Introduction
Lung cancer ranks among the most rapidly growing
types of cancer with a high morbidity and mortality rate. Also, it
is one of the most serious malignancies for population health and
life (1,2). Non-small cell lung cancer (NSCLC)
accounts for 70–80% among all types of lung cancer and the survival
rate is approximately 11% (3).
Patient relapse may lead to disease metastasis and consequently,
death. Therefore, identification of novel treatments for human
NSCLC is required.
MicroRNAs (miRNAs), endogenous non-coding
single-stranded RNAs bound to the 3′-untranslated region (3′-UTR)
of mRNAs lead to the proliferation, metastasis and invasion of
cancer (4–6). Previous studies have shown that miR-196
has an aberrant expression in various tumors and has been involved
in the progression of cancers. For example, Mueller and Bosserhoff
showed that silencing miR-196a may deregulate several genes in
melanoma cells ito nfluence melanoma progression (7). miR-196a has been found to be
highly-expressed in colorectal cancer cells and it may enhance
cancer cell migration and invasion (8). In addition, a study showed that the
re-expression of miR-196b significantly contributed to leukemia
development (9). The effect of
miR-196 on the malignant progression of gliomas has also been
demonstrated (10). Moreover,
miR-196s acted as potent suppressors in the cell migration and
metastasis of breast tumors (11). It
has been previously reported that miR-196b is significantly
upregulated in lung cancer and is considered to be a potential
marker of survival (12). However,
the underlying mechanism of miR-196b in the NSCLC cell process has
yet to be explored.
Transcription factor GATA-6 (GATA6) has been
identified to be involved in the development, proliferation, and
differentiation of several organs (13,14). GATA6
promoted cholangiocarcinoma cell invasion and metastasis and acted
as a potential oncogene in cholangiocarcinoma (15,16). GATA6
has been shown to be misregulated in colon cancer cells, suggesting
a relevant role in the progression of colon cancer (17,18).
Moreover, GATA6 was verified to inhibit lung adenocarcinoma
metastasis (19).
In our study, we showed the upregulation of miR-196b
and downregulation of the GATA6 expression in NSCLC. GATA6 has been
characterised as a direct target gene of miR-196b in NSCLC. The
relationship between miR-196b and GATA6 protein level was found to
be negatively correlated in NSCLC tissues. Overall, we drew a
conclusion that miR-196b may enhance NSCLC cell migration and
invasion by targeting GATA6.
Materials and methods
Tissue sample collection
NSCLC tissues and normal lung tissues were obtained
from 40 paired patients all of whom signed written informed consent
after surgery at The Eastern Medical District of Linyi People's
Hospital (Linyi, China). Then, we stored the samples in a −80°C
refrigerator. All the experiments were approved by the Ethics
Committee of The Eastern Medical District of Linyi People's
Hospital (Linyi, China).
Cell culture
The NSCLC cell lines (A549, H226, H1650, H1299,
SPC-A1) were purchased from the American Type Culture Collection
(ATCC, Manassas, VA, USA). The cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA)
containing 10% fetal bovine serum, penicillin (100 U/ml) and
streptomycin (100 U/ml), which was incubated at 37°C in a 5%
CO2 environment.
Transfection of miR-196b
miR-196b mimic, miR-196b inhibitor, control mimic
and control inhibitor were transfected to A549 cells or H226 cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) the next day when the cells were 70–80% confluent.
Overexpression of GATA6
The GATA6 plasmid was purchased from GeneCopoeia
(Germantown, MD, USA), and transfected in A549 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.).
Western blot analysis
After transfection for 48 h, RIPA lysis containing
proteinase inhibitors (Beyotime Institute of Biotechnology, Haimen,
China) was used to extract total protein. The protein
concentrations were tested with the BCA kit (Beyotime Institute of
Biotechnology). The total proteins (50 µg) were added into the well
with SDS-PAGE and electrophoresis at 60 V was performed when
bromophenol blue ran out of the bottom. The proteins were then
transferred to nitrocellulose filter (NC) membranes, and skimmed
milk (5–10%) was used to block he membranes at room temperature for
2 h. Subsequently, primary antibodies (GATA6, Abcam, Cambridge, UK;
GAPDH, Cell Signaling Technology, Inc., Danvers, MA, USA) were
added in to incubate the samples at 4°C overnight. After being
washed with 1X TBST (pH 7.4) three times, the secondary antibodies
were added in and incubated at room temperature for 2 h. Protein
bands were detected using the chemiluminescence method (ECL;
Millipore Billerica, MA, USA). GAPDH served as a loading
control.
