Introduction
Gastric cancer (GC), the fourth most common type of
cancer in humans, is the second leading cause of cancer-related
deaths worldwide (1). The prognosis
in GC patients is dismal, especially for those with advanced TNM
stages (2). The main reason for the
unsatisfactory prognosis in GC patients is the occurrence of local
and systemic metastasis. However, the molecular mechanisms
underlying GC metastasis remain largely unknown. Investigating the
mechanisms and regulatory networks of GC metastasis will contribute
to the identification of novel biomarkers and therapeutic targets
of GC.
MicroRNAs (miRNAs) are a group of short noncoding
RNA sequences that can post-transcriptionally regulate the
expression of target genes (3,4). Previous
studies have demonstrated that miRNAs are involved in regulating
several cellular functions, including cell proliferation,
apoptosis, motility and differentiation (5,6). Abnormal
expression and function of miRNAs has been confirmed to serve
important roles in different types of human cancers, including GC
(7,8).
Multiple miRNAs are regarded as attractive biomarkers and potential
therapeutic targets for GC (9,10).
miR-449c was recently reported to have important
roles in human cancer, such as non-small cell lung cancer (11) and liver cancer (12). miR-449c was demonstrated to inhibit
the proliferation and invasion ability of non-small cell lung
cancer cells by targeting MYC proto-oncogene, bHLH transcription
factor (c-Myc) (11). In liver
cancer, miR-449c was demonstrated to target SYR-box 4 (SOX4) and to
inhibit the epithelial-mesenchymal transition and metastasis of
liver cancer cells (12). However,
the expression and function of miR-449c in GC remains unknown.
In the present study, miR-449c was demonstrated to
be downregulated in GC tissues and cell lines compared with normal.
Decreased miR-449c levels were associated with poor prognosis in GC
patients. Overexpression of miR-449c inhibited, while knockdown of
miR-449c promoted, the migration and invasion of GC cells.
Furthermore, the results demonstrated that 6-phosphofructo-2-kinase
(PFKFB3) was a direct downstream target of miR-449c in GC cells,
and that overexpression of PFKFB3 could abrogate the inhibitory
effect of miR-449c mimic on the migration and invasion of GC
cells.
Materials and methods
Clinical specimens and cell
culture
GC tissues and adjacent non-tumor tissues were
collected from GC patients who received surgical treatments between
2004 and 2012 in the Department of Digestive Disease, The Second
Affiliated Hospital of Xinjiang Medical University (Xinjiang,
China). The age range of the patients with GC was 24–79 years old
(mean age, 36.8 years old), with a male/female ratio of 1.5 (males,
36; females, 24). None of the patients received chemotherapy prior
to surgical treatment. The clinical samples were collected with
informed consent from all patients. The specimens were stored in
liquid nitrogen prior to being subjected to further experiments.
The protocols for collecting and using the clinical specimens were
approved by the Institutional Research Ethics Committee of the
Second Affiliated Hospital of Xinjiang Medical University (Urumqi,
China).
The normal gastric epithelial GES-1 cell line and
four GC cell lines (AGS, SGC-7901, MKN-45 and MGC-803) were
purchased from American Type Culture Collection (Manassas, VA, USA)
and the Cell Bank of Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). The cells were cultured in RPMI1640
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 mg/ml penicillin and 100 mg/ml streptomycin.
All cell cultures were maintained in a humidified atmosphere with
5% CO2 at 37°C.
Cell transfection
miR-449c mimic (cat no. miR10013771-1-5), miR-449c
inhibitor (cat no. miR20010251-1-5) and corresponding control
vectors (cat nos. miR01201-1-5 and miR02201-1-5) were purchased
from Guangzhou RiboBio Co., Ltd (Guangzhou, China). For miRNA
transfection, the final concentration transfected was 50 nM.
pcDNA3.1-PFKFB3 and empty control vector were bought from GeneChem
Co. Ltd. (Shanghai, China). For plasmid transfection, the final
concentration transfected was 1.3 µg/ml. Transfection into GC cells
was performed with lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Following 48 h after transfection, the cells were used for further
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from clinical tissues was extracted using
an RNA isolation kit (cat. no. AP-MN-MS-RNA; Corning Inc., Corning,
NY, USA). Total RNA and miRNAs from GC cells were extracted using
TRIzol Reagent (Thermo Fisher Scientific, Inc.) and microRNA
purification kit (Norgen Biotek Corp., Thorold, ON, Canada)
respectively, according to the manufacturer's protocol. cDNA
reverse transcription was performed with High-Capacity cDNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). qPCR for miR-449c was performed using Taqman microRNA assay
primers (Applied Biosystems; Thermo Fisher Scientific, Inc.), while
qPCR for PFKFB6 mRNA was performed using SYBR Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 and GAPDH
were used as internal controls for miR-449c and PFKFB6,
respectively. The thermocycling conditions were as follows: 95°C
for 30 sec, followed by 40 cycles at 95°C for 5 sec, then 60°C for
30 sec. Relative fold changes in mRNA expression were calculated
using the 2−ΔΔCq method (13). Primers for miR-449c, PFKFB3, U6 and
GAPDH were purchased from GenePharma Co., Ltd. (Shanghai, China).
