Introduction
Breast cancer (BRCA) is the most prevalent cancer
type in women, with 74.1 new cases and 8.0 mortalities per 100,000
reported cases in developing countries in 2012 (1–3). Based on
statistical estimation in China, 268,600 new breast cancer cases
and 69,500 mortalities occurred due to BRCA in 2015 (4). Although the molecular subtyping of
breast cancer is well developed and treatments are relatively
abundant, many patients succumb to disease due to distant
metastasis.
The estrogen receptor positive (ER+)
subtype is currently the most curable breast cancer subtype.
Tamoxifen, which binds to the ER and disrupts the ER signaling
pathway, is the most popular treatment for the
ER+-breast cancer. However, a large proportion of
tamoxifen-treated ER+-patients still succumb to breast
cancer (5). The current clinical
staging system is insufficient for predicting the survival of the
patients with ER+-breast cancer and treated with
tamoxifen (6). Therefore, gene
expression biomarkers of breast cancer are urgently needed for
prognosis and treatment optimization.
According to previous studies, a single biomarker
often fails to predict the outcome of the ER+-breast
cancer across datasets, while models that are based on multiple
genes significantly are more accurate (7). Therefore, in the present study, using
Cox multivariate regression and data on gene expression levels, a
risk score model for the prognosis of the ER+-breast
cancer outcome in patients taking tamoxifen was developed using the
GSE17005 dataset. Patients in the high-risk group exhibited a
significantly higher 5-year survival rate compared with those in
the low-risk group. Furthermore, this result was also replicated in
four other independent cohorts (GSE26971, GSE22219, GSE42568 and
GSE56884). According to the 5-year survival receiving operating
characteristic (ROC), the area under the curve (AUC) of the risk
score was higher compared with the other clinicopathological
indicators in predicting the 5-year survival rate of the patients
with ER+-breast cancer. The association between the
clinicopathological indicators and the risk score was evaluated,
and a nomogram describing the 5-year survival rate was also
plotted. In summary, the risk score is a robust prognostic
indicator for the survival of the patients with
ER+-breast cancer and treated with tamoxifen.
Materials and methods
Data pre-processing and sample
selection
Samples in all datasets were filtered, and those
without clear records of the ER+ diagnosis or tamoxifen
treatment were excluded from the dataset. The raw data containing
the GSE17005, GSE26971, GSE22219, GSE42568 and GSE56884 datasets
were downloaded using the raw data format from Gene Expression
Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) according to the
corresponding accession number. The samples that were either not
from patients with ER+-breast cancer or treated with
tamoxifen for five years were excluded from the present study.
Subsequent to pre-processing, including normalization using Robust
Multi-array Average (RMA), the data were used for further analysis.
The probes and the gene names were matched, and the average values
were calculated for genes that match the multiple probes. The
Cancer Genome Atlas (TCGA) datasets were not used because the TCGA
datasets were generated using RNA-seq platform while the other
datasets were from microarray. The formula was dependent on the
expression values (fragments per kb of transcript per million for
RNA-seq and intensity for microarray), therefore the TCGA dataset
was not used as training or test dataset in the present study.
Feature selection and model
development
Cox univariate regression was implemented on
GSE17005 and GSE26971 datasets. Genes that significantly associated
with the survival rates in both datasets were selected as
candidates. After 100 repeats and 100 iterations, the frequencies
of genes were counted. Genes with the highest frequency were used
for model development, and these 10 genes were identified.
Multivariate Cox regression was then used to construct the linear
risk score model in GSE17005. During the risk score calculation in
the validation datasets, the coefficient for each gene was set to a
constant value.
Statistical analysis
Statistical analyses were implemented using the R
software (https://www.r-project.org/; version
3.0.1) and its packages. RMA normalization of the raw datasets was
performed using the R function ‘rma’ in the R package ‘affy’. Cox
univariate regression and multivariate regression was implemented
using the package ‘survival’, and the ROC curve was plotted using
functions in package ‘pROC’ (8).