RNA isolation and RT-qPCR
We used TRIzol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) to extract total RNA from the cell lines or
tissue samples (Invitrogen; Thermo Fisher Scientific, Inc.), then,
we used the BioPhotometer (Eppendorf, Hamburg, Germany) to
determine the concentration of the total RNA. All-in-One™ miRNA
First-Strand cDNA synthesis kit was used to synthesize cDNA, and
RT-qPCR was performed using the TaqMan PCR probes. The of the
primer sequences were as follows: miR-196b-F: TAG
GTACCACTTTATCCCGTTCACCA, miR-196b-R: ATC TCGAGGCAGGGAGAGAGGAATAA;
GATA6-F: CTC CAACTTCCACCTCTTCTAAC, GATA6-R: TGGTGTGGT GGAGTCG;
U6-F: GCCCATCTTGACCCGAAT, U6-R: AACGCTTCACGAATTTGCGT; GAPDH-F:
ACAGTCAG CCGCATCTTCTT, GAPDH-R: ACGACCAAATCCGT TGACTC. GAPDH and U6
were used as endogenous controls. Relative expression levels of
genes analysed were calculated using the 2−ΔΔCq
method.
Cell migration assay
A549 and H226 cell migration was measured using a
Transwell chamber with a polycarbonic membrane with 8-mm pore size
in vitro. NSCLC cells were treated with different
transfection for 24 h. Then, 1×105 cells were seeded
into the upper chamber and a complete culture medium with 20% FBS
was added to the lower chamber as an attractant. After incubation
for 24 h at 37°C, in a 5% CO2 environment, the media
were removed. The cotton swabs were used to remove the cells from
the top chamber. Moreover, the 100% methanol was used to fix the
cells migrated to the lower membrane and 0.1% crystal violet was
used to stain the migrating cells. The number of cells was
quantified by a microscope (Olympus, Tokyo, Japan).
Cell invasion assay
A549 and H226 cells invasion was measured using a
millicell invasion chamber with 8-μm pore size polycarbonate
membranes (Neuro Probe Inc., Gaithersburg, MD, USA) pre-coated with
2 mg/ml Matrigel (BD Bioscience, San Jose, CA, USA). Then
1×105 cells were added to the upper chamber coated with
Matrigel and a complete culture medium with 20% FBS was added to
the lower chamber as an attractant and invaded for 24 h. The cotton
swabs were used to remove the cells in the top chamber. Moreover,
100% methanol was used to fix the invaded cells to the lower
membrane and 0.1% crystal violet was used to stain the invading
cells. The number of cells was quantified by microscope
(Olympus).
Dual luciferase reporter assay
The wild-type and mutated miR-196b putative targets
