in adult women (26). The
standard treatment for EOC is surgical resection of the tumor mass,
followed by a combination of paclitaxel and platinum. Although
paclitaxel is effective as a first-line drug for advanced ovarian
cancer, progression of the disease and mortality remain problems
that originate from drug resistance. The main cause of paclitaxel
resistance is thought to be the heterogeneity of the tumor tissue
(27). EOC is biologically and
morphologically heterogeneous, and it is possible to divide cases
into several subtypes, which are then prescribed different
treatments with different clinical outcomes (28). In the present study, an in
vitro ATP-TCA, which has been widely used to determine the drug
sensitivity of solid tumors, was used to assess heterogeneity in
EOC. There was noticeable heterogeneity in chemosensitivity among
the EOC samples examined: Highly- to mildly-differentiated or
early-stage (I/II) EOC specimens had lower chemosensitivity to
paclitaxel when compared with specimens with low differentiation or
an advanced-stage (III), respectively. These results were
consistent with those of a previous study, and implied that
chemotherapy was not effective at preventing the recurrence of
early-stage ovarian cancer (29).
In order to further screen the suitable biomarkers
for predicting chemosensitivity to paclitaxel in ovarian cancer,
the quantitative proteomic technique iTRAQ was performed to analyze
the proteins from paclitaxel-resistant and paclitaxel-sensitive
tissues. A total of 496 significantly differentially expressed
proteins were identified, including 233 proteins which were
upregulated and 263 proteins which were downregulated in
paclitaxel-resistant tissues compared with paclitaxel-sensitive
tissues. Two proteins of interest (Plxdc2 and CK7) were selected
from among the upregulated proteins, which may be associated with
paclitaxel resistance in EOC. The expression of Plxdc2 and CK7 in
EOC tissues with different chemosensitivities (sensitive, weakly
sensitive and resistant) was further detected by western blotting.
The two proteins were revealed to be upregulated in the EOC tissues
with paclitaxel resistance, consistent with the results from the
iTRAQ analysis.
Plxdc2 has the ability to alter normal neurogenesis
patterns, and is a novel mitogen for neural progenitors, and is
present in the developing neural tube (30). Miller et al (31) were interested in Plxdc2 due to its
protein architecture and expression pattern, and described the
expression pattern of Plxdc2 in the developing mouse embryo.
Notable similarities between the Plxdc2 expression multiple Wnt
family members (Wnt1, Wnt3a, Wnt5a and Wnt8b) have been identified
(32). In addition, Cheng et
al (33) revealed that Plxdc2 is
a cell-surface receptor for pigment epithelium derived factor
(PEDF). PEDF is a secreted factor with multiple biological
functions. It was initially considered to be a neurotrophic factor,
but its recognized functions later expanded to include a stem cell
niche factor, an inhibitor of cancer cell growth and, notably, the
most potent natural antiangiogenic factor (34–36). A
number of animal models have demonstrated the therapeutic value of
PEDF in the treatment of blinding diseases and multiple types of
cancer. Even in the presence of strong proangiogenic factors, PEDF
is able to inhibit endothelial cell migration and angiogenesis.
Furthermore, PEDF is a non-inhibitory member of the serine protease
inhibitors (serpin) superfamily, which possesses potent
physiological anti-angiogenic functions. PEDF decreases abnormal
neovascularization by exerting anti-angiogenic effects which
inhibit pro-angiogenic factors, including vascular endothelial
growth factor, and this function has been investigated primarily in
the eye and in cancer (37). In the
present study, Plxdc2 expression was revealed to be upregulated in
paclitaxel-resistant EOC tissues. Therefore, elucidating the
associations between Plxdc2 and PEDF may lead to an improved
understanding of the mechanisms and the development of novel
therapeutic strategies for chemoresistant EOC.
CK7 is a simple, ~55 kDa epithelial cytokeratin
which is primarily expressed in single-layered simple epithelia
(38). Cytokeratins are intermediate
cytoskeletal structural proteins present in the epithelial cells of
the majority of organs, and are involved in mechanical support.