The primer sequences were as follows: MiR-449c forward,
5′-CGCGGATCCTAATGCAATCGTTTGCATCTG-3′ and reverse,
5′-CCGGAATTCTGGGTTTGGTCTTTCAAGGAG-3′; U6 forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGT-3′;
PFKFB3 forward, 5′-CCTCACTCGCAGCCACTTCT-3′ and reverse,
5′-CAGTTCCTACTCAATTCCAA-3′; GAPDH forward,
5′-TGGGTGTGAACCACGAGAA-3′ and reverse,
5′-GGCATGGACTGTGGTCATGA-3′.
Protein extraction and western blot
analysis
Total cellular protein was extracted using
radioimmunoprecipitation assay lysis buffer containing 50 mmol/l
Tris-Cl (pH 7.5), 0.2 mmol/l EDTA, 150 mmol/l NaCl, 1 mmol/l PMSF
and 1% Nonidet-P40 supplemented with protease inhibitor. The
bicinchoninic acid kit (Pierce; Thermo Fisher Scientific, Inc.) was
used for quantification of protein concentration. A total of 30 µg
cellular proteins were loaded on 4–20% SDS-PAGE for protein
separation. The separated protein samples were transferred to
polyvinylidene fluoride membranes. Then, the membranes were blocked
with 5% non-fat milk for 1 h at room temperature, and then
incubated with the following antibodies overnight at 4°C: PFKFB3
(1:1,000; cat no. #9645; Cell Signaling Technologies, Inc.,
Danvers, MA, USA) and GAPDH (1:2,000; cat no. #5174; Cell Signaling
Technologies, Inc.). The membranes were then incubated with
secondary mouse anti-rabbit (cat. no., 3678) or goat anti-mouse
(cat. no., 58802) antibodies (1:2,000) conjugated with horseradish
peroxidase (Cell Signaling Technologies, Inc.) at room temperature
for 1 h. The signal intensity of protein bands was visualized with
enhanced chemiluminescence reagents (GE Healthcare, Chicago, IL,
USA). Image J software (version 1.41; National Institutes of
Health, Bethesda, MD, USA) was used to quantify signal
intensity.
Migration and invasion assays
Boyden chambers/transwell assays were used to
measure the migratory ability of GC cells, following the
manufacturer's protocol. Matrigel-coated transwell inserts with 8
µm pores (Nalge Nunc International, Penfield, NY, USA) were used
for measuring the invasive ability of GC cells. The same 8 µm pore
inserts were used for migration as well, with the addition of
Matrigel coating. Transwell inserts were coated with 100 µl
Matrigel (1:8 dilution in RPMI1640 medium). GC cells were suspended
in serum-free RPMI-1640 medium (10×104 cells per 500 µl
serum-free media) and were seeded in the upper chamber of the
transwell inserts. A total of 800 µl RPMI1640 containing 10% serum
was added in the lower chamber. Following 24–48 h of incubation,
crystal violet staining (5 min at room temperature) was used for
the identification of the invaded GC cells. A Leica light
microscope was used to count the migrated or invaded cells at the
magnification of ×200. A total of nine optical fields were counted
per sample, with at least three samples per group.
Luciferase reporter assay
Wild type (wt) or mutant (mt) PFKFB3 3′-UTR was
cloned into the pmiR-RB-REPORT™ luciferase vector (Promega
Corporation, Madison, WI, USA). The luciferase reporting vectors
containing the interacting sequences of PFKFB3 were co-transfected
with miR-449c mimic or miR-449c inhibitor into GC cells in 6-well
plates using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, the luciferase
activity was measured using the dual-luciferase reporter assay
system (Promega Corporation). Renilla luciferase activity was
normalized to firefly luciferase activity.
Bioinformatics analysis
Targetscan database (http://www.targetscan.org/vert_71/) was searched to
identify the potential downstream target of miR-449c on November
2016. The search term used was ‘miR-449c’. The complimentary
sequences mediating the binding between miR-449c and PFKFB3 were
identified using the Targetscan database.
Statistical analysis
All statistical analyses in the present study were
performed using the GraphPad Prism 5 software (GraphPad Software,
Inc., La Jolla, CA, USA). Quantitative data were presented as mean
± standard error of the mean, from replicate experiments (n>3).
