Introduction
Prostate cancer (PCa) remains the most commonly
diagnosed cancer and the third most common cause of
cancer-associated mortality among males in western countries
(1). In China, the incidence rate of
PCa is increasing year by year and has already exceeded the
incidence of bladder cancer (2). The
majority of patients with middle/late stage or recurrence of PCa
exhibit androgen-independent growth and resistance to multiple
therapies, which leads to the failure of conventional treatment and
a high mortality rate (3). Therefore,
there is an urgent requirement to identify effective biomarkers to
strengthen the efficiency of early diagnosis and improve the
therapeutic strategies used to treat PCa (4).
MicroRNAs (miRNAs/miRs) are endogenously expressed,
small non-coding RNAs that regulate target gene expression, cell
proliferation, cell differentiation and cell apoptosis through
combination with the 3′-untranslated region (3′-UTR) of target
mRNAs to induce mRNA degradation. Previous studies have revealed an
association between the expression of various miRNAs and
tumorigenesis, invasion, metastasis, resistance to chemotherapy and
poor prognosis (5,6).
There is evidence linking the reduced expression of
miR-139 to cancer progression and carcinogenesis (7). For instance, miR-139 inhibits cell
proliferation and invasion by targeting the expression of
insulin-like growth factor 1 receptor in non-small cell lung cancer
(8). miR-139 also exerts a tumor
suppressor function by targeting Notch1 in colorectal and breast
cancer (9,10). To the best of our knowledge, the
effects of miR-139 in PCa, and the molecular mechanisms underlying
these effects, remain elusive. Therefore, the present study aimed
to investigate the biological functions of miR-139 on the cell
cycle, proliferation, apoptosis, migration and invasion in PCa.
Materials and methods
Cell lines and tissue samples
The PCa PC-3, C4-2B and LNCaP cell lines were grown
in a 37°C, 5% (v/v) CO2 growth chamber. Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml
streptomycin solution. All cell culturing reagents were obtained
from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA,
USA).
Tissue samples [10 samples of benign prostatic
hyperplasia (BPH) and 10 samples of PCa] were obtained from
patients who underwent surgical treatment by transurethral
resection of the prostate or radical prostatectomy between January
2012 and October 2013. The age of the patients was 65.6±3.5 years.
The tissue samples were collected by the Department of Pathology,
Tongji Hospital, Tongji Medical College, Huazhong University of
Science and Technology (Wuhan, China). The histopathological
diagnosis was reviewed by at least two specialists in pathology.
Tissue samples were obtained and handled in accordance with a
protocol approved by the Institutional Review Board for Human
Research of Tongji Hospital (Wuhan, China). Written informed
consent was obtained from all patients.
Reverse transcription-quantitative
polymerase chain reaction
miRNAs were isolated using the miRNeasy Mini kit
(Qiagen Inc.) according to the manufacturer's protocol. miRNAs were
assayed using TaqMan MicroRNA assays (Thermo Fisher Scientific,
Inc.) in accordance with the manufacturer's protocol. Samples were
normalized to U6 small nuclear RNA (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The comparative Cq method (11) was used to calculate the relative
changes in gene expression on the Applied Biosystems®
7500 Fast Real Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the following thermocycler program for all
genes: 5 min of pre-incubation at 95°C followed by 40 cycles of 15
sec at 95°C, 15 sec at 60°C, and 30 sec at 72°C. The primers used
were as follows: miR-139-5p, forward, 5′-CCTCTACAGTGCACGTGTCTC-3′,
and reverse, 5′-CGCTGTTCTCATCTGTCTCGC-3′; and U6, forward,
5′-TGCTCGCTTCGGCAGC-3′, and reverse,
5′-AAAAATATGGAACGCTTCACG-3′.
miRNA transfection
Cells were plated in DMEM without antibiotics ~24 h
prior to transfection. Transient transfection of a human miR-139-5p
precursor (has-miR-139-5P mimic; cat no. MIMAT0000250; Ambion;
Thermo Fisher Scientific, Inc.) or miRNA mimic negative control
(miR-CN; cat no. AM7110; Ambion, Thermo Fisher Scientific, Inc.)
was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Apoptosis assays
Apoptosis was assessed by measuring the membrane
redistribution of phosphatidylserine using an Annexin V-FITC
Apoptosis Detection kit (BD Pharmingen; BD Biosciences, San Jose,
CA, USA). A total of 1×106 cells were collected, washed
twice with PBS and resuspended in 500 µl of the kit's staining
solution, containing fluorescein isothiocyanate-conjugated Annexin
V antibodies and propidium iodide (PI). Following incubation on ice
for 30 min, the cells were analyzed using a BD FACSCalibur flow
cytometer and CellQuest Pro software (version 5.1; BD Biosciences).
Basal apoptosis and necrosis rates were determined in untranfected
cells.
Cell cycle analysis by fluorescence
activated cell sorting
Cells were trypsinized, washed twice in ice-cold
PBS, and fixed with cold 70% ethanol for 24 h. Propidium iodide
solution (40 µg/ml) and RNase A (cat. no. EN0531; Thermo Fisher
Scientific, Inc.) were added to the cells, which were then
incubated for 45 min in the dark at 4°C prior to analysis.
Cell proliferation assay
Cells were seeded at a density of 2х103
per well in 96-well plates and incubated in DMEM containing 10%
FBS. Following seeding, cells were transfected as aforementioned.
The tetrazolium salt MTT was added to the wells, cells were
incubated for 4 h at 37°C, then the medium was changed with 150 µl
dimethyl sulfoxide at room temperature to dissolve the formazan
crystals, the optical density (OD) was measured at 490 nm using a
microplate reader. The growth inhibitory rate=(1-OD value of the
observed group/OD value of the control group) ×100%.