They are also crucial for epithelial function, as cytokeratins are
involved in signal transduction, cell polarity and gene regulation
(39). They are maintained during
carcinogenesis (40,41). CK7 is expressed by a number of ductal
and glandular epithelial cells (mainly gallbladder, hepatic ducts,
and pancreatic ducts), by female genital tract tissues (ovary,
endometrium, fallopian tube, and cervix) and by breast, lung, and
urinary tract tissues (42). Chu
et al (43) conducted
immunohistochemistry to assess CK7 and cytokeratin 20 expression in
435 epithelial malignancy specimens, and 5% stained cells was
considered to be positive. Overall, 100% of lung, ovary, uterine
and salivary gland cancers were CK7-positive. In addition, CK7 is a
low molecular weight cytokeratin and its expression has been used
to assess the differentiation of human primary and metastatic
tumors of unknown origin (44,45). In
the present study, CK7 was revealed to be upregulated in
paclitaxel-resistant EOC tissues, which may be involved in tumor
metastasis and chemoresistance.
In conclusion, the mechanisms underlying paclitaxel
resistance in ovarian cancer remain to be fully elucidated.
Although further studies are required for large-scale validation of
the candidate biomarkers identified by the present study, to the
best of our knowledge the present study is the first to identify
these candidate markers for paclitaxel-resistance in EOC. These
results improve our understanding of the mechanisms underlying
chemotherapy resistance and may help predict responses to targeted
therapeutic agents. Furthermore, the identified proteins may aid
further studies of the molecular mechanisms underlying paclitaxel
treatment and resistance in EOC.
Acknowledgements
The present study was sponsored by the Capital
Health Research and Development Projects of China (grant no.
2011-2008-05).
References
|
1
|
Cornelison R, Llaneza DC and Landen CN:
Emerging Therapeutics to overcome chemoresistance in epithelial
ovarian cancer: A mini-review. Int J Mol Sci. 18:E21712017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duda K, Cholewa H, Łabuzek K,
Boratyn-Nowicka A and Okopień B: Novel strategies of ovarian cancer
treatment. Pol Merku Lekarski. 39:337–342. 2015.
|
|
3
|
Ramalingam P: Morphologic,
immunophenotypic, and molecular features of epithelial ovarian
cancer. Oncology (Williston Park). 30:166–176. 2016.PubMed/NCBI
|
|
4
|
Bookman MA: Optimal primary therapy of
ovarian cancer. Ann Oncol. 27 Suppl 1:i158–i162. 2016. View Article : Google Scholar
|
|
5
|
Matsumoto K, Onda T and Yaegashi N:
Pharmacotherapy for recurrent ovarian cancer: Current status and
future perspectives. Jpn J Clin Oncol. 45:408–410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O'Donnell RL, Kaufmann A, Woodhouse L,
McCormick A, Cross PA, Edmondson RJ and Curtin NJ: Advanced ovarian
cancerdisplays functional intratumor heterogeneity that correlates
to ex vivo drug sensitivity. Int J Gynecol Cancer. 26:1004–1011.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tatar B, Boyraz G, Selçuk İ, Doğan AK,
Usubütün A and Tuncer ZS: In vitro chemosensitivity in ovarian
carcinoma: Comparison of three leading assays. J Turk Ger Gynecol
Assoc. 17:35–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang H, Zhang L, Wei L, Gao X, Tang LI,
Gong W, Min NA, Zhang LI and Yuan Y: Knockdown of cathepsin L
sensitizes ovarian cancer cells to chemotherapy. Oncol Lett.
11:4235–4239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee M, Kim SW, Nam EJ, Cho H, Kim JH, Kim
YT and Kim S: ATP-based chemotherapy response assay in primary or
recurrent ovarian and peritoneal cancer. Yonsei Med J.
55:1664–1671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han SS, Choi SH, Lee YK, Kim JW, Park NH,
Song YS, Lee HP and Kang SB: Predictive value of individualized
tumor response testing by ATP based chemotherapy response assay in
ovarian cancer. Cancer Invest. 26:426–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao D, Zhang W, Li XG, Wang XB, Zhang LF,
Li M, Li YF, Tian HM, Song PP, Liu J, et al: Predicting clinical
chemo-sensitivity of primary ovarian cancer using adenosine
triphosphate-tumor chemo-sensitivity assay combined with detection
of drug resistance genes. Zhonghua Fu Chan Ke Za Zhi. 46:193–198.