Kaplan-Meier analysis was performed to evaluate the prognostic
significance of miR-449c in GC. Statistical significance was
analyzed using the Chi-square and Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
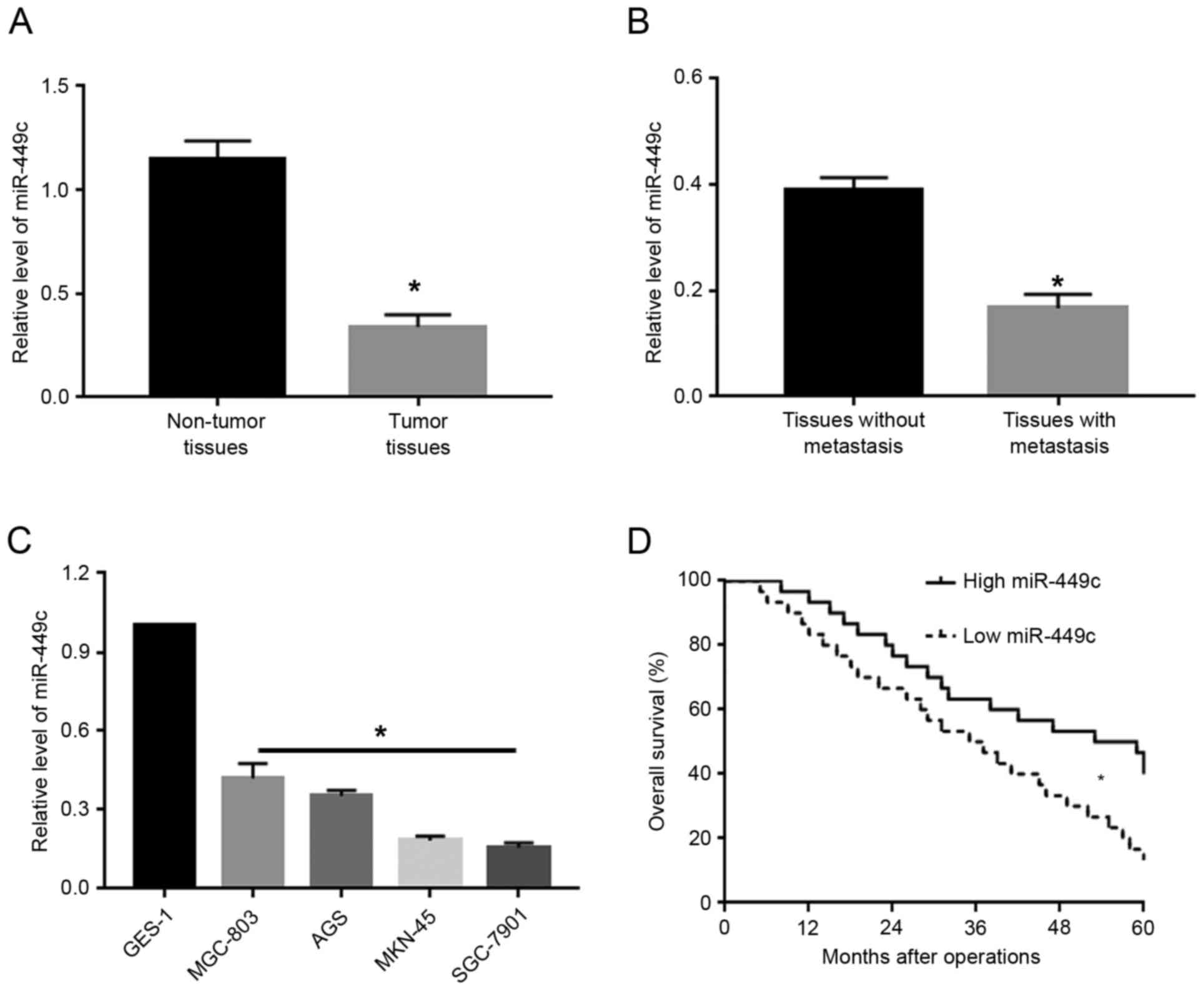
miR-449c expression is downregulated
in GC
To investigate the expression levels of miR-449c in
GC, qPCR was used to measure miR-449c levels in GC tissues and
adjacent non-tumor tissues. The results from the qPCR analysis
demonstrated that, compared with adjacent non-tumor tissues, GC
tissues exhibited significantly decreased levels of miR-449c
(P<0.05; Fig. 1A). Next, the GC
tissues were subdivided into two groups: cases with metastasis and
cases without metastasis. Compared with the non-metastatic cancer
tissues, tissues from patients with metastasis exhibited
significantly decreased levels of miR-449c (P<0.05; Fig. 1B). Furthermore, the expression levels
of miR-449c were measured in several GC cell lines, including
MGC-803, AGS, MKN-45 and SGC-7901. Compared with the normal gastric
epithelial cell line GES-1, the miR-449c expression levels were
significantly reduced (P<0.05; Fig.
1C). Of note, Kaplan-Meier analysis revealed that decreased
miR-449c expression was significantly associated with decreased
overall survival rate in GC patients (P<0.05; Fig. 1D).