Wound healing assay
Cells were grown on 6-well plates. At 24 h after
transfection, the confluent cells were carefully scratched using a
200 µl pipette tip. Images were captured in 8 different regions per
well at 0 and 48 h using a Zeiss Axiovert 200 M (Zeiss AG,
Oberkochen, Germany) inverted light microscope. For each region,
the distance migrated by the cells was measured at 3 different
points. The migrated distance was recorded as a mean of 24 points
every hour. Then, the formula [mean speed (µm/min)=migration
distance/migration time] was used to assess the migration speed of
tumor cells.
Cell invasion assay
The cell invasion assay was performed using Boyden
Transwell chambers (8.5 mm in diameter) with Matrigel added to the
filters (pore size, 8 µm; Costar; Corning Incorporated, Corning,
NY, USA). Briefly, 500 µl DMEM containing 10% FBS was added to the
bottom well. At 24 h after transfection, cells were re-suspended in
DMEM without FBS at a concentration of 1×106 cells/ml,
and 500 µl cell suspension was added to the top well. Following
incubation at 37°C for 36 h, the cells that had not migrated were
removed from the upper surface of the filters using cotton swabs,
and those that had invaded to the lower surface of the filters were
fixed in 100% methanol at room temperature for 15 min, followed by
staining with 0.05% crystal violet at room temperature for 2 h.
Invasiveness was determined by counting the mean number of cells
within five random fields of view using an inverted light
microscope (magnification, ×100).
Dual luciferase reporter assay
Potential miR-139-5p binding sites were predicted
using TargetScan (www.targetscan.org). The sequence of 42 bp segments
with the wild-type or mutant 3′UTR seed region of Notch1 (prepared
by the deletion of 10 nucleotides in the seed region) was
synthesized and cloned into a pMIR-REPORT luciferase vector
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
synthesized oligonucleotides were as follows: Wild-type Notch1:
5′-CTAGTGACTTTAAAAGTGATCTACATGAGGAACTGTAGATGATGTGAGCT-3′;
5′-CACATCATCTACAGTTCCTCATGTAGATCACTTTTAAAGTCA-3′; Mutant Notch1:
5′-CTAGTGACTTTAAAAGTGATCTACATGAGTGATGTGAGCT-3′;
5′-CACATCACTCATGTAGATCACTTTTAAAGTCA-3′. The Notch1 construct or
control construct was co-transfected into C4-2B and PC-3 cells
along with miR-139 or miR-CN using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.). Firefly luciferase activity was
measured 48 h following transfection and normalized against Renilla
luciferase activity, using the Dual Luciferase Reporter Assay
system (Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol.
Western blot analysis
Cells were lysed on ice for 30 min with a lysis
buffer containing 150 mmol/l NaCl, 50 mmol/l Tris (pH 7.4), 1%
Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and protease
inhibitor cocktail (cat. no. S8830; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), then the product were centrifuged at 14,000 ×
g at 4°C for 30 min. A total of 100 µg protein were denatured in
SDS sample buffer [2% SDS, 62.5 mM Tris-base (pH 6.8), 10%
glycerol, 5% β-mercaptoethanol, and 0.005% bromophenol blue] and
loaded onto a 10% SDS-PAGE gel. The separated proteins were
transferred onto nitrocellulose membranes and the membranes were
blocked overnight at 4°C in TBS with 5% (w/v) powdered skimmed
milk, and were stained with the recommended dilution of primary
antibodies against Notch1 (cat. no. 3447; dilution. 1:1,000),
cyclin D1 (cat. no. 2978; dilution, 1:1,500), MMP7 (cat. no. 3801;
dilution, 1:1,000), MMP9 (cat. no. 13667; dilution, 1:1,000) and
β-actin (cat. no. 4970; dilution, 1:1,500) at room temperature for
2 h (all from Cell Signaling Technology, Inc., Danvers, MA, USA).
Following washing, the blots were incubated with a 1:2,000 dilution
of goat-anti-rabbit (cat. no. 7074; Cell Signaling Technology Inc.)
or goat-anti-mouse (cat. no. 7076; Cell Signaling Technology Inc.)
immunoglobulin G antibodies conjugated to horseradish peroxidase at
room temperature for 1 h. The blots were developed with the
Enhanced Chemiluminescence Western Blot detection kit (Pierce;
Thermo Fisher Scientific).
Transmission electron microscopy
Cell pellets were fixed in 2% paraformaldehyde and
2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at
room temperature for 5 min. Firstly, 10 ml 1× PBS was added to a
Petri dish containing the cells to briefly wash them. Next, the
supernatant was discarded and 1 ml 1× fixative was added into the
dish at room temperature. Following this, cells were scraped and
transferred into a 1.5-ml Eppendorf tube. The cells were then
centrifuged at 500–800 × g at room temperature for 5 min. The
pellet was embedded in epoxy resin at 60°C for 48 h. Ultra-thin
sections (100 nm) were cut with an ultramicrotome and stained with
5% uranyl acetate in 50% ethanol, followed by 2% aqueous lead
citrate at 37°C for 30 min. Finally, the ultra-thin sections were
observed on a Philips CM12 Microscope (FEI Inc., Eindhoven,
Netherlands).
Statistical analysis
Results are reported from at least three different
experiments. Statistical analyses were performed with SPSS software
version 13.0 (SPSS, Inc., Chicago, IL, USA). For the comparisons
between 2 groups (cell cycle assay, cell invasion assay, wound
healing assay and dual luciferase reporter assay), Student's t-test
was used for analysis. For combination studies (miR-139-5P
expression data), one-way analysis of variance followed by
least-significant difference post-hoc test was used for analysis.
All data are reported as the mean ± standard error of the mean.