2011.(In Chinese). PubMed/NCBI
|
|
12
|
Fehm T, Zwirner M, Wallwiener D, Seeger H
and Neubauer H: Antitumor activity of zoledronic acid in primary
breast cancer cells determined by the ATP tumor chemosensitivity
assay. BMC Cancer. 12:3082012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Neubauer H, Stefanova M, Solomayer E,
Meisner C, Zwirner M, Wallwiener D and Fehm T: Predicting
resistance to platinum-containing chemotherapy with the ATP tumor
chemosensitivity assay in primary ovarian cancer. Anticancer Res.
28:949–955. 2008.PubMed/NCBI
|
|
14
|
Safinya CR, Chung PJ, Song C, Li Y, Ewert
KK and Choi MC: The effect of multivalent cations and Tau on
paclitaxel-stabilized microtubule assembly, disassembly, and
structure. Adv Colloid Interface Sci. 232:9–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian Y, Tan AC, Sun X, Olson MT, Xie Z,
Jinawath N, Chan DW, Shih IeM, Zhang Z and Zhang H: Quantitative
proteomic analysis of ovarian cancer cells identified mitochondrial
proteins associated with paclitaxel resistance. Proteomics Clin
Appl. 3:1288–1295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gagné JP, Ethier C, Gagné P, Mercier G,
Bonicalzi ME, Mes-Masson AM, Droit A, Winstall E, Isabelle M and
Poirier GG: Comparative proteome analysis of human epithelial
ovarian cancer. Proteome Sci. 5:162007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian Y, Yao Z, Roden RB and Zhang H:
Identification of glycoproteins associated with different
histological subtypes of ovarian tumors using quantitative
glycoproteomics. Proteomics. 11:4677–4687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling ZQ, Qi CJ, Lu XX, Qian LJ, Gu LH,
Zheng ZG, Zhao Q, Wang S, Fang XH, Yang ZX, et al: Heterogeneity of
chemosensitivity in esophageal cancer using ATP-tumor
chemosensitivity assay. Acta Pharmacol Sin. 33:401–406. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Konecny G, Crohns C, Pelgram M, Felber M,
Lude S, Kurbacher C, Cree IA, Hepp H and Untch M: Correlation of
drug response with the ATP tumorchemosensitivity assay in primary
FIGO stage III ovarian cancer. Gynecol Oncol. 77:258–263. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang H, Wu K, Liu Y, Wu Y and Wang X:
Integrative proteomics to understand the transmission mechanism of
Barley yellow dwarf virus-GPV by its insect vector Rhopalosiphum
padi. Sci Rep. 5:109712015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Perkins DN, Pappin DJ, Creasy DM and
Cottrell JS: Probability-based protein identification by searching
sequence databases using mass spectrometry data. Electrophoresis.
20:3551–3567. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong XW, Zou Y, Liu SP, Yi QY, Hu CM,
Wang C, Xia QY and Zhao P: Proteomic-based insight into Malpighian
tubules of silkworm Bombyx mori. PLoS One. 8:e757312013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shan N, Zhou W, Zhang S and Zhang Y:
Identification of HSPA8 as a candidate biomarker for endometrial
carcinoma by using iTRAQ-based proteomic analysis. Onco Targets
Ther. 9:2169–2179. 2016.PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
The Gene Ontology Consortium: Expansion of
the Gene Ontology knowledgebase and resources. Nucleic Acids Res.
45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Christie M and Oehler MK: Molecular
pathology of epithelial ovarian cancer. J Br Menopause Soc.
12:57–63. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giordano S, Zucchetti M, Decio A, Cesca M,
Nerini Fuso I, Maiezza M, Ferrari M, Licandro SA, Frapolli R,
Giavazzi R, et al: Heterogeneity of paclitaxel distribution in
different tumor models assessed by MALDI mass spectrometry imaging.