Results
Candidate gene selection and model
development
The workflow of the present study is illustrated in
the Fig. 1A. Using the univariate Cox
regression analysis, the association between the gene expression
and the overall survival was calculated in 2 independent datasets:
GSE17005 and GSE26971. A total of 48 genes that significantly
associated with the survival rates in both datasets were identified
as the candidate genes. Random forest variable hunting was
conducted to retrieve the best combination of biomarkers, and ten
genes were selected (Fig. 1B). The
multivariate Cox regression analysis was implemented in GSE17005
instead of GSE26971 as the sample size of GSE17005 is bigger. The
risk score was calculated using the following formula: Risk
score=(−0.382802602) × NEK2 + 1.057608407 × STC2 + 0.216378311 ×
CCNA2 + 0.196075897 × AK5 + 0.553253489 × CTDSP1 + (−0.544558722) ×
FOXD1 + (−0.606202483) × KCNK1 + (−0.245868702) × TNNC2 +
(−0.093556696) × CENPE + (−0.212704033) × STAT6. The coefficients
are shown in Fig. 1C. The negative
coefficient values indicate the tumor suppressor genes, and the
positive values indicate the oncogenes for cancer development.
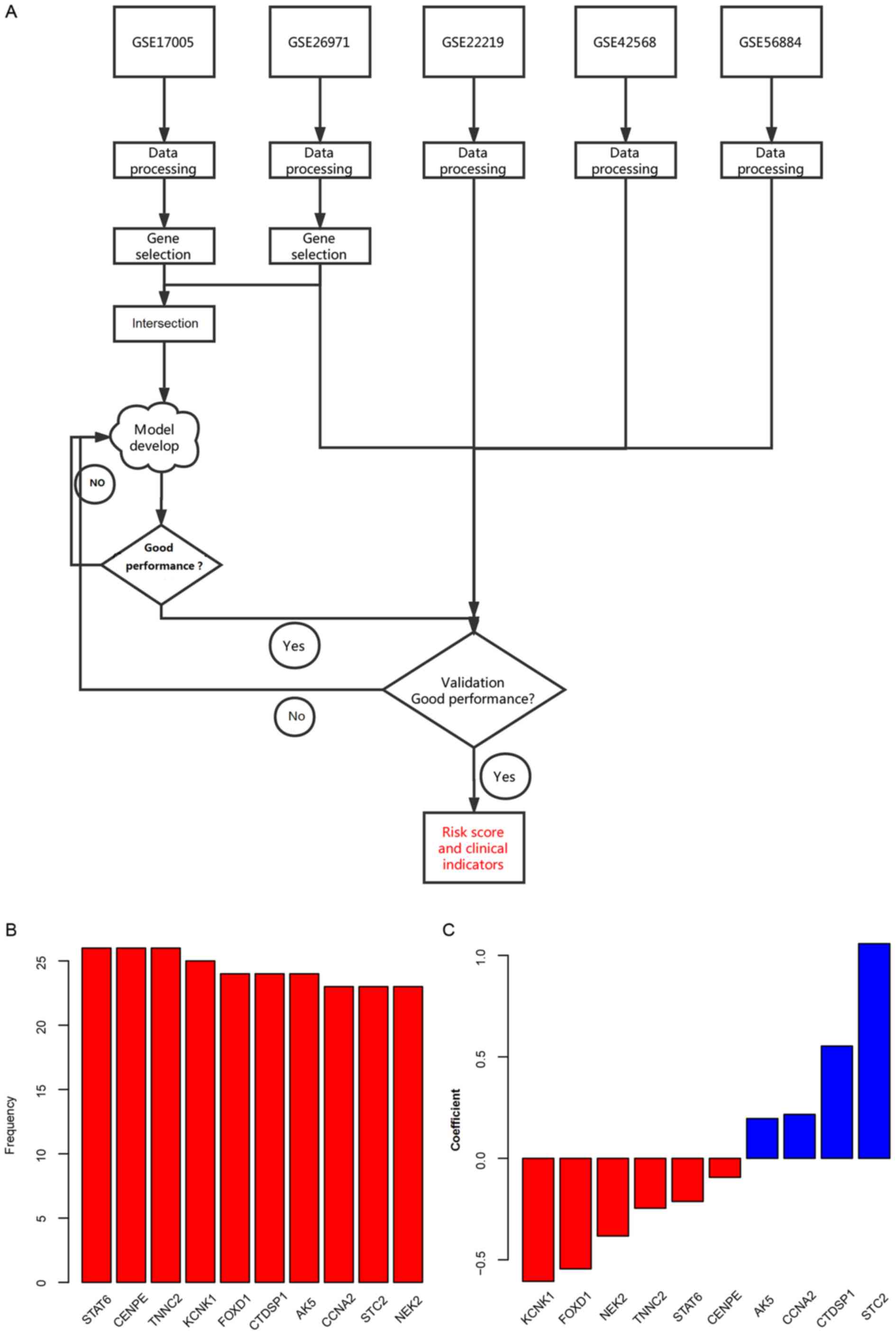 | Figure 1.Workflow of the present study and
candidate genes used for model development. (A) Workflow of the
present study. (B) Frequency of genes in random forest variable
hunting. (C) Coefficient for each gene in the risk score
calculation formula. AK5, adenylate kinase 5; CCNA2, cyclin A2;
CENPE, centromere protein E; CTDSP1, carboxy-terminal domain RNA
polymerase II polypeptide A small phosphatase 1; FOXD1, forkhead
box D1; KCNK1, potassium two pore domain channel subfamily K member
1 and troponin C type 1 (Slow); NEK2, NIMA related kinase 2; STAT6,
signal transducer and activator of transcription 6; STC2,
stanniocalcin 2; TNNC2, troponin C, skeletal muscle. |
Prognostic value of the risk score in
GSE17005
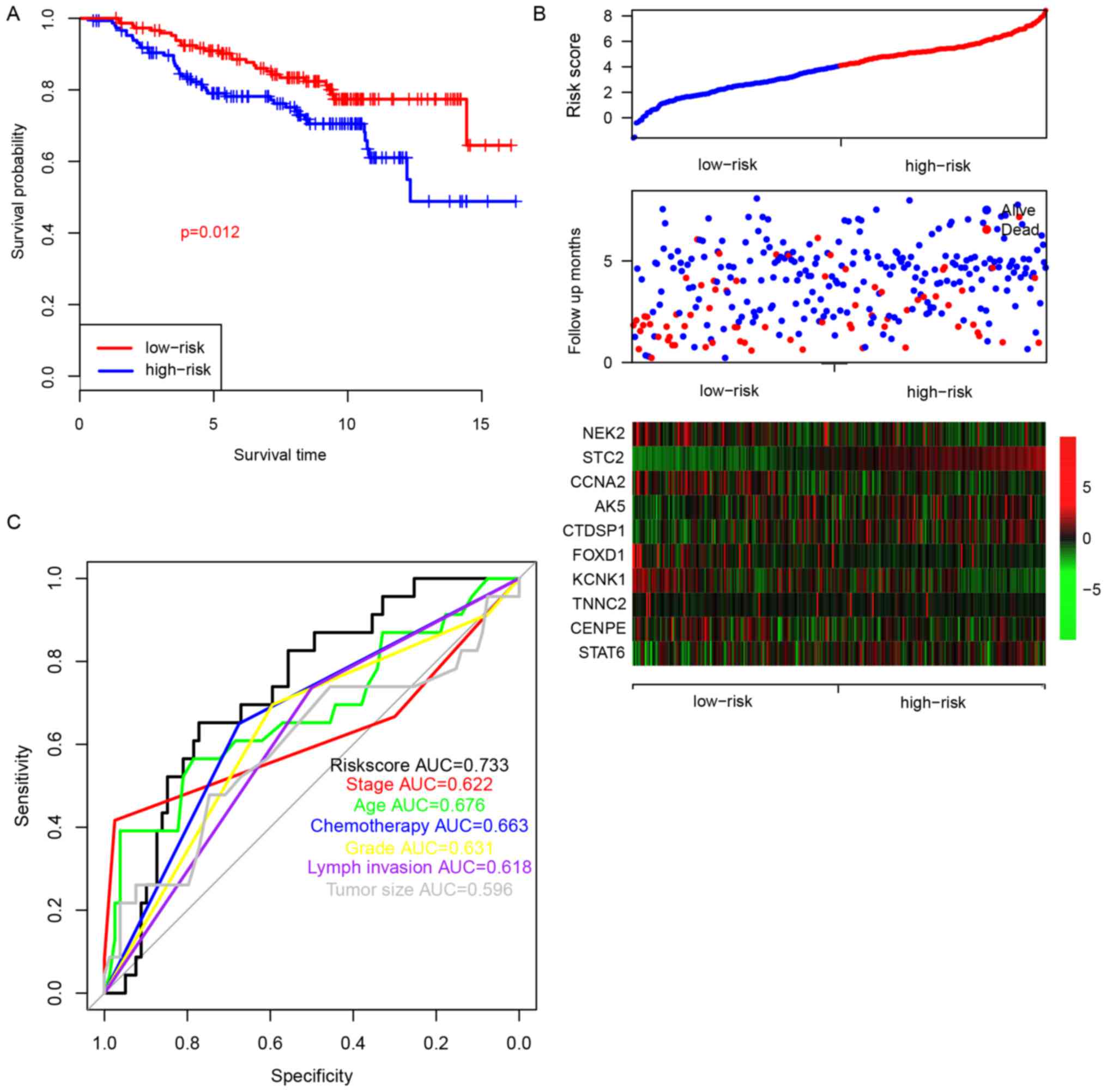
To evaluate the prognostic significance of the risk
score for patients with ER+-breast cancer and treated
with tamoxifen, the survival difference in patients from the
high-risk and low-risk groups was analyzed. The median value of the