on GATA6 3′UTR were cloned into pGL3-promoter vector, a final
concentration of 100 nM of cells of different transfection
treatments were collected 48 h after transfection using
Lipofectamine 2000, and luciferase activity was assayed with the
dual-luciferase reporter assay system (Promega, Madison, WI,
USA).
Statistical analysis
The experiments were repeated three times. SPSS
v.19.0 software (IBM Corp., Armonk, NY, USA) was used to perform
the statistical analyses as well as GraphPad Prism 5.02 software
(GraphPad Software, Inc., La Jolla, CA, USA) to complete graph
presentations. All data were presented as mean ± SD. Students
t-test or post hoc test after one-way analysis of variance (ANOVA)
in SPSS were used to analyze the differences between the groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Detection of miR-196b mRNA and GATA6
protein expression in NSCLC tissues
First, we investigated the miR-196b expression in
eight NSCLC tissue specimens and their corresponding non-cancerous
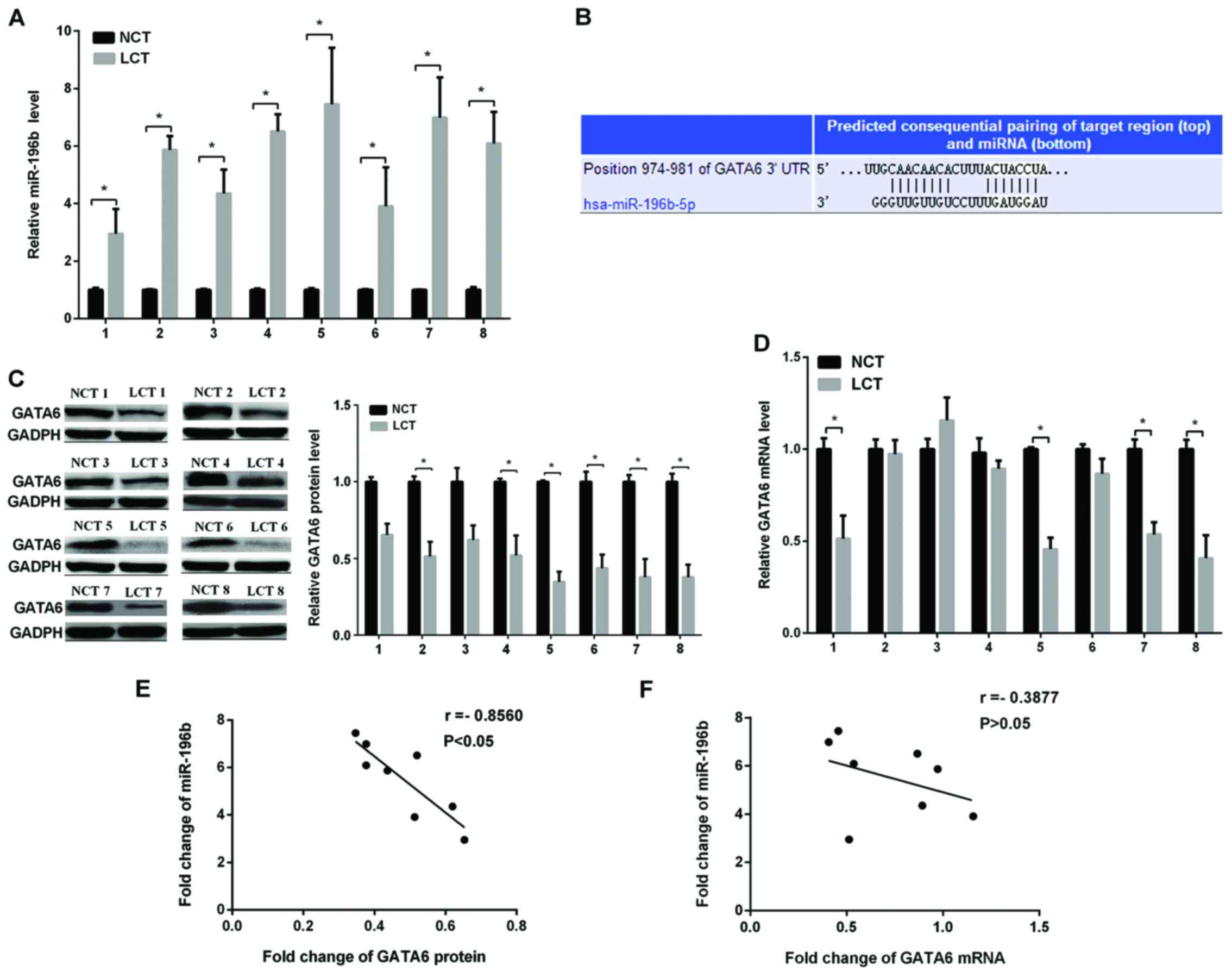
tissue (NCT) specimens using RT-qPCR. The results showed that the
miR-196b expression was markedly higher in NSCLC samples (Fig. 1A). The role of miRNA via targeting the
3′-UTR of mRNA is involved in the regulation of gene expression.
Then, we used the TargetScan and miRanda to verify the direct
target of miR-196b and it was found that GATA6 may be the target of
miR-196b. The predicted target sites between miR-196b and GATA6 are
shown in Fig. 1B. It is generally
considered that the expression level of miRNAs and their target
genes are opposite. Next, we investigated the GATA6 protein level
in the same eight NSCLC and non-cancerous tissues, as shown in
Fig. 1C. The GATA6 protein expression
level was conspicuously lower in the NSCLC than in normal tissues,
while the GATA6 mRNA level varied randomly (Fig. 1D). Regression analysis of correlation
was used to show the relationship between the miR-196b and GATA6
protein levels (Fig. 1E) and miR-196b
and GATA6 mRNA levels (Fig. 1F). We
found that the inverse correlation coefficient (r=−0.7394) between
the miR-196b expression and GATA6 protein level was higher than
that of the miR-196b expression and GATA6 mRNA level (r=−0.3375).
The results strongly suggested that miR-196b enhanced NSCLC cell
migration and invasion by downregulating the GATA6 protein level
and not the mRNA level.