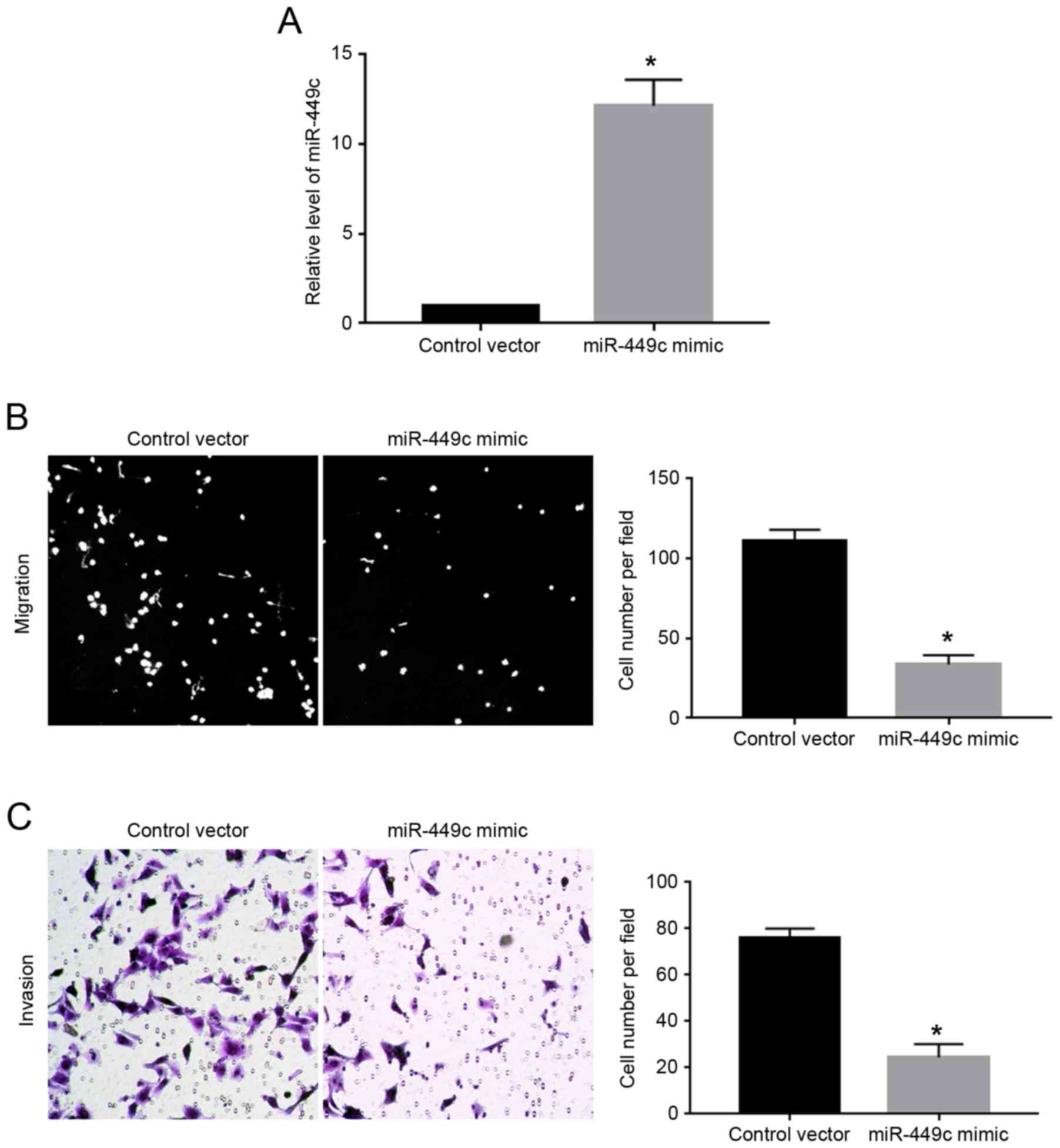
Overexpression of miR-449c inhibits
migration and invasion in GC cells
As illustrated in Fig.
1C, the expression levels of miR-449c were the lowest in
SGC-7901 cells. Therefore, SGC-7901 cells were transfected with
miR-449c mimic. Overexpression of miR-449c mimic significantly
increased the expression level of miR-449c in SGC-7901 cells
(P<0.05; Fig. 2A). Then, transwell
assays were performed to evaluate the effect of miR-449c
overexpression on the migration and invasion ability of the
SGC-7901 cells. The results demonstrated that overexpression of
miR-449c significantly decreased both the migration (P<0.05;
Fig. 2B) and the invasion of SGC-7901
cells (P<0.05, Fig. 2C), compared
with cells transfected with control vector.
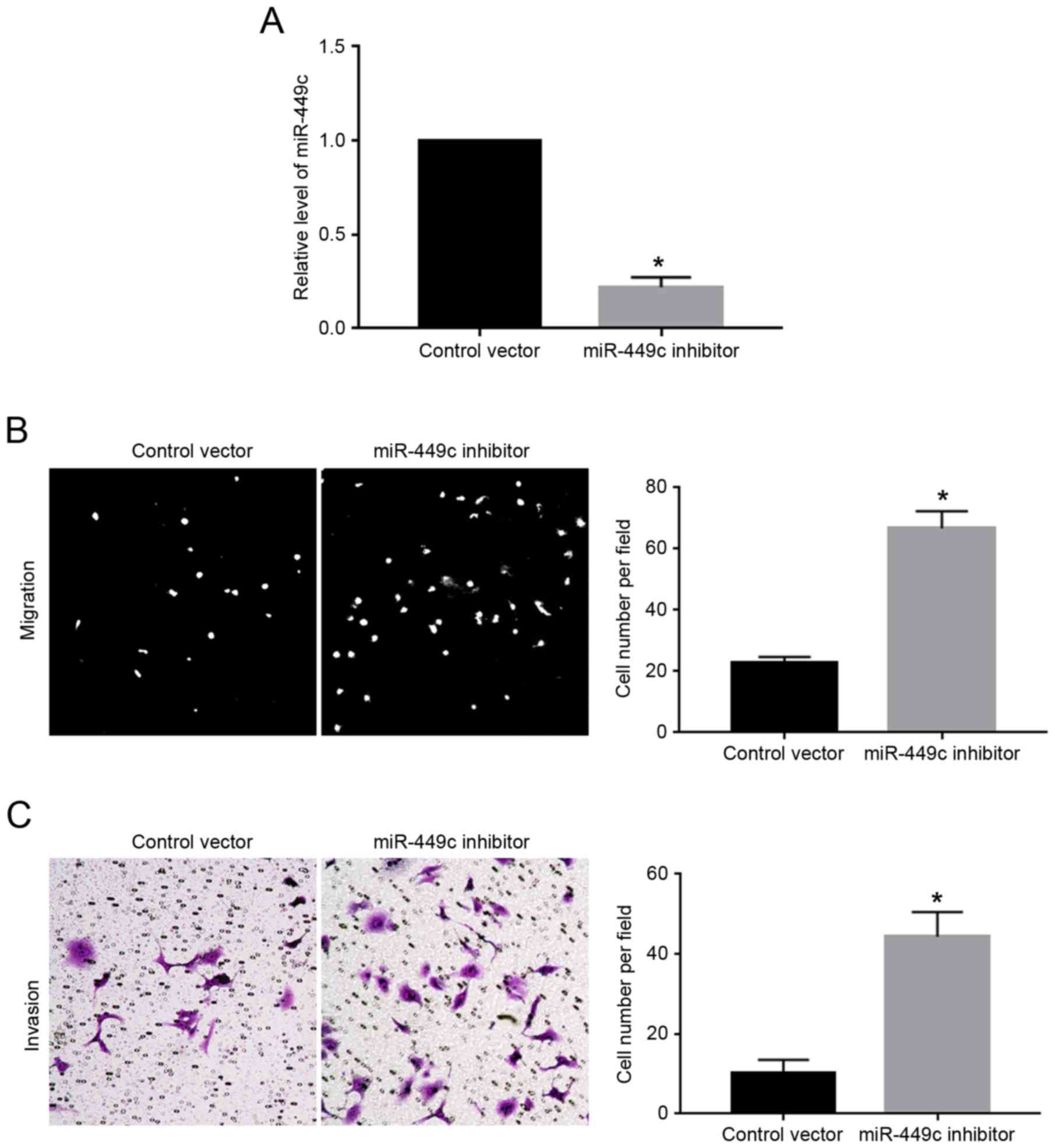
Knockdown of miR-449c inhibits
migration and invasion in GC cells
As illustrated in Fig.
1C, the expression levels of miR-449c were the highest in
MGC-803 cells. Therefore, MGC-803 cells were transfected with