risk score was used as a cutoff. The overall survival (OS) of
patients in the high-risk group was significantly lower compared
with the low-risk group (Fig. 2A,
P=0.012). As shown in Fig. 2B,
patients in the high-risk group were characterized with early
mortality, low expression levels of NIMA related kinase 2, cyclin
A2 (CCNA2), forkhead box D1 (FOXD1), potassium two pore domain
channel subfamily K member 1 and troponin C type 1 (slow), and high
expression levels of stanniocalcin 2 (STC2), adenylate kinase 5
(AK5), carboxy-terminal domain RNA polymerase II polypeptide A
small phosphatase 1 (CTDSP1) and signal transducer and activator of
transcription 6 (STAT6). The ROC curve of 5-year survival was also
plotted according to age, stage, grade, chemotherapy, lymph
invasion, tumor size and risk score (Fig.
2C), and the area under the receiving operating characteristic
curve was 0.676, 0.622, 0.631, 0.663, 0.618, 0.596 and 0.733,
respectively. Collectively, these results indicate that the risk
score is a clinically important predictor of the 5-year survival of
patients with ER+-breast cancer and treated with
tamoxifen.
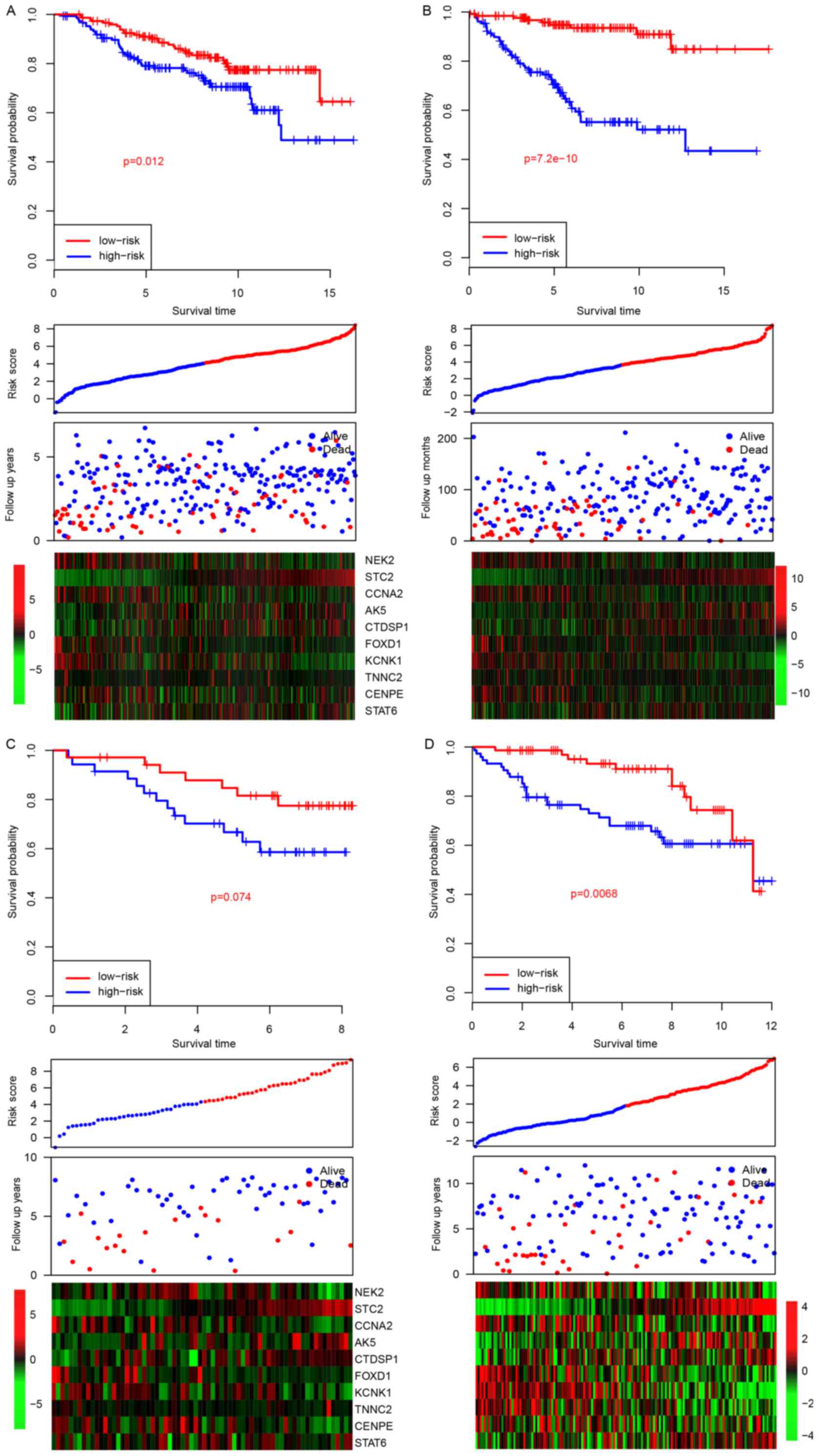
Validation of risk score
performance
The good performance of the model in the training
dataset may be due to over-fitness of the model to this dataset,
particularly in multivariate analysis (9). Therefore, in order to evaluate this
possibility, the risk scores for samples in four other datasets
(GSE26971, GSE22219, GSE42568 and GSE56884) were calculated after
the coefficients were fixed (Fig. 3).
Similar to the survival profile in the training dataset, the
survival of patients in the high-risk group was significantly
poorer compared with the patients in the low-risk group in three of
the four datasets (P=0.012, 7.2×10−10 and 0.0068 for
GSE26971, GSE22219 and GSE56884, respectively; Fig. 3A, B and D, top panel). The survival
profile of the patients in the GSE42568 dataset was not
significantly different between the two groups, which may be due to
a smaller sample size. Similar expression trends were also observed
in all four datasets (Fig. 3A, middle
and bottom panel).