Corroboration of GATA6 as a direct
target of miR-196b
To further examine the relationship between miR-196b
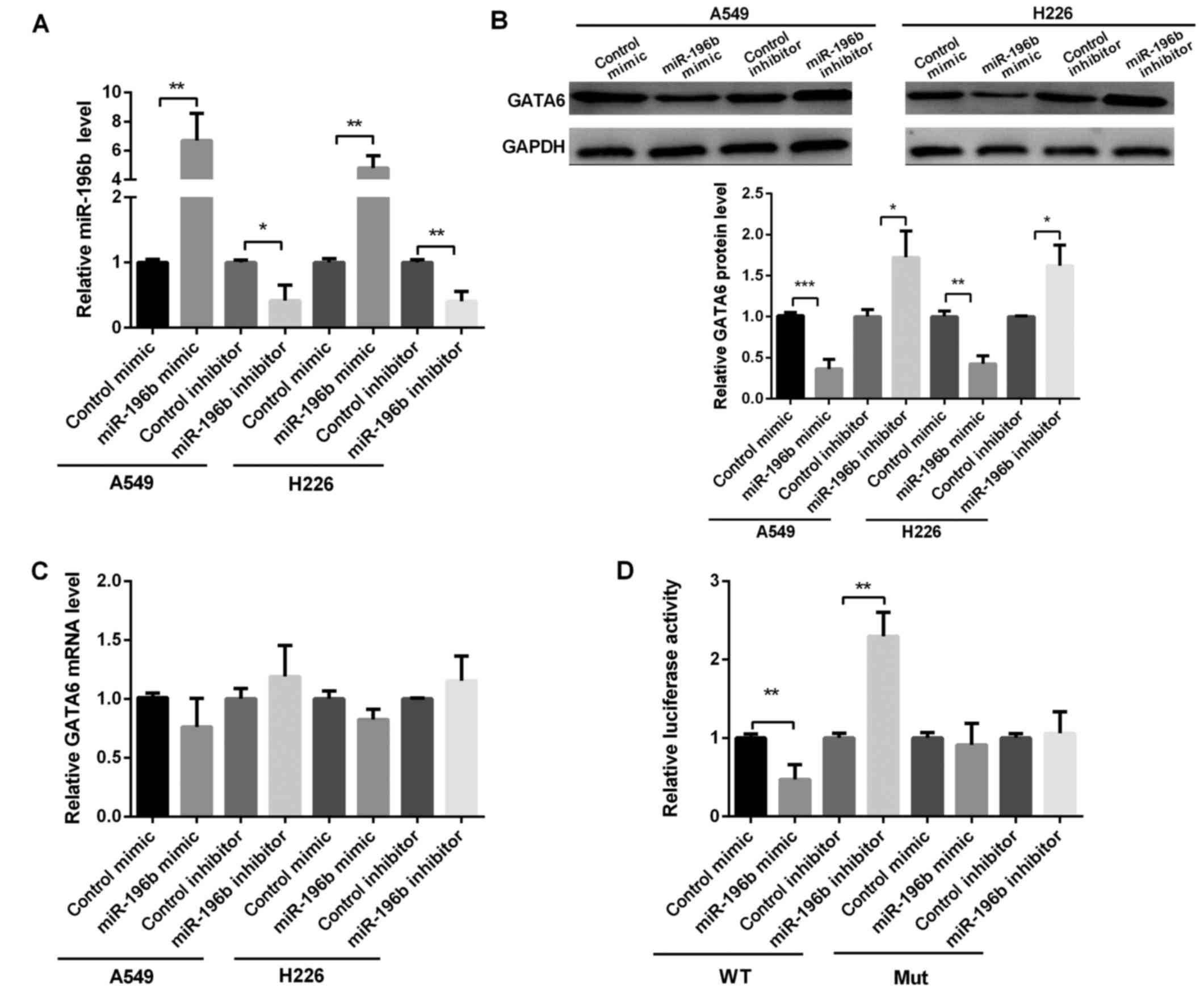
and GATA6, we evaluated the GATA6 expression in A549 and H226 cells
after transfection with miR-196b mimic or inhibitor (Fig. 2A). It was shown that GATA6 protein
level was markedly reduced after the overexpression of miR-196b but
significantly increased when silencing miR-196b in A549 and H226
cells (Fig. 2B). However, the GATA6
mRNA level did not change for overexpressed or silenced miR-196b
(Fig. 2C), suggesting that miR-196b
regulated GATA6 expression at the post-transcriptional level.
We used dual luciferase reporter assay to detect the
predicted sequence binding sites of miR-196b and GATA6. As shown in
Fig. 2D, the luciferase reporter
activity in the miR-196b mimic group was obviously lower, whereas
the luciferase reporter activity in the miR-196b inhibitor group
was obviously higher than that in the control group (Fig. 2D). Then, we detected miR-196b binding
ability in the mutated type of miR-196b. The results showed that
miR-196b mimic or miR-196b inhibitor group exert no effect on the
luciferase reporter activity (Fig.