miR-449c inhibitor, which resulted in a significant decrease of
miR-449c expression levels in MGC-803 cells (P<0.05; Fig. 3A). Subsequently, miR-449c knockdown
significantly enhanced the migration (P<0.05; Fig. 3B) and invasion (P<0.05; Fig. 3C) of MGC-803 cells, compared with
cells transfected with control vector.
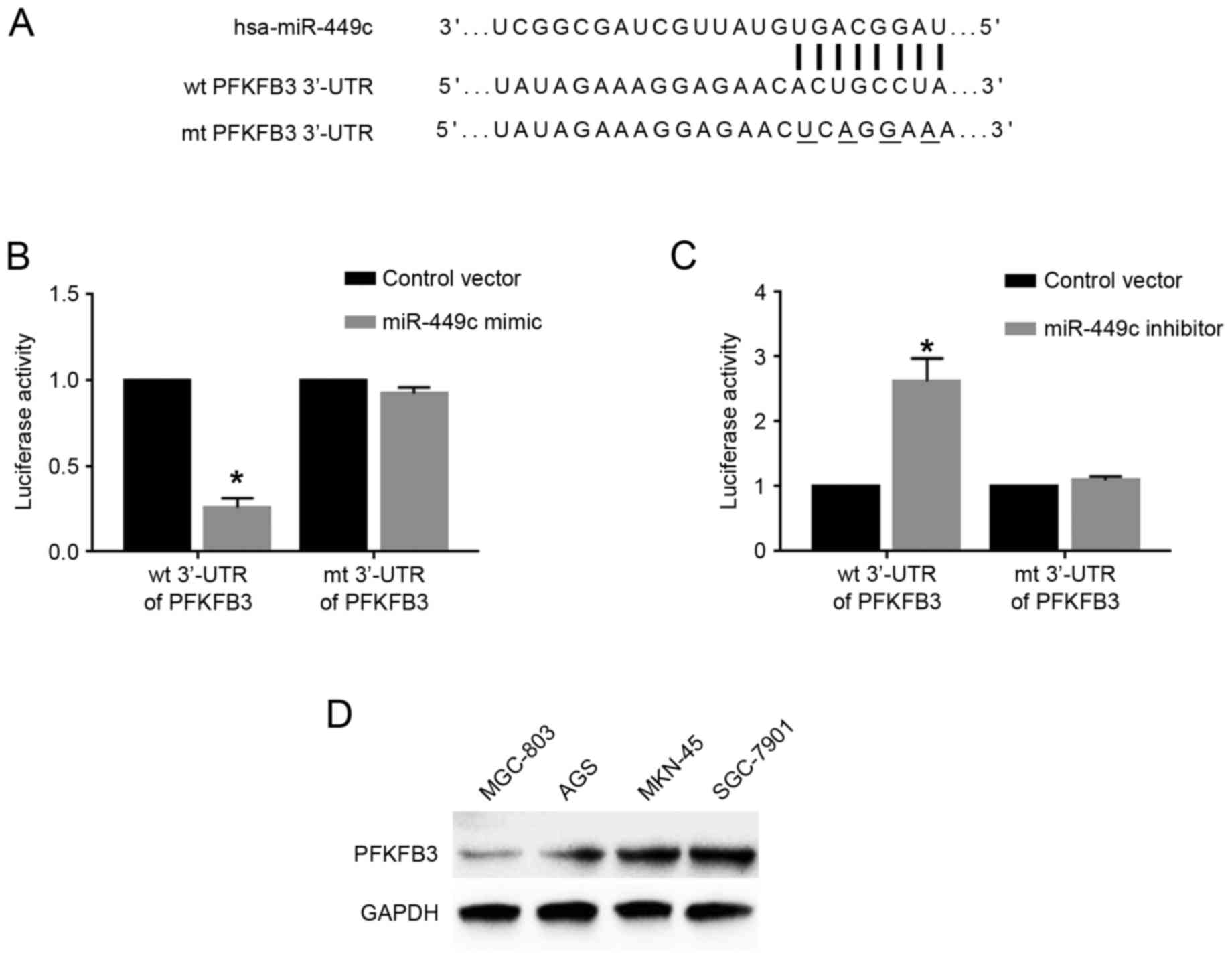
PFKFB3 is a direct downstream target
of miR-449c in GC cells
Following demonstrating the functional role of
miR-449c in GC cells, the molecular mechanisms underlying these
functions were further explored. Based on predictions using the
online bioinformatics database Targetscan (http://www.targetscan.org/vert_71/), the 3′-UTR of the
gene PFKFB3 was revealed to contain the binding sequences for
miR-449c (Fig. 4A), indicating that
PFKFB3 may be a downstream target of miR-449c. To determine whether
miR-449c could interact with the PFKFB3 3′-UTR, luciferase assays
were performed. Overexpression of miR-449c in SGC-7901 cells
significantly reduced the luciferase activity of wt PFKFB3 3′-UTR,