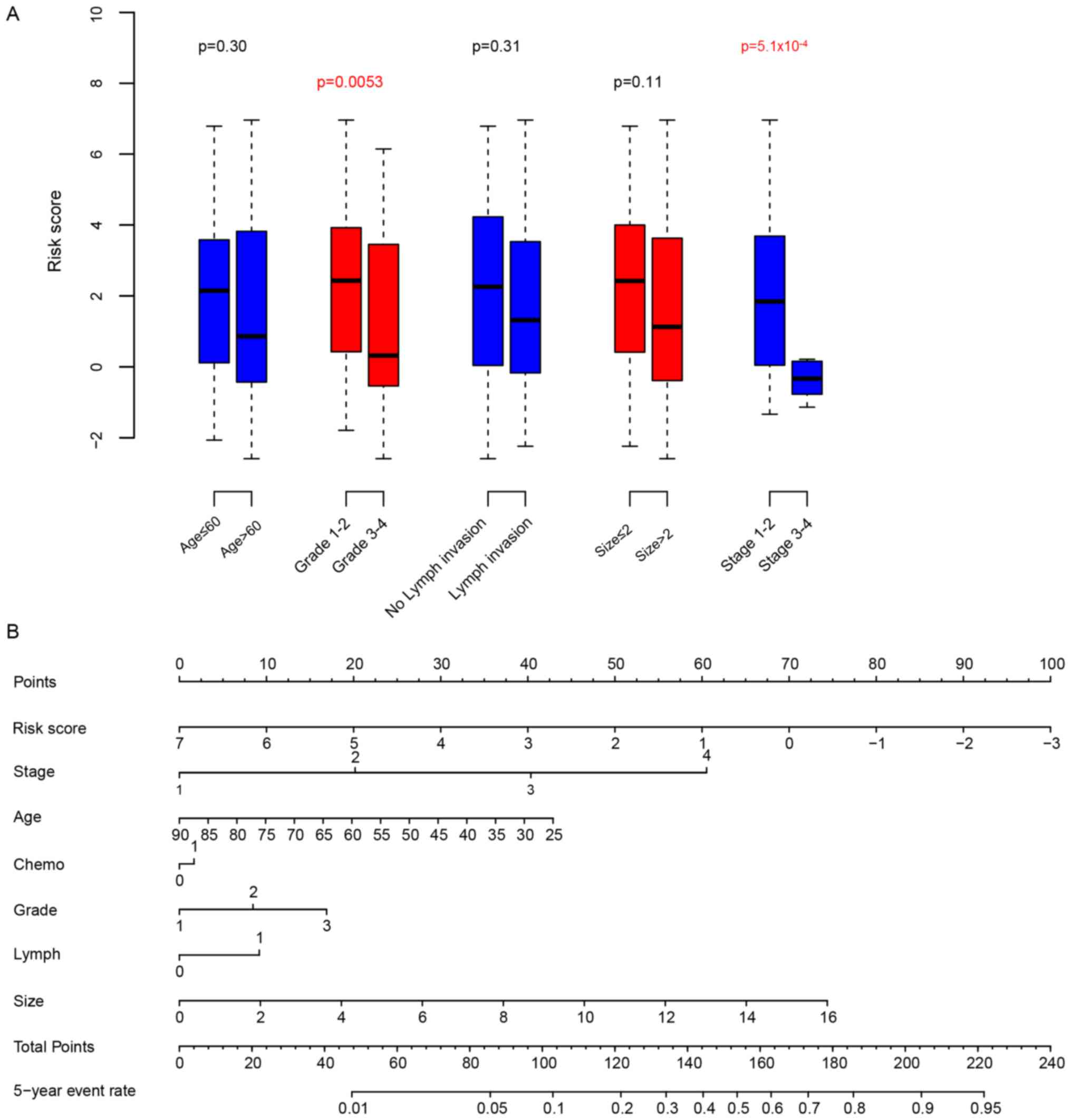
Risk score and clinicopathological
indicators
Next, the association between the clinical
observations and the risk score was calculated. The risk score was
significantly associated with the breast cancer grade and the TNM
stage. The risk score was independent from other clinical
parameters (Fig. 4A). In order to
compare the clinical significance and survival prediction of
clinicopathological observations and the risk score, a nomogram of
the 5-year survival rate was plotted (Fig. 4B). According to the nomogram, the risk
score exhibited the widest range, indicating that it is an
important prognostic indicator.
Discussion
Although the therapy of the ER+-breast
cancer is relatively well-established to date and mostly consists
of prescribing tamoxifen, the survival prognosis for patients with
ER+-breast cancer and treated with tamoxifen varies, and
clinicopathological observations remain insufficient (10,11).