Results
miR-139 inhibits the proliferative
activity of PCa cells
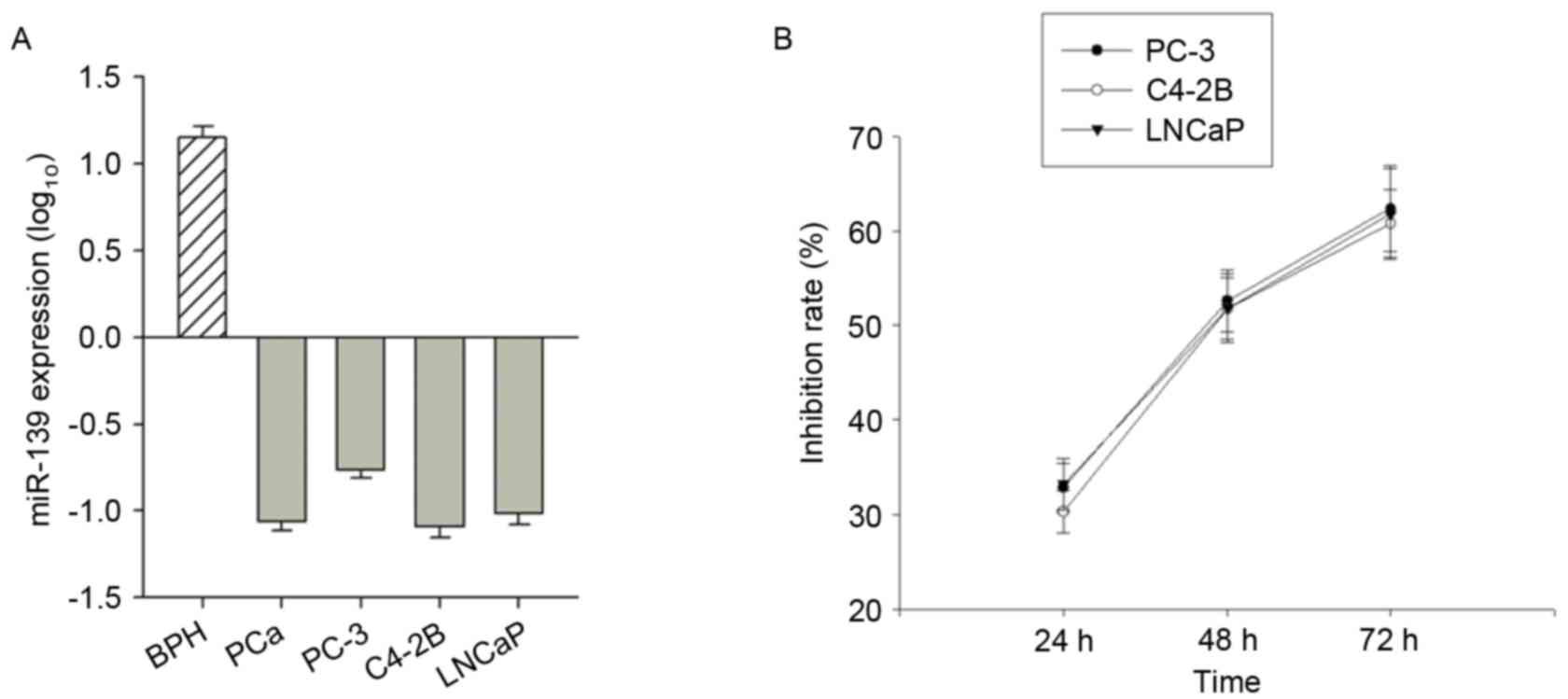
miR-139-5P expression data were detected for BPH
(n=10) and PCa (n=10) patient samples, and the PC-3, C4-2B and
LNCaP PCa cell lines. miR-139-5P expression was higher in BPH
tissue compared with the prostate cancer tissue, suggesting that
there was a significant decrease in the expression level of
miR-139-5P in PCa (P<0.001; Fig.
1A). Additionally, the expression of miR-139-5P in the three
PCa cell lines was lower than BPH tissue (Fig. 1A).
The MTT assay results revealed that the growth
inhibitory rate [(1-OD value of the observed group/OD value of the
control group) ×100%] of miR-139 transfected cells at 24, 48 and 72
h after transfection were 32.83±2.61, 52.58±3.29 and 62.36±4.55% in
PC-3 cells, respectively; 30.28±2.25, 51.74±3.27 and 60.80±3.58% in
C4-2B cells, respectively; and 33.20±2.67, 51.83±3.59, 61.79±4.85%
in LNCaP cells (Fig. 1B). The
inhibition rate of the three transfected PCa cell lines increased
over time, and it was concluded that miR-139 may inhibit PCa cell
proliferation.