2D). Thus, miR-196b inhibited GATA6 translation by binding to
the 3′-UTR of GATA6.
miR-196b promotes NSCLC migration by
targeting GATA6
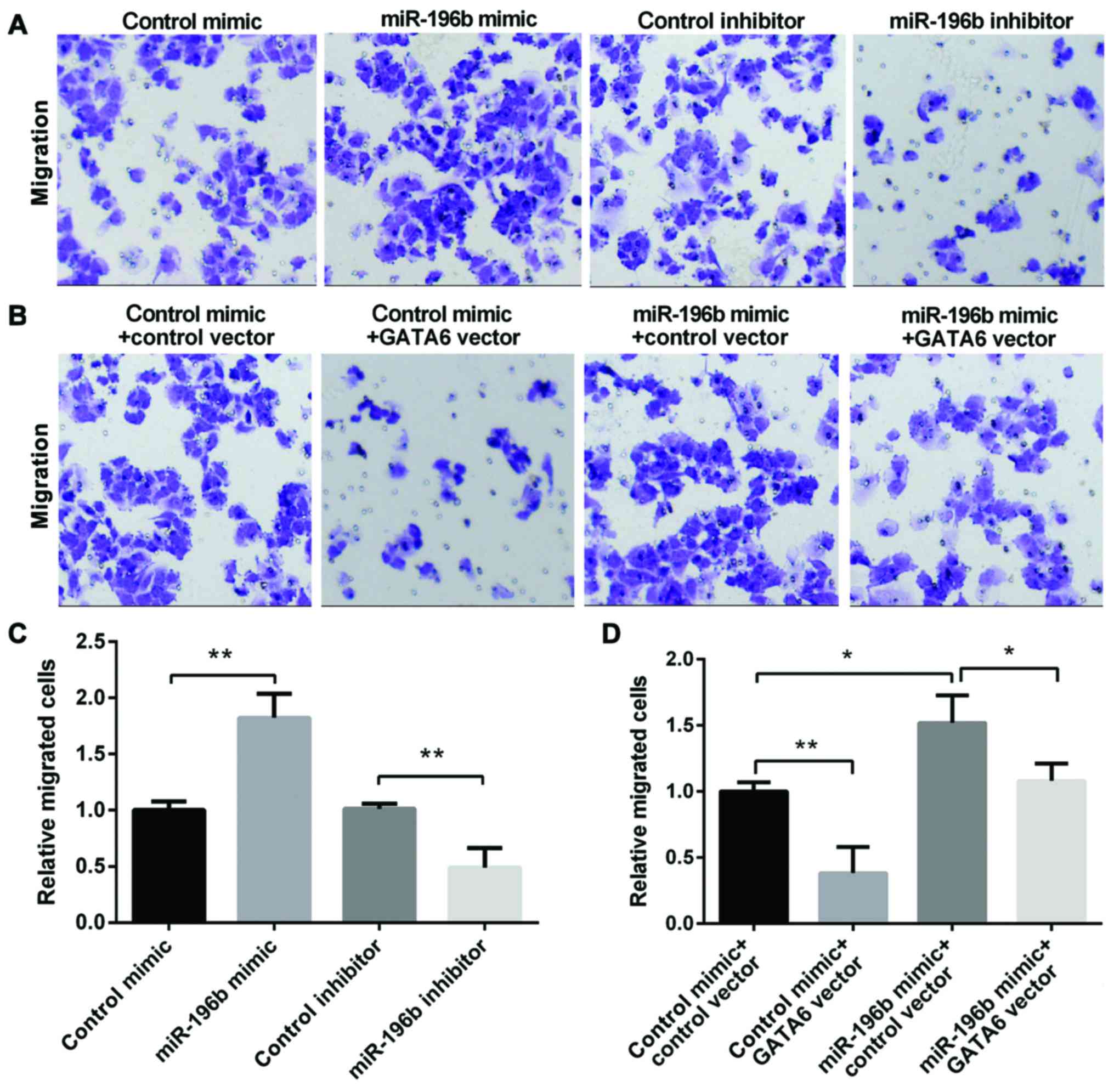
We used the Transwell assay to investigate the role
that miR-196b and GATA6 played on NSCLC migration. The miR-196b
mimic group showed an enhanced migration, whereas the miR-196b
inhibitor group showed a decreased migration (Fig. 3A and B). Moreover, the overexpression
of GATA6 decreased cell migration, while miR-196b mimic promoted
cell migration. However, re-expression of both miR-196b and GATA6
showed lower migration than the cell overexpression of miR-196b
alone (Fig. 3C and D), suggesting
that GATA6 may attenuate the promotion effect of miR-196b on NSCLC
migration. In conclusion, miR-196b may promote NSCLC cell migration
by targeting GATA6.