Therefore, gene biomarkers for prognosis, drug selection and
follow-up are urgently needed. Although many single molecular
biomarkers for breast cancer prognosis have been studied in the
past years, the clinical effect across datasets was observed to be
limited, a multiple gene-based model is now preferred (8,12–15). In the present study, Cox multivariate
regression and random forest variable hunting were used on the
GSE17005 dataset, and a risk score model for survival prediction
was constructed. Patient groups with high and low risk scores were
significantly different in terms of survival. Furthermore, this
result was validated in three independent datasets. Compared with
other clinicopathological indicators, the risk score is also an
important prognostic indicator. Consistent with this, the value of
5-year survival ROC of the risk score is 0.733, which is
considerably higher compared with other clinical parameters (age,
tumor stage, tumor grade, chemotherapy, lymph invasion and tumor
size). In a previous study, tamoxifen-resistant biomarkers were
identified using transcriptomic signatures. Among these genes, the
highest 5-year relapse ROC was 0.64 for a single biomarker
(16), while in the present study, it
reached 0.733, which indicates the high performance of the
model.
Among these genes, STAT6 was previously shown to be
associated with mortality and metastasis in breast cancer (17,18). CENPE
was reported to be associated with cell cycle (19), and FOXD1 was indicated to be
associated with proliferation and drug resistance in breast cancer
(20). CTDSP1 was reported to inhibit
the migration and invasion of breast cancer cells (21). Aberrant methylation of AK5 was
identified in breast cancer, although the mechanism is unclear
(22). CCNA2 expression was
associated with resistance to tamoxifen in ER+-breast
cancer (23). STC2 expression was
associated with patient prognosis across different cancer types
(24,25). These reports indicate that the genes
used for the development of this model are relatively reliable.
The present study has several limitations. The
platform used in the four datasets is a microarray, which may limit
the utilization of the risk score. Additionally, since the present
study is a retrospective study, other epidemiological
characteristics, clinical manifestations, pathological features and
treatment methods of the samples were not assessed, and therefore a
comprehensive analysis of the correlation scores between
clinicopathological observations and risk score cannot be
performed. Finally, the risk score formula was developed using the
GSE17005 dataset, and a different formula was generated using the
other datasets. It is difficult to justify which formula is better.
In this article, the GSE17005 dataset was used for model
development to minimize prediction error, and therefore bias may
also exist.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data is available on Gene Expression Omnibus
(https://www.ncbi.nlm.nih.gov/geo/).
Author contributions
HH, QC, WS and ML performed the experiments. YY, ZZ
and PL analyzed the data. HH and PL were major contributors in the
writing of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All the experimental procedures were approved by the
Ethics Committee of Wenzhou Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huo Z, Gao Y, Yu Z, Zuo W and Zhang Y:
Metastasis of breast cancer to renal cancer: Report of a rare case.
Int J Clin Exp Pathol. 8:15417–15421. 2015.PubMed/NCBI
|
|
3
|
Oztas E, Kara H, Kara ZP, Aydogan MU, Uras
C and Ozhan G: Association between human telomerase reverse
transcriptase gene variations and risk of developing breast cancer.
Genet Test Mol Biomarkers. 20:459–464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Miller K and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takahashi M, Hayashida T, Okazaki H, Miyao
K, Jinno H and Kitagawa Y: Loss of B-cell translocation gene 2
expression in estrogen receptor-positive breast cancer predicts
tamoxifen resistance. Cancer Sci. 105:675–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salomaa V, Havulinna A, Saarela O, Zeller
T, Jousilahti P, Jula A, Muenzel T, Aromaa A, Evans A, Kuulasmaa K
and Blankenberg S: Thirty-one novel biomarkers as predictors for
clinically incident diabetes. PLoS One. 5:e101002010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z: Too much covariates in a
multivariable model may cause the problem of overfitting. J Thorac
Dis. 6:E196–E197. 2014.PubMed/NCBI
|
|
10
|
Apuri S: Neoadjuvant and adjuvant
therapies for breast cancer. South Med J. 110:638–642. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Glassman D, Hignett S, Rehman S, Linforth
R and Salhab M: Adjuvant endocrine therapy for hormone-positive
breast cancer, focusing on ovarian suppression and extended
treatment: An update. Anticancer Res. 37:5329–5341. 2017.PubMed/NCBI
|
|
12
|
Gogalic S, Sauer U, Doppler S, Heinzel A,
Perco P, Lukas A, Simpson G, Pandha H, Horvath A and Preininger C:
Validation of a protein panel for the non-invasive detection of
recurrent non-muscle invasive bladder cancer. Biomarkers.