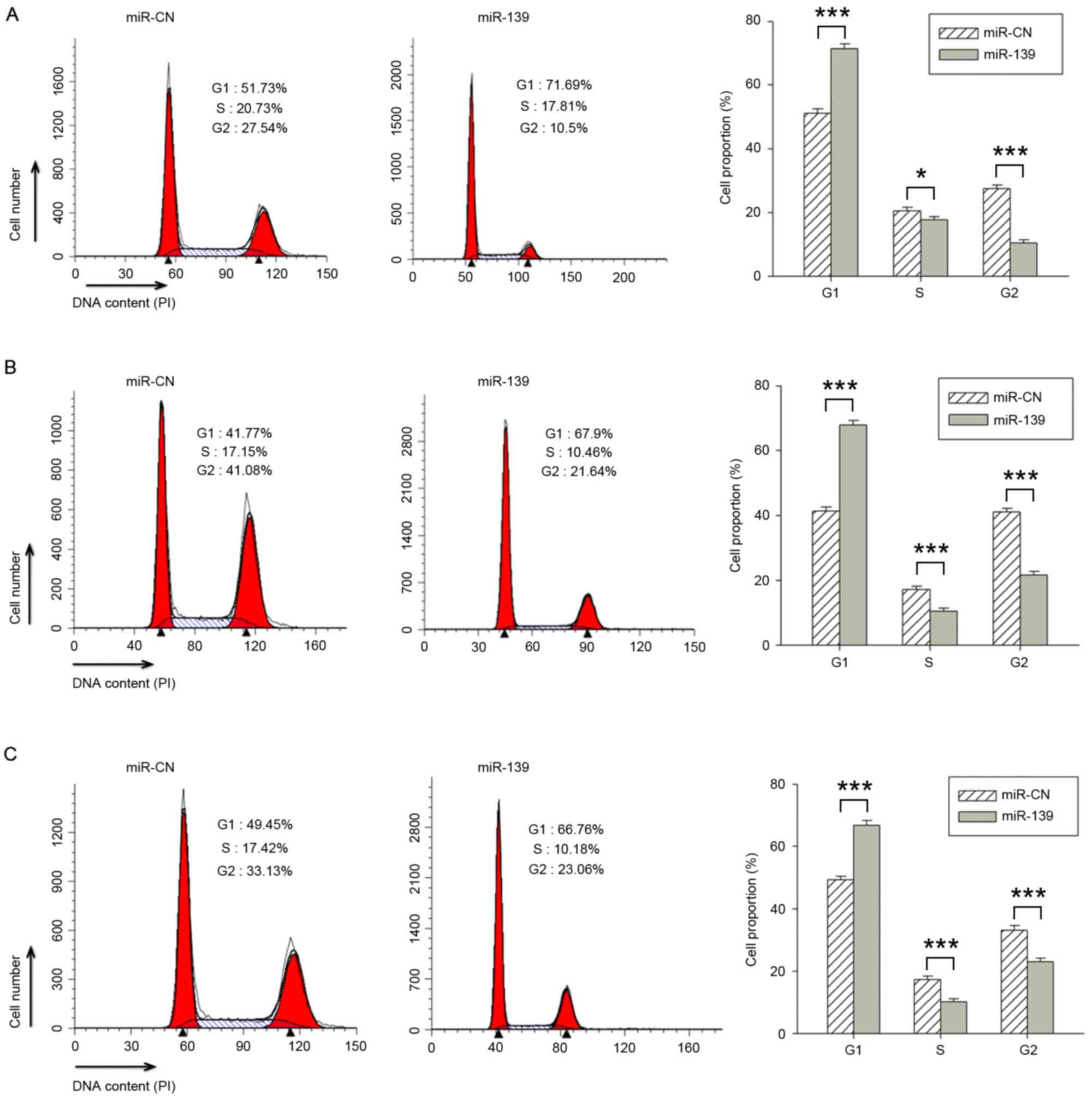
miR-139 induces G0/1-phase
cell cycle arrest in PCa cells
The three aforementioned PCa cell lines were
transfected with miR-139 or the miR-CN, and the cell cycle
distribution was assessed using flow cytometry. The distribution
results differed between transfected and control cells. The
proportion of miR-139 transfected cells in G1, S and
G2 phase were 71.38±1.52, 17.63±1.04 and 10.43±0.96%,
respectively, in PC-3 cells (Fig.
2A); 67.86±1.46, 10.45±0.98 and 21.62±1.08%, respectively, in
C4-2B cells (Fig. 2B); and
66.74±1.52, 10.18±0.93 and 23.04±1.08% in LNCaP cells (Fig. 2C), respectively. The cells transfected
with miR-139 exhibited a significantly increased percentage of
cells in the G1 phase, and a decreased percentage in the
S and G2 phases, compared with cells transfected with
miR-CN (P<0.001). It was therefore concluded that miR-139
inhibited the proliferation of PCa cells by inducing
G0/1-phase cell cycle arrest.
Effect of miR-139 on PCa cell
apoptosis
The early apoptosis rates as determined by flow
cytometry at 48 h after the transfection of the three PCa cell
lines with miR-139 were 5.48% in PC-3 cells, 4.76% in C4-2B cells
and 6.40% in LNCaP cells (data not shown). Compared with the
control groups, the early apoptosis rates exhibited no significant
difference. Therefore, miR-139 may have no effect on PCa cell
apoptosis.