but had no effect on that of mt PFKFB3 3′-UTR (P<0.05; Fig. 4B). By contrast, miR-449c knockdown in
MGC-803 cells significantly increased the luciferase activity of wt
PFKFB3 3′-UTR, but had no effect on that of mt PFKFB3 3′-UTR
(P<0.05; Fig. 4C). Western blot
analysis for the protein expression levels of PFKFB3 in GC cell
lines demonstrated that MGC-803 cells, which had the highest
miR-449c levels, exhibited the lowest PFKFB3 levels, while SGC-7901
cells, which had the lowest miR-449c levels, exhibited the highest
PFKFB3 levels (Fig. 4D). Furthermore,
RT-qPCR and western blot analyses were performed to investigate
whether miR-449c could regulate the expression of PFKFB3 in GC
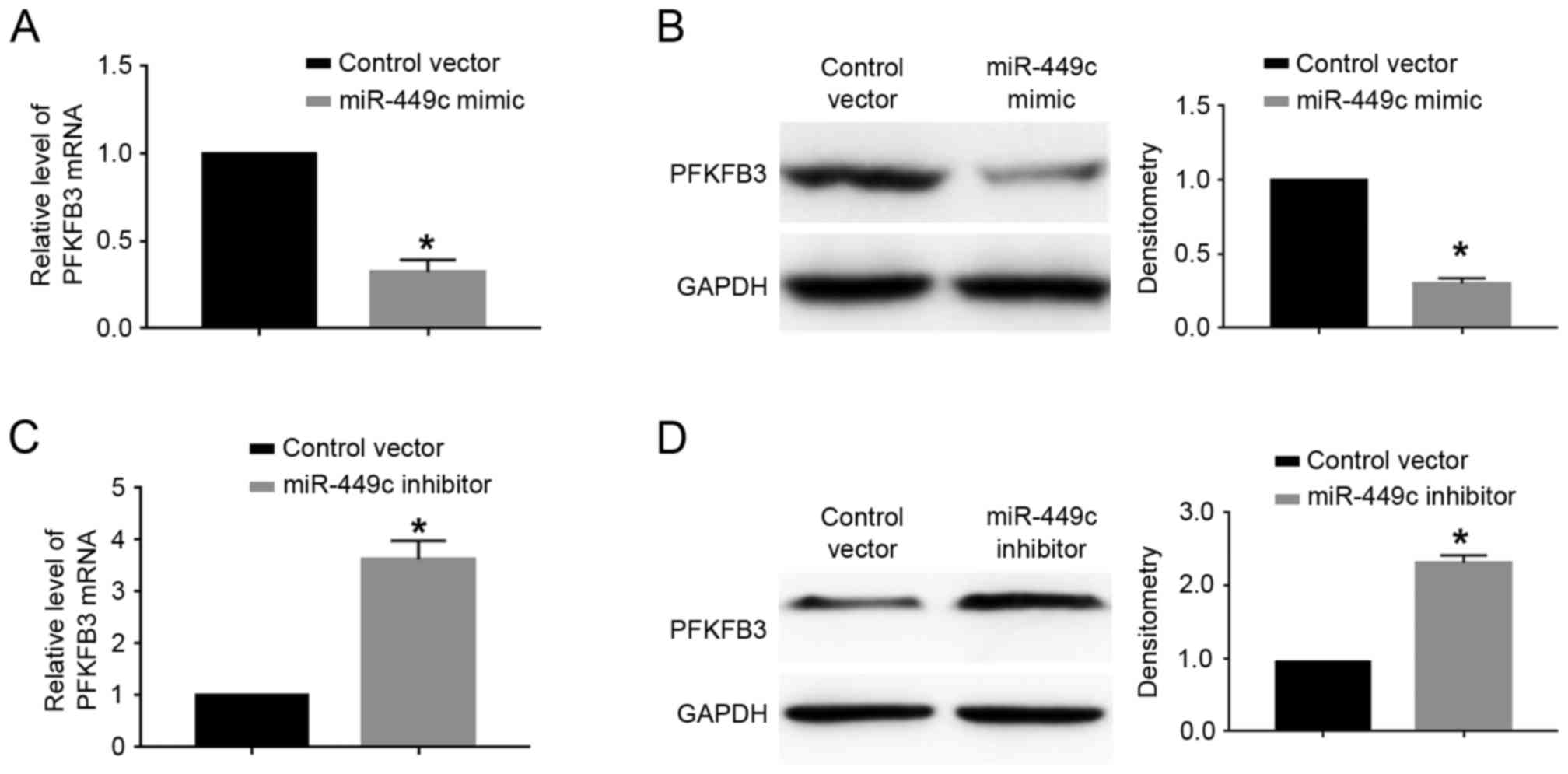
cells. The results demonstrated that miR-449c overexpression
significantly decreased the mRNA (P<0.05; Fig. 5A) and protein (P<0.05; Fig. 5B) expression levels of PFKFB3 in
SGC-7901 cells. By contrast, miR-449c knockdown significantly
increased the mRNA (P<0.05; Fig.