miR-196b promotes invasion of NSCLC
via targeting GATA6
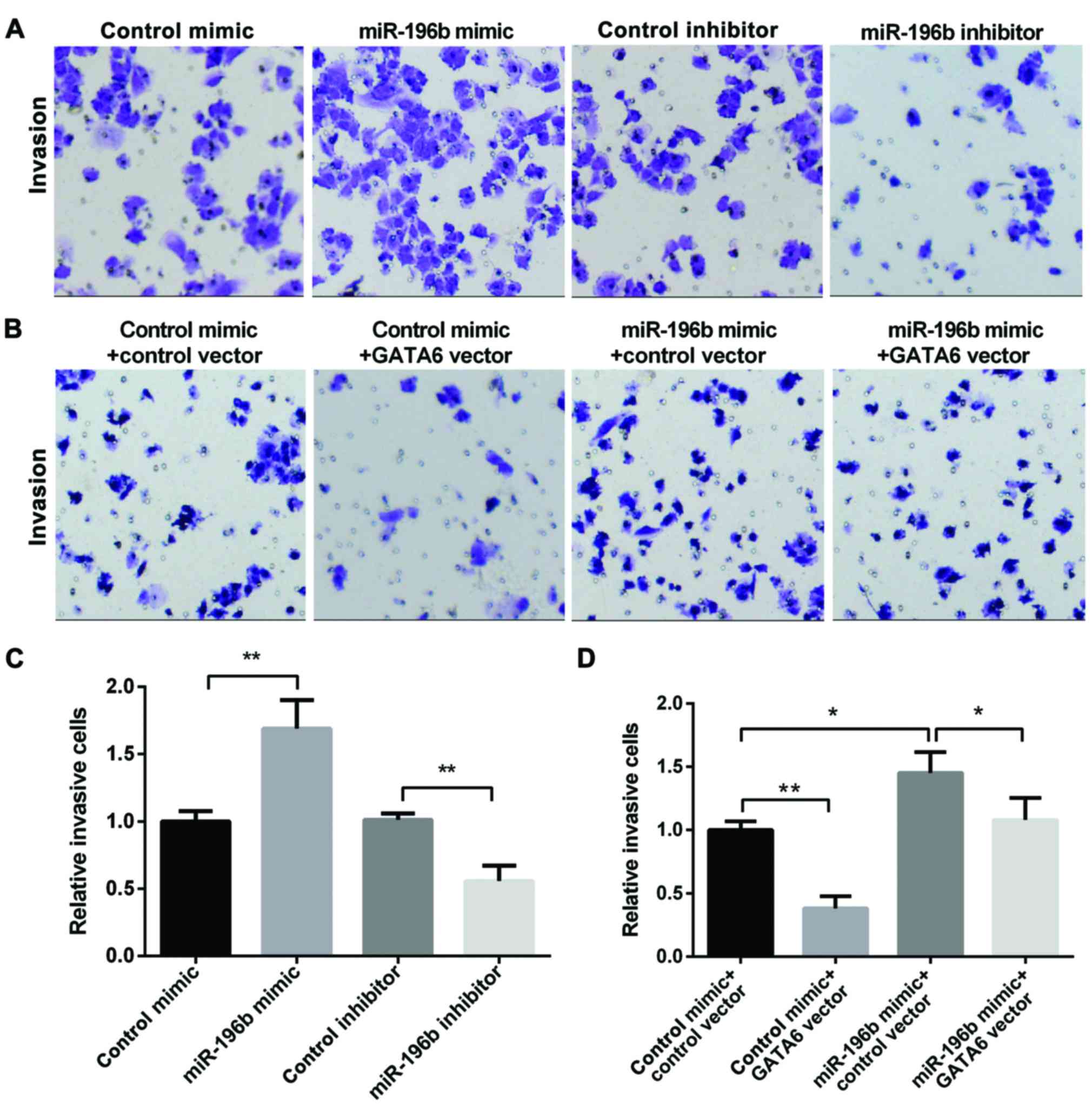
Next, we used the Transwell assay to investigate the
role that miR-196b and GATA6 played in NSCLC invasion. The miR-196b
mimic group showed increased invasion, while, the miR-196b
inhibitor group showed decreased invasion (Fig. 4A and B). Moreover, the overexpression
of GATA6 decreased cell invasion, while the miR-196b mimic promoted
cell invasion. However, re-expression of both miR-196b and GATA6
showed lower invasion than that of the cell overexpression of
miR-196b alone (Fig. 4C and D),
suggesting that GATA6 may attenuate the promotion effect of
miR-196b on NSCLC invasion. In conclusion, miR-196b may promote
NSCLC cell invasion by targeting GATA6.