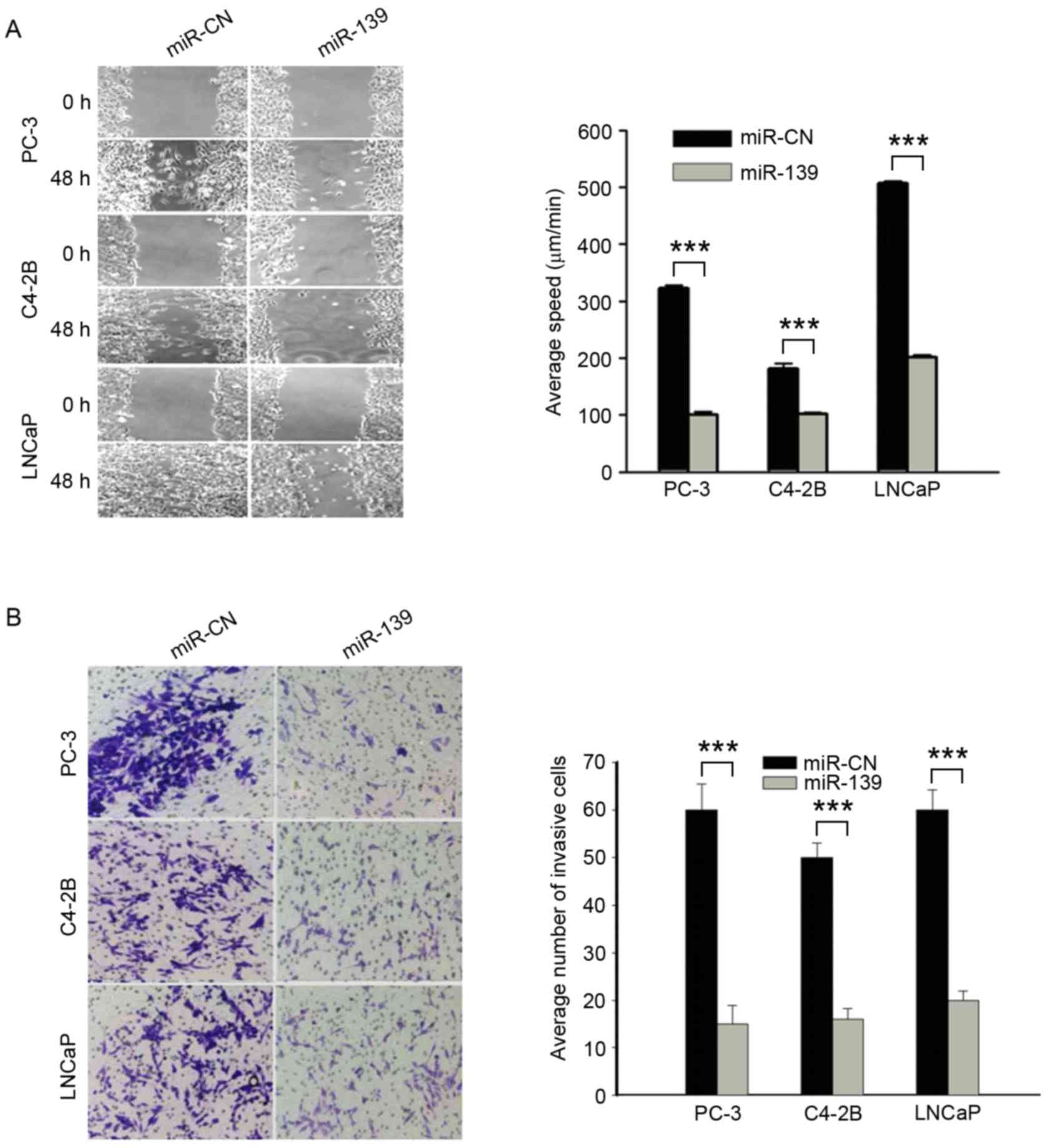
miR-139 reduces cell migration and
invasiveness
The three PCa cell lines were transfected with
miR-139 or the miR-CN, and the degree of cell migration was
measured after 48 h. Wound healing was only observed in the miR-CN
group. The mean migration speeds of the negative control groups
were 322.47±5.01, 181.33±8.79 and 506.72±4.22 µm/min in the PC-3,
C4-2B and LNCaP cell lines, respectively; those of the
miR-139-transfected cells were 100.5±4.77, 102.2±2.86 and
201.2±4.58 µm/min in PC-3, C4-2B and LNCaP cell lines,
respectively. The migration speed was significantly lower in the
groups transfected with miR-139 compared with the control groups
(P<0.001; Fig. 3A). Similarly,
cell invasiveness for each cell line following transfection with
miR-139 or the negative control was also observed using a Matrigel
transwell assay. The results revealed a significant difference
between the miR-139 and miR-NC groups (P<0.001; Fig. 3B). Consequently, it was concluded that
cell migration and invasion were reduced by transfection with
miR-139.
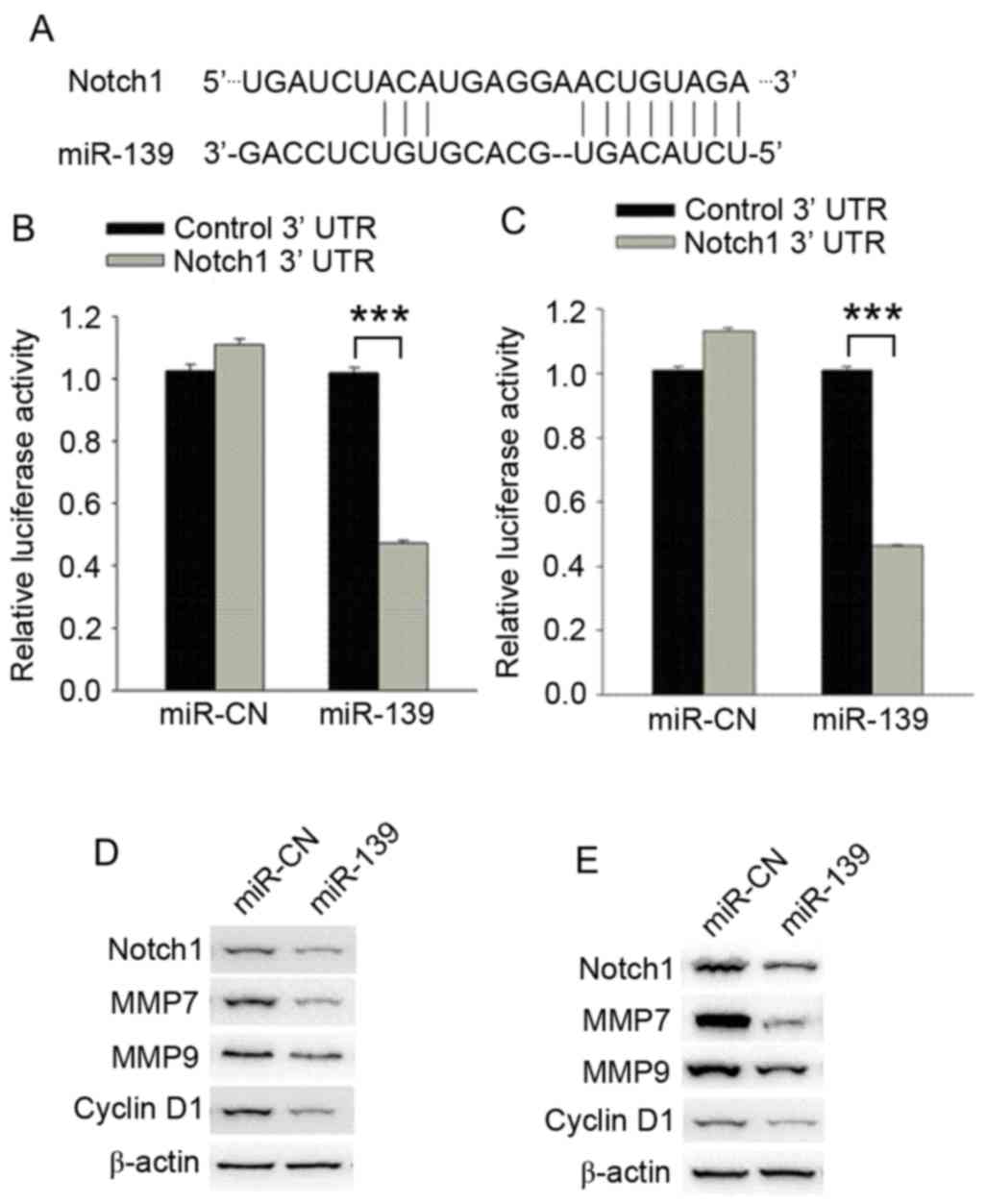
miR-139 decreases the expression of
the target genes Notch1 and cyclin D1
Complementary base pairing was found between the
Notch1 3′UTR and miR-139, as discovered by computational analysis
(Fig. 4A), and a dual luciferase
reporter assay revealed that luciferase activity was significantly
lower in C4-2B and PC-3 cells when co-transfected with a Notch1