5C) and protein (P<0.05; Fig.
5D) expression levels of PFKFB3 in MGC-803 cells.
Overexpression of PFKFB3 abrogates the
effect of miR-449c in GC cells
To further confirm whether PFKFB3 is the mediator of
the biological function of miR-449c in GC cells, SGC-7901 cells
overexpressing miR-449c were transfected with PFKFB3 expression
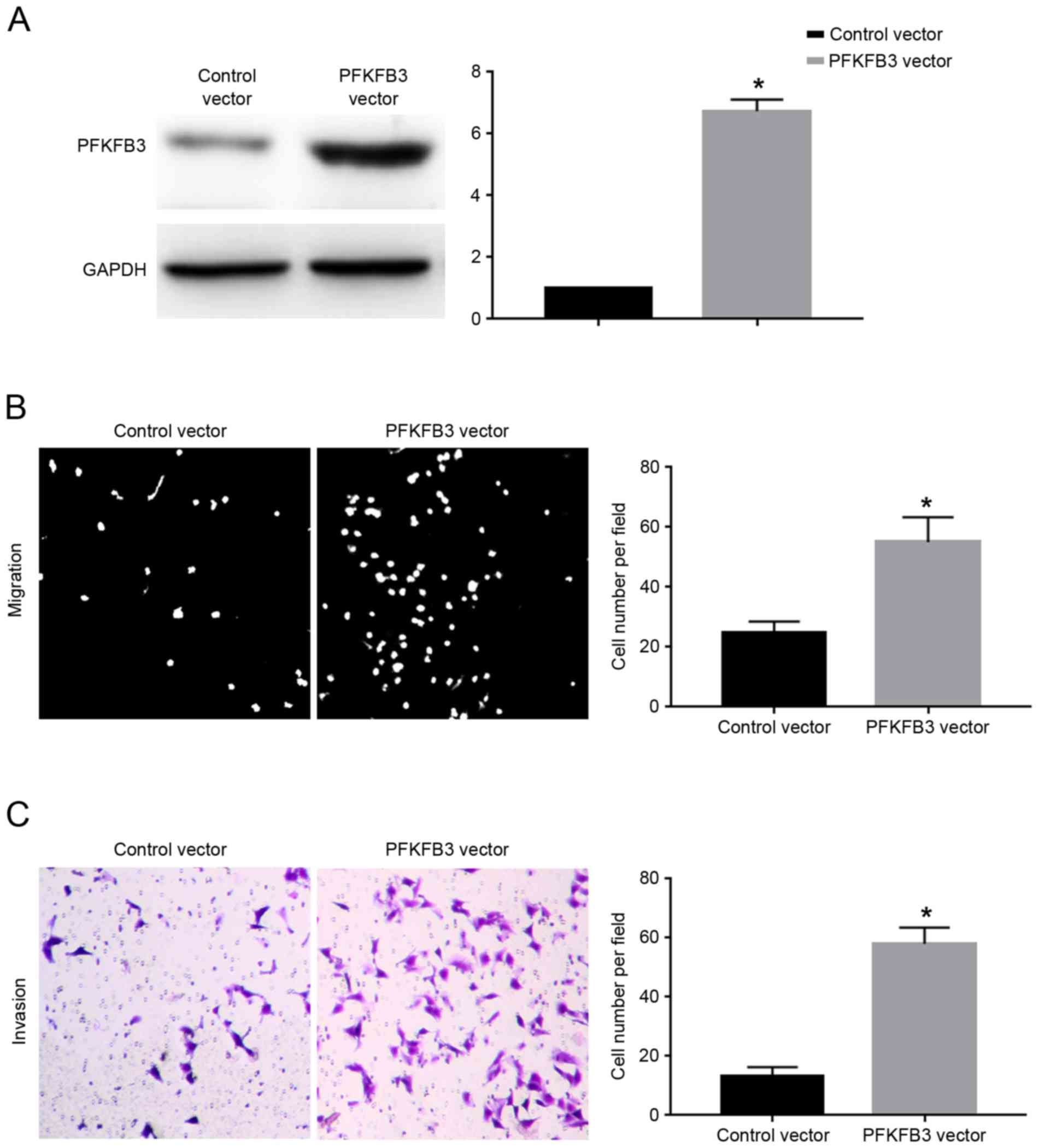
vector. Transfection of the PFKFB3 vector significantly increased
the protein expression levels of PFKFB3 in SGC-7901 cells
overexpressing miR-449c (P<0.05; Fig.