Discussion
It has been proven that miR-196 family members
participate in tumorigenesis, and they are misexpressed in various
malignancies (20,21). For instance, miR-196b could promote
tumor progression in lung cancers as an oncogene (22). In this study, we stated that miR-196b
expression was markedly increased in NSCLC tissues and cell lines,
which was consistent with a previous study where miR-196b was
upregulated in the early-stage of NSCLC patients (23), and acted as a potential marker in lung
cancer patients (12). We also
confirmed that the miR-196b mimic may promote NSCLC cell migration
and invasion, while the miR-196b inhibitor suppresses migration and
invasion.
miRNAs regulated post-transcriptional gene
expression by binding to the 3′UTR of target mRNAs (24). miR-196b was proven to regulate the
GATA6 expression in colorectal cancer cells (25). However, our results have shown that
GATA6 was regulated by miR-196b in NSCLC. miR-196b overexpression
may inhibit the GATA6 expression in NSCLC cells, while the
inhibition of miR-196b stimulated the GATA6 expression.
GATA6 is a member of the GATA family of Zn-finger
transcription factors which are involved in the development of
several tissues and organs (15,26). A
recent study suggests that modulating the expression of GATA6 may
affect the growth of NSCLC cells (27). Mehta et al verified GATA6 and
NKX2-1 as diagnostic biomarkers of non-invasive lung cancer
(28). BMP4 regulation by GATA6
showed that it can inhibit tumorigenesis and metastasis of lung
adenocarcinoma cells (29). In this
study, we found that the GATA6 expression in NSCLC tumor was
remarkably lower than that in the normal tissues. Pearson's
correlation scatter plot showed that the relationship between the
miR-196b expression and GATA6 was negatively correlated in NSCLC
tissues. Furthermore, we confirmed that the overexpression of GATA6
may inhibit NSCLC cell migration and invasion. This is consistent
with a previous study showing that GATA6 may inhibit lung
adenocarcinoma metastasis (19).
Additionally, we found that the overexpression of GATA6 may
partially reverse the function of miR-196b.
In conclusion, we have demonstrated that miR-196b
was markedlly increased and negatively associated with GATA6 in
NSCLC cells. Moreover, the overexpression of miR-196b enhanced
NSCLC cell migration and invasion via direct targeting GATA6. The
results strongly suggest that GATA6 plays a significant role in
regulating the progression of lung cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL wrote the manuscript and collected tissue
samples. CF was responsible for cell culture. SS performed PCR. All
authors have read and approved the final study.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The Eastern Medical District of Linyi People's Hospital (Linyi,
China) and all patients signed written informed consent after
surgery.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fassina A, Cappellesso R and Fassan M:
Classification of non-small cell lung carcinoma in transthoracic
needle specimens using microRNA expression profiling. Chest.
140:1305–1311. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoo AS, Staahl BT, Chen L and Crabtree GR:
MicroRNA-mediated switching of chromatin-remodelling complexes in
neural development. Nature. 460:642–646. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Madhyastha R, Madhyastha H, Pengjam Y,
Nakajima Y, Omura S and Maruyama M: NFkappaB activation is
essential for miR-21 induction by TGFβ1 in high glucose conditions.
Biochem Biophys Res Commun. 451:615–621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han H, Sun D, Li W, Shen H, Zhu Y, Li C,
Chen Y, Lu L, Li W, Zhang J, et al: A c-Myc-MicroRNA functional
feedback loop affects hepatocarcinogenesis. Hepatology.
57:2378–2389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mueller DW and Bosserhoff AK: MicroRNA
miR-196a controls melanoma-associated genes by regulating HOX-C8
expression. Int J Cancer. 129:1064–1074. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schimanski CC, Frerichs K, Rahman F,
Berger M, Lang H, Galle PR, Moehler M and Gockel I: High miR-196a
levels promote the oncogenic phenotype of colorectal cancer cells.