wild-type 3′UTR and miR-139 compared with transfection with a
mutant Notch1 3′UTR and miR-139 (P<0.001; Fig. 4B and C). These results indicated that
miR-139 was bound to the Notch1 3′UTR sequence. In addition,
western blot analysis revealed that the levels of Notch1, matrix
metalloproteinase (MMP)7, MMP9 and cyclin D1 in miR-139-transfected
cells were decreased compared with those in the negative control
group (Fig. 4D and E).
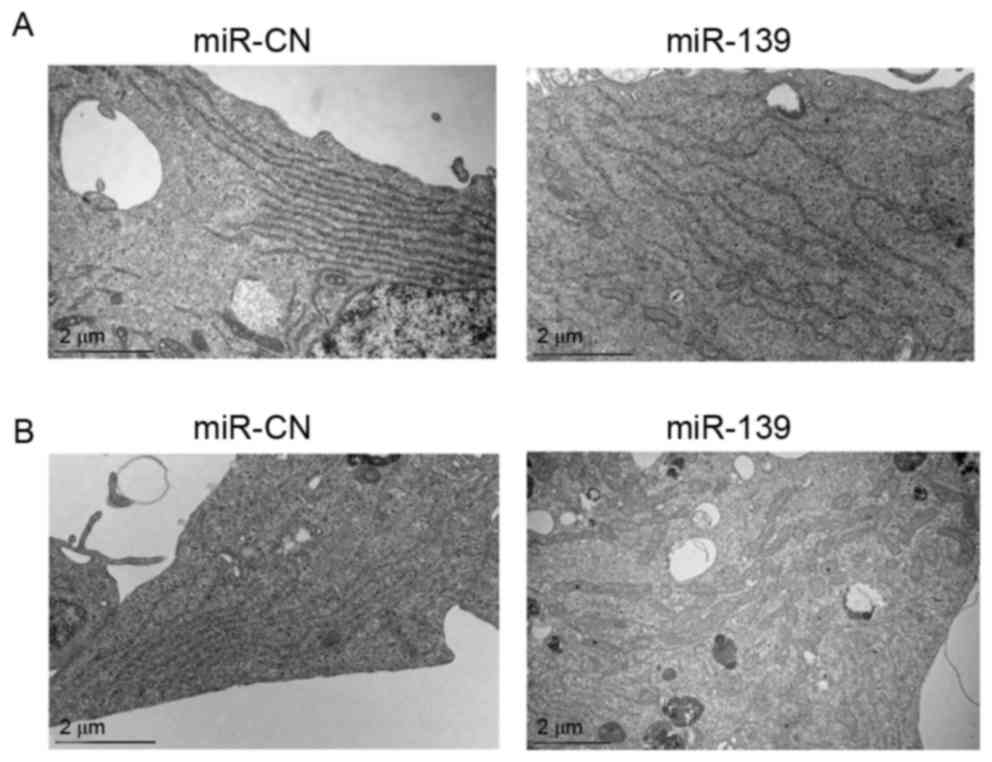
Ultrastructural observation by transmission electron microscopy
revealed the degranulation of the rough endoplasmic reticulum (ER)
and mitochondrial swelling in PC-3 and C4-2B cells transfected with
miR-139 (Fig. 5A and B).
Discussion
The majority of early-stage PCa tumors are
hormone-dependent, so androgen deprivation therapy is the
conventional treatment. However, >50% of patients will
experience recurrence and metastasis or develop
androgen-independent disease. The failure of endocrine therapy or
the development of chemotherapy resistance leads to a reduction in
quality of life and a decrease in survival time for the affected
patients (3). A number of previous
studies have demonstrated that miRNA dysregulation is a common
event in tumor tissues. miRNAs are highly conserved,
non-protein-coding, small, single-stranded RNAs that widely occur
in eukaryotic genomes. Genetic variants in miRNA genes may alter
miRNA expression and function, affecting human disease (12,13).