Sci Rep. 6:392842016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Symeonides S and Gourley C: Ovarian cancer
molecular stratification and tumor heterogeneity: A necessity and a
challenge. Front Oncol. 5:2292015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J and Li H: Heterogeneity of tumor
chemosensitivity in ovarian epithelial cancer revealed using
theadenosine triphosphate-tumor chemosensitivity assay. Oncol Lett.
9:2374–2380. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miller-Delaney SF, Lieberam I, Murphy P
and Mitchell KJ: Plxdc2 is a mitogen for neural progenitors. PLoS
One. 6:e145652011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miller SF, Summerhurst K, Rünker AE,
Kerjan G, Friedel RH, Chédotal A, Murphy P and Mitchell KJ:
Expression of Plxdc2/TEM7R in the developing nervous system of the
mouse. Gene Expr Patterns. 7:635–644. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi W, Yang C, Dai Z, Che D, Feng J, Mao Y,
Cheng R, Wang Z, He X, Zhou T, et al: High levels of pigment
epithelium-derived factor in diabetes impair wound healing through
suppression of Wnt signaling. Diabetes. 64:1407–1419. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheng G, Zhong M, Kawaguchi R, Kassai M,
Al-Ubaidi M, Deng J, Ter-Stepanian M and Sun H: Identification of
PLXDC1 and PLXDC2 as the transmembrane receptors for the
multifunctional factor PEDF. Elife. 3:e054012014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sagheer U, Gong J and Chung C: Pigment
Epithelium-Derived Factor (PEDF) is a determinant of stem cell
fate: Lessons from an Ultra-Rare disease. J Dev Biol. 3:112–128.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang S, Zhai G, Shi W, Wang Y, Zhu L, Dai
Y and Chen C: Pigment Epithelium-Derived factor inhibits
oxygen-induced retinal neovascularization in a murine model. Fetal
Pediatr Pathol. 35:173–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Belkacemi L and Zhang SX: Anti-tumor
effects of Pigment Epithelium-Derived Factor (PEDF): Implication
for cancer therapy. A mini-review. J Exp Clin Cancer Res. 35:42016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chuderland D, Ben-Ami I, Bar-Joseph H and
Shalgi R: Role of pigment epithelium-derived factor in the
reproductive system. Reproduction. 148:R53–R61. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sandilands A, Smith FJ, Lunny DP, Campbell
LE, Davidson KM, MacCallum SF, Corden LD, Christie L, Fleming S,
Lane EB and McLean WH: Generation and characterisation of Keratin 7
(K7) knockout mice. PLoS One. e644042013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Windoffer R, Beil M, Magin TM and Leube
RE: Cytoskeleton in motion: The dynamics of keratin intermediate
filaments in epithelia. J Cell Biol. 194:669–678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bayrak R, Yenidunya S and Haltes H:
Cytokeratin 7 and Cytokeratin 20 expression in colorectal
adenocarcinoma. Pathol Res Pract. 207:156–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gurzu S and Jung I: Aberrant pattern of
the cytokeratin 7/cytokeratin 20 immunophenotype in colorectal
adenocarcinomas with BRAF mutation. Pathol Res Pract. 208:163–166.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Toyoshima M, Momono Y, Makino H, Kudo T,
Oka N, Sakurada J, Suzuki H, Kodama H and Yoshinaga K: Cytokeratin
7-positive/cytokeratin 20-negative cecal adenocarcinoma
metastaticto the uterine cervix: A case report. World J Surg Oncol.
14:222016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chu P, Wu E and Weiss LM: Cytokeratin 7
and cytokeratin 20 expression in epithelial neoplasms: A survey of
435 cases. Mod Pathol. 13:962–972. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shin JH, Bae JH, Lee A, Jung CK, Yim HW,
Park JS and Lee KY: CK7, CK20, CDX2 and MUC2 Immunohisto-chemical
staining used to distinguish metastatic colorectal carcinoma
involving ovary from primary ovarian mucinous adenocarcinoma. Jpn J
Clin Oncol. 40:208–213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moll R, Divo M and Langbein L: The human
keratins: Biology and pathology. Histochem Cell Biol. 129:705–733.
2008. View Article : Google Scholar : PubMed/NCBI
|