6A). Overexpression of PFKFB3 significantly inhibited the
migration (P<0.05; Fig. 6B) and
invasion (P<0.05; Fig. 6C) of
SGC-7901 cells overexpressing miR-449c. The present results suggest
that PFKFB3 overexpression abrogated the inhibitory effects of
miR-449c on the migration and invasion of SGC-7901 cells.
Discussion
miRNAs have been demonstrated as important
regulators of the development and progression of human cancer types
(4). miRNAs have been reported to
regulate the growth, metastasis, stem cell biology and drug
resistance of cancer cells (9,14,15). Among numerous miRNAs, miR-449c was
identified as a novel cancer-related microRNA, and it was reported
to inhibit the proliferation and invasion ability of non-small cell
lung cancer cells (11). In liver
cancer, miR-449c was reported to inhibit the epithelial-mesenchymal
transition and metastasis of liver cancer cells (12). However, the expression and function of
miR-449c in GC remained unknown. The present study demonstrated
that miR-449c is downregulated in GC tissues and cells compared
with normal tissues and cells. Functionally, both gain and loss-of
function assays demonstrated that miR-449c inhibited the migration
and invasion of GC cells. The present data indicate that miR-449c
may exert tumor suppressive roles in GC by inhibiting the migration
and invasion of GC cells.
6-phosphofructo-2-kinase (PFKFB3) has been reported
to be a versatile protein in human cancers (16,17). It
has been demonstrated to regulate the glycolysis, oxidative stress,
proliferation, apoptosis and metastasis of cancer cells (16). However, the underlying mechanisms for
the elevated expression of PFKFB3 in GC remain unknown. A previous
study has reported that PFKFB3 is under the regulation of miR-26b
in osteosarcoma cells. In the present study, PFKFB3 was
demonstrated to be the downstream target of miR-449c in GC cells.
PFKFB3 overexpression abrogated the inhibitory effects of miR-449c
on the migration and invasion of GC cells. These data suggest that
PFKFB3 may have mediated the inhibitory effect of miR-449c on the
migration and invasion of GC cells. Notably, previous studies of
non-small cell lung cancer and liver cancer cells have reported
that c-Myc and SOX4 were the downstream targets of miR-449c,
respectively, suggesting that miR-449c may have different
downstream targets in different types of human cancers.
In summary, the present study demonstrated that
miR-449c was downregulated in GC tissues and cells. Decreased
expression of miR-449c was associated with poor prognosis in GC
patients. Overexpression of miR-449c inhibited migration and
invasion in SGC-7901 cells, while knockdown of miR-449c promoted
these biological behaviors in MGC-803 cells. Finally, PFKFB3 was
revealed as a novel downstream target and functional mediator of
miR-449c in GC cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XC performed the experiments in the present study,
AW performed the data analysis, and XC and XY designed this study
and wrote the manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Institutional
Research Ethics Committee of the Second Affiliated Hospital of
Xinjiang Medical University. Informed consent to participate in the
study was obtained from participants.
Consent for publication
All participants agreed to the publication of this
study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rugge M, Fassan M and Graham DY:
Epidemiology of gastric cancer. Gastric Cancer Springer. 23–34.
2015. View Article : Google Scholar
|
|
2
|
Chang WJ, Du Y, Zhao X, Ma LY and Cao GW:
Inflammation-related factors predicting prognosis of gastric
cancer. World J Gastroenterol. 20:4586–4596. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong H, Lei J, Ding L, Wen Y, Ju H and
Zhang X: MicroRNA: Function, detection, and bioanalysis. Chem Rev.
113:6207–6233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wen D, Danquah M, Chaudhary AK and Mahato
RI: Small molecules targeting microRNA for cancer therapy: Promises
and obstacles. J Control Release. 219:237–247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miao LJ, Huang SF, Sun ZT, Gao ZY, Zhang
RX, Liu Y and Wang J: MiR-449c targets c-Myc and inhibits NSCLC
cell progression. FEBS Lett. 587:1359–1365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sandbothe M, Buurman R, Reich N, Greiwe L,
Vajen B, Gürlevik E, Schäffer V, Eilers M, Kühnel F, Vaquero A, et
al: The microRNA-449 family inhibits TGF-β-mediated liver cancer
cell migration by targeting SOX4. J Hepatol. 66:1012–1021. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. MicroRNA Cancer Regulation Springer.
1–20. 2012.
|
|
16
|
Clem BF, O'Neal J, Tapolsky G, Clem AL,
Imbert-Fernandez Y, Kerr DA II, Klarer AC, Redman R, Miller DM,
Trent JO, et al: Targeting 6-phosphofructo-2-kinase (PFKFB3) as a
therapeutic strategy against cancer. Mol Cancer Ther. 12:1461–1470.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Imbert-Fernandez Y, Clem A, Clem B,
Tapolsky G, Telang S and Chesney J: Suppression of
6-Phosphofructo-2-Kinase (PFKFB3) for the treatment of breast
cancer. AACR. 76:2016.
|