Previous studies have revealed that miRNAs participate in a number
of physiological and pathological processes by degrading target
mRNAs. For example, in tumorigenesis and cancer development,
different miRNAs can either promote or inhibit the expression of
oncogenic genes (5,14). In addition to functioning as
biomarkers, miRNAs may become direct targets for cancer treatments,
potentially providing a novel direction for malignant tumor
diagnosis, treatment and prognosis. Previous miRNA microarray
analyses have revealed that miR-139 expression is downregulated in
gastric cancer (15), colon cancer
(16), breast cancer (17), liver cancer (18) and glioma (19), evidence of the association between
reduced miR-139 expression and tumorigenesis; however, the
expression of miR-139-5p in PCa remains unclear. Therefore, the
present study aimed to investigate expression and biological
functions of miR-139 in prostate cancer cells.
In the present study, the analysis of the cell cycle
of cells transfected with miR-139 revealed a significantly
increased percentage of cells in the G1 phase and a
decreased percentage of cells in the S and G2 phases.
miR-139 appeared to inhibit the proliferation of PCa cells by
inducing G0/1-phase cell cycle arrest. Cell growth
experiments in vitro confirmed that miR-139 significantly
inhibited PCa cell proliferation. A previous study also revealed
that miR-139 inhibited cell proliferation and induction of
G0/1 arrest in colorectal cancer (9). Previous studies suggested that the
expression of miR-139 induced apoptosis in colorectal cancer and
glioma cells (9,18). By contrast, the results of the present
study demonstrated that miR-139 had no effect on apoptosis in all
three PCa cell lines tested.
Mechanistic investigations by Zhang et al
(9) and Song et al (20) revealed that miR-139 suppresses
colorectal cancer proliferation by targeting Notch1 mRNA. Another
previous study revealed that Notch signaling serves complicated
functions in PCa (21). The
luciferase reporter assay conducted in the present study revealed
that luciferase activity was significantly decreased in C4-2B and
PC-3 cells co-transfected with the wild-type Notch1 3′UTR and
miR-139. The results indicated that miR-139 bound Notch1 directly
in PCa cells. In addition, western blot analysis revealed that the
levels of Notch1 and cyclin D1 protein in miR-139 transfected cells
were markedly lower. As cyclin D1 has been demonstrated as a direct
target of Notch1 in breast cancer (22), we hypothesized that miR-139 also
targeted Notch1 and regulated the expression of cyclin D1 in PCa.
MMP7 and MMP9 are involved in wound healing and tumor malignancy
(23,24), so the decreased levels of MMP7 and
MMP9 in miR-139-transfected cells supported the conclusion that
transfection with miR-139 reduced cell migration and
malignancy.
A previous study suggested that the
mitochondria-associated ER membrane functions as a platform for
various intracellular stress responses, including apoptotic
signaling, inflammatory signaling, the autophagic response and the
unfolded protein response, and dysregulation of these signaling
pathways may be associated with cancer cell metabolism (25). ER-associated protein degradation may
act as a key regulatory factor that decides cell fate in breast
cancer (26). The present study
observed rough ER degranulation and mitochondrial swelling in
miR-139-transfected PCa cells, although the mechanism by which this
occurred is unknown. The ultrastructural changes may be associated
with protein interactions between Notch1 and cyclin D1.
Evidence suggests that other miRNAs also serve an
important function in PCa. Cohort research has suggested that it is
possible to use the measurement of 14 miRNAs as a combined ‘miR
Score’ to identify low-risk aggressive PCa (27). For instance, the decreased expression
of miRNA-128 in the serum and PCa tissue may be associated with the
malignant progression of tumors and a decreased recurrence-free
survival rate (28). miRNA-195
suppresses tumor cell proliferation and metastasis by directly
modulating the expression of breast cancer-overexpressed gene 1
(29), while suppressing cell
migration and invasion by targeting FOS-like 1 expression in PCa
(30). By contrast, miRNA-556-5p
functions as an onco-miRNA and promotes prostate cancer cell growth
by suppressing protein phosphatase 2 regulatory subunit B-α
(PPP2R2A) expression. Previous experimental data have demonstrated
that the ectopic expression of miRNA-556-5p results in the
downregulation of PPP2R2A protein, which in turn results in the
downregulation of cyclin dependent kinase inhibitor 1B and the
upregulation of cyclin D1 (31). The
molecular interaction networks between different miRNAs, their
respective target proteins and the complete cancer-associated
mechanisms underlying the effect of miRNAs remain to be
clarified.
In summary, to the best of our knowledge, the
present study revealed for the first time that miR-139 reduces
cyclin D1 expression and inhibits cell proliferation through
targeting Notch1 in PCa. Furthermore, MMP7 and MMP9 expression was
downregulated in miR-139-transfected PCa cells. These data
suggested that this pathway may be a potential therapeutic target
for PCa treatment.
Acknowledgements
The authors would like to thank Dr Qi-Lin Ao (Tongji
Medical College, Huazhong University of Science and Technology,
Wuhan, China) for reviewing histology data.
Funding
The present study was supported by the National
Science Foundation of China (grant nos. 81772775, 81472783,
81630060, 81572571 and 81372801).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QS and JW conceived and designed the study. QS, DW,
DL, PW, GC performed the literature search, data extraction and
statistical analysis and drafted the paper. KL, SL, XB, CF
performed the experiments. JW supervised the literature search,
data extraction and analysis, and reviewed the paper. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Tissue samples were obtained and handled in
accordance with a protocol approved by the Institutional Review
Board for Human Research of Tongji Hospital (Wuhan, China). Written
informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wei J, Wang Z, Makarov D and Li X: Current
treatments and novel therapeutic targets for castration resistant
prostate cancer with bone metastasis. Am J Clin Exp Urol. 1:30–38.
2013.PubMed/NCBI
|
|
4
|
Kelly BD, Miller N, Sweeney KJ, Durkan GC,
Rogers E, Walsh K and Kerin MJ: A circulating MicroRNA signature as
a biomarker for prostate cancer in a high risk group. J Clin Med.