World J Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Popovic R, Riesbeck LE, Velu CS, Chaubey
A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et
al: Regulation of mir-196b by MLL and its overexpression by MLL
fusions contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guan Y, Mizoguchi M, Yoshimoto K, Hata N,
Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al:
miRNA-196 is upregulated in glioblastoma but not in anaplastic
astrocytoma and has prognostic significance. Clin Cancer Res.
16:4289–4297. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zhang M, Chen H, Dong Z, Ganapathy
V, Thangaraju M and Huang S: Ratio of miR-196s to HOXC8 messenger
RNA correlates with breast cancer cell migration and metastasis.
Cancer Res. 70:7894–7904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng R and Blobel GA: GATA transcription
factors and cancer. Genes Cancer. 1:1178–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aronson BE, Stapleton KA and Krasinski SD:
Role of GATA factors in development, differentiation, and
homeostasis of the small intestinal epithelium. Am J Physiol
Gastrointest Liver Physiol. 306:G474–G490. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian F, Li D, Chen J, Liu W, Cai L, Li J,
Jiang P, Liu Z, Zhao X, Guo F, et al: Aberrant expression of GATA
binding protein 6 correlates with poor prognosis and promotes
metastasis in cholangiocarcinoma. Eur J Cancer. 49:1771–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian F, Chen J, Zheng S, Li D, Zhao X,
Jiang P, Li J and Wang S: miR-124 targets GATA6 to suppress
cholangiocarcinoma cell invasion and metastasis. BMC Cancer.
17:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Haveri H, Westerholm-Ormio M, Lindfors K,
Mäki M, Savilahti E, Andersson LC and Heikinheimo M: Transcription
factors GATA-4 and GATA-6 in normal and neoplastic human
gastrointestinal mucosa. BMC Gastroenterol. 8:92008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Belaguli NS, Aftab M, Rigi M, Zhang M,
Albo D and Berger DH: GATA6 promotes colon cancer cell invasion by
regulating urokinase plasminogen activator gene expression.
Neoplasia. 12:856–865. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheung WK, Zhao M, Liu Z, Stevens LE, Cao
PD, Fang JE, Westbrook TF and Nguyen DX: Control of alveolar
differentiation by the lineage transcription factors GATA6 and HOPX
inhibits lung adenocarcinoma metastasis. Cancer Cell. 23:725–738.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Zhang Y, Zhang L, Weakley SM and
Yao Q: MicroRNA-196: Critical roles and clinical applications in
development and cancer. J Cell Mol Med. 15:14–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hamamoto J, Soejima K, Yoda S, Naoki K,
Nakayama S, Satomi R, Terai H, Ikemura S, Sato T, Yasuda H, et al:
Identification of microRNAs differentially expressed between lung
squamous cell carcinoma and lung adenocarcinoma. Mol Med Rep.
8:456–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Võsa U, Vooder T, Kolde R, Fischer K, Välk
K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A and Annilo T:
Identification of miR-374a as a prognostic marker for survival in
patients with early-stage nonsmall cell lung cancer. Genes
Chromosomes Cancer. 50:812–822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fantini S, Salsi V, Reggiani L, Maiorana A
and Zappavigna V: The miR-196b miRNA inhibits the GATA6 intestinal
transcription factor and is upregulated in colon cancer patients.
Oncotarget. 8:4747–4759. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kwei KA, Bashyam MD, Kao J, Ratheesh R,
Reddy EC, Kim YH, Montgomery K, Giacomini CP, Choi Y-L, Chatterjee
S, et al: Genomic profiling identifies GATA6 as a candidate
oncogene amplified in pancreatobiliary cancer. PLoS Genet.
4:e10000812008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zito G, Naselli F, Saieva L, Raimondo S,
Calabrese G, Guzzardo C, Forte S, Rolfo C, Parenti R and Alessandro
R: Retinoic acid affects lung adenocarcinoma growth by inducing
differentiation via GATA6 activation and EGFR and Wnt inhibition.
Sci Rep. 7:47702017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mehta A, Cordero J, Dobersch S,
Romero-Olmedo AJ, Savai R, Bodner J, Chao CM, Fink L, Guzmán-Díaz
E, Singh I, et al: Non-invasive lung cancer diagnosis by detection
of GATA6 and NKX2-1 isoforms in exhaled breath condensate. EMBO Mol
Med. 8:1380–1389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim JS, Kurie JM and Ahn YH: BMP4
depletion by miR-200 inhibits tumorigenesis and metastasis of lung
adenocarcinoma cells. Mol Cancer. 14:1732015. View Article : Google Scholar : PubMed/NCBI
|