22:674–681. 2017.PubMed/NCBI
|
|
13
|
Urquidi V, Netherton M, Gomes-Giacoia E,
Serie DJ, Eckel-Passow J, Rosser CJ and Goodison S: A microRNA
biomarker panel for the non-invasive detection of bladder cancer.
Oncotarget. 7:86290–86299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Huang J, Sun J, Xiang S, Yang D,
Ying X, Lu M, Li H and Ren G: The transcription levels and
prognostic values of seven proteasome alpha subunits in human
cancers. Oncotarget. 8:4501–4519. 2017.PubMed/NCBI
|
|
15
|
Kavalieris L, O'Sullivan P, Frampton C,
Guilford P, Darling D, Jacobson E, Suttie J, Raman JD, Shariat SF
and Lotan Y: Performance characteristics of a multigene urine
biomarker test for monitoring for recurrent urothelial carcinoma in
a multicenter study. J Urol. 197:1419–1426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mihaly Z, Kormos M, Lanczky A, Dank M,
Budczies J, Szász MA and Győrffy B: A meta-analysis of gene
expression-based biomarkers predicting outcome after tamoxifen
treatment in breast cancer. Breast Cancer Res Treat. 140:219–232.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Slattery ML, Lundgreen A, Hines LM,
Torres-Mejia G, Wolff RK, Stern MC and John EM: Genetic variation
in the JAK/STAT/SOCS signaling pathway influences breast
cancer-specific mortality through interaction with cigarette
smoking and use of aspirin/NSAIDs: The breast cancer health
disparities study. Breast Cancer Res Treat. 147:145–158. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Papageorgis P, Ozturk S, Lambert AW,
Neophytou CM, Tzatsos A, Wong CK, Thiagalingam S and Constantinou
AI: Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress
breast cancer lung metastasis. Breast Cancer Res. 17:982015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou S, Li N, Zhang Q, Li H, Wei X, Hao T,
Li Y, Azam S, Liu C, Cheng W, et al: XAB2 functions in mitotic cell
cycle progression via transcriptional regulation of CENPE. Cell
Death Dis. 7:e24092016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao YF, Zhao JY, Yue H, Hu KS, Shen H,
Guo ZG and Su XJ: FOXD1 promotes breast cancer proliferation and
chemotherapeutic drug resistance by targeting p27. Biochem Biophys
Res Commun. 456:232–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun T, Fu J, Shen T, Lin X, Liao L, Feng
XH and Xu J: The small c-terminal domain phosphatase 1 inhibits
cancer cell migration and invasion by dephosphorylating
ser(p)68-twist1 to accelerate twist1 protein degradation. J Biol
Chem. 291:11518–11528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miyamoto K, Fukutomi T, Akashi-Tanaka S,
Hasegawa T, Asahara T, Sugimura T and Ushijima T: Identification of
20 genes aberrantly methylated in human breast cancers. Int J
Cancer. 116:407–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao T, Han Y, Yu L, Ao S, Li Z and Ji J:
CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen
resistance. PLoS One. 9:e917712014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arigami T, Uenosono Y, Ishigami S,
Yanagita S, Hagihara T, Haraguchi N, Matsushita D, Hirahara T,
Okumura H, Uchikado Y, et al: Clinical significance of
stanniocalcin 2 expression as a predictor of tumor progression in
gastric cancer. Oncol Rep. 30:2838–2844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jansen MP, Sas L, Sieuwerts AM, Van
Cauwenberghe C, Ramirez-Ardila D, Look M, Ruigrok-Ritstier K,
Finetti P, Bertucci F, Timmermans MM, et al: Decreased expression
of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer
and endocrine therapy resistance in advanced disease. Mol Oncol.
9:1218–1233. 2015. View Article : Google Scholar : PubMed/NCBI
|