4:1369–1379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang HD, Jiang LH, Sun DW, Li J and Tang
JH: MiR-139-5p: Promising biomarker for cancer. Tumour Biol.
36:1355–1365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu W, Hang M, Yuan CY, Wu FL, Chen SB and
Xue K: MicroRNA-139-5p inhibits cell proliferation and invasion by
targeting insulin-like growth factor 1 receptor in human non-small
cell lung cancer. Int J Clin Exp Pathol. 8:3864–3870.
2015.PubMed/NCBI
|
|
9
|
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu
CW, Wang K, Zheng S, Ng SS, Chan FK, et al: microRNA-139-5p exerts
tumor suppressor function by targeting NOTCH1 in colorectal cancer.
Mol Cancer. 13:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang HD, Sun DW, Mao L, Zhang J, Jiang
LH, Li J, Wu Y, Ji H, Chen W, Wang J, et al: MiR-139-5p inhibits
the biological function of breast cancer cells by targeting Notch1
and mediates chemosensitivity to docetaxel. Biochem Biophys Res
Commun. 465:702–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cammaerts S, Strazisar M, de Rijk P and
Del Favero J: Genetic variants in microRNA genes: Impact on
microRNA expression, function, and disease. Front Genet. 6:1862015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Du WD, Chen G, Ruan J, Xu S, Zhou
FS, Zuo XB, Lv ZJ and Zhang XJ: Association analysis of genetic
variants in microRNA networks and gastric cancer risk in a Chinese
Han population. J Cancer Res Clin Oncol. 138:939–945. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao
X and Liang S: Roles of microRNA on cancer cell metabolism. J
Transl Med. 10:2282012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo
ZY, Zhao J, Meng YL, Ren XL, Wang T, et al: HER2 interacts with
CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139
in gastric cancer cells. Gastroenterology. 141:2076–2087.e6. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D,
Le V, Yin P, Ni L and Liu J: Post-transcriptional regulation of the
tumor suppressor miR-139-5p and a network of miR-139-5p-mediated
mRNA interactions in colorectal cancer. FEBS J. 281:3609–3624.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li T, Yin J, Yuan L, Wang S, Yang L, Du X
and Lu J: Downregulation of microRNA-139 is associated with
hepatocellular carcinoma risk and short-term survival. Oncol Rep.
31:1699–1706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li RY, Chen LC, Zhang HY, Du WZ, Feng Y,
Wang HB, Wen JQ, Liu X, Li XF, Sun Y, et al: MiR-139 inhibits Mcl-1
expression and potentiates TMZ-induced apoptosis in glioma. CNS
Neurosci Ther. 19:477–483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song M, Yin Y, Zhang J, Zhang B, Bian Z,
Quan C, Zhou L, Hu Y, Wang Q, Ni S, et al: MiR-139-5p inhibits
migration and invasion of colorectal cancer by downregulating AMFR
and NOTCH1. Protein Cell. 5:851–861. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng G, Ma L, Meng Q, Ju X, Jiang K, Jiang
P and Yu Z: Notch signaling in the prostate: Critical roles during
development and in the hallmarks of prostate cancer biology. J
Cancer Res Clin Oncol. 142:531–547. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cohen B, Shimizu M, Izrailit J, Ng NF,
Buchman Y, Pan JG, Dering J and Reedijk M: Cyclin D1 is a direct
target of JAG1-mediated Notch signaling in breast cancer. Breast
Cancer Res Treat. 123:113–124. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsu TI, Lin SC, Lu PS, Chang WC, Hung CY,
Yeh YM, Su WC, Liao PC and Hung JJ: MMP7-mediated cleavage of
nucleolin at Asp255 induces MMP9 expression to promote tumor
malignancy. Oncogene. 34:826–837. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong VW, Garg RK, Sorkin M, Rustad KC,
Akaishi S, Levi K, Nelson ER, Tran M, Rennert R, Liu W, et al: Loss
of keratinocyte focal adhesion kinase stimulates dermal proteolysis
through upregulation of MMP9 in wound healing. Ann Surg.
260:1138–1146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kato H and Nishitoh H: Stress responses
from the endoplasmic reticulum in cancer. Front Oncol. 5:932015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fan P, Cunliffe HE, Maximov PY, Agboke FA,
McDaniel RE, Zou X, Ramos P, Russell ML and Jordan VC: Integration
of downstream signals of insulin-like growth factor-1 receptor by
endoplasmic reticulum stress for estrogen-induced growth or
apoptosis in breast cancer cells. Mol Cancer Res. 13:1367–1376.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mihelich BL, Maranville JC, Nolley R,
Peehl DM and Nonn L: Elevated serum microRNA levels associate with
absence of high-grade prostate cancer in a retrospective cohort.
PLoS One. 10:e01242452015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun X, Yang Z, Zhang Y, He J, Wang F, Su
P, Han J, Song Z and Fei Y: Prognostic implications of tissue and
serum levels of microRNA-128 in human prostate cancer. Int J Clin
Exp Pathol. 8:8394–8401. 2015.PubMed/NCBI
|
|
29
|
Guo J, Wang M and Liu X: MicroRNA-195
suppresses tumor cell proliferation and metastasis by directly
targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res.
34:912015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y,
Liu Y, Li S, Liang Z, Xu X, et al: MicroRNA-195-5p, a new regulator
of Fra-1, suppresses the migration and invasion of prostate cancer
cells. J Transl Med. 13:2892015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao W, Cao L, Zeng S, Qin H and Men T:
Upregulation of miR-556-5p promoted prostate cancer cell
proliferation by suppressing PPP2R2A expression. Biomed
Pharmacother. 75:142–147. 2015. View Article : Google Scholar : PubMed/NCBI
|