Introduction
Uterine cancer is the second most common malignancy
in females and the incidence of this disease is only lower than
that of breast cancer (1). According
to a report released by the International Agency for Research on
Cancer, >300,000 women die from uterine cancer each year, and
>80% of them are from developing countries (2). In recent years, the incidence of uterine
cancer shows an increasing trend in developing countries,
especially in young women, incidence of this disease increases by
3% every year (3). So far, the
mechanism of uterine cancer has not been completely elucidated.
Therefore, the development of new biomarkers and effective
treatment targets is the key to further improve the prognosis of
uterine cancer. miRNAs are a class of highly conserved endogenous
regulatory RNA molecules with a length of 18–26 bp. miRNAs bind to
mRNA molecules transcribed from target genes and inhibit the its
translation, ultimately regulating the expression of target genes
(3,4).
Many miRNAs, such as miR-16, miR-155 and miR-223, were upregulated
in uterine cancer tissues (5),
expression of some other miRNAs, such as miR-143 and miR-145,
decreased in uterine cancer tissues (6), indicating that miRNAs may be key players
in uterine cancer. miR-222 is highly expressed in a variety of
tumors, including breast (7), bladder
(8), colon (9), glioblastoma (10), and pancreatic cancer (11). However, the role of miR-222 in uterine
cancer is still unclear. Therefore, in this study, the expression
of miR-222 in uterine cancer and adjacent tissues was detected by
stem-loop RT-PCR to investigate the relationship between miR-222
expression and clinicopathological features of this disease, so as
to explore the role of miR-222 in the occurrence and development of
uterine cancer and its clinical significance.
Patients and methods
Patients
Experimental data
A total of 66 patients with uterine cancer diagnosed
by gynecological and pathological examination in Dongying People's
Hospital (Dongying, China) were enrolled from March 2014 to October
2016. During surgical resection, uterine cancer and adjacent
tissues were collected. Ages of patients ranged from 33 to 75
years. Among those patients, 27 patients had tumors <4 cm in
diameter and the remaining patients all had a tumor diameter ≥4 cm.
No patients received radiotherapy or chemotherapy before admission.
Uterine cancer histopathological types: squamous cell carcinoma in
53 cases, adenocarcinoma in 9 cases, adenosquamous carcinoma in 4
cases. Uterine cancer tissue differentiation degree: 9 cases in
grade I, 19 cases in grade II and 38 cases in grade III. FIGO
stage: 6 cases in stage I, 25 cases in stage II, 18 cases in stage
III and 17 cases in stage IV. All patients or their families signed
informed consent. This study was approved by the Ethics Committee
of Dongying People's Hospital (Dongying, China).
Reagents and instruments
Tissue total RNA extraction kit (BioLabs Technology
Co., Ltd., Beijing, China), miRNA reverse transcription kit
(GeneCopoeia, Inc., Rockville, MD, USA), fluorescence quantitative
PCR kit SYBR-Green (Huaxia Yuanyang Technology Co., Ltd., Beijing,
China), urea (Healthdream Biological Technology Co., Ltd., Wuhan,
China), ABI 7500 fluorescence quantitative PCR instrument (Thermo
Fisher Scientific, Inc., Waltham, MA, USA), quantitative
spectrophotometer (Jiang Xue Technology Co., Ltd., Chongqing,
China).
Method
miR-222 expression analysis
Total RNA was extracted according to the
manufacturer's instructions, and A260/A280 ratio was meaured using
a quantitative spectrophotometer. Total RNA was reverse transcribed
into cDNA in accordance with the instructions of miRNA reverse
transcription kit. Reverse transcription system: 1 µg total RNA, 50
µM stem-loop RT primer, 2 U RNase inhibitor, 5 U M-MLV reverse
transcriptase, and 0.5 µM dNTPs. Reaction conditions: 16°C for 30
min, 42°C for 30 min and 75°C for 15 min. All cDNA samples were
stored at −20°C. This experiment was performed 3 times (Table I).
 | Table I.Primers used in stem-loop RT-PCR. |
Table I.
Primers used in stem-loop RT-PCR.
| Primer | Sequence |
|---|
| miR-222 |
|
| RT stem-loop
primer |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCAGTA-3′ |
| Forward primer |
5′-GTTCGTFFFAFCTACATCTGGC-3′ |
| let-7a |
|
| RT stem-loop
primer |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTATAC-3′ |
| Forward primer |
5′-ATGGTTCGTGGGTGAGGTAGTAGGTTGT-3′ |
| Common reverse
primer |
5′-GTGTCGTGGAGTCGGCAATTC-3′ |
RT-qPCR reaction system (15 µl): 1 µl cDNA, 1X
SYBR-Green I Mastermix, 0.5 µM miRNA specific forward primer, 0.5
µM universal reverse primer. Reaction conditions: 94°C for 10 min,
followed by 40 cycles of 95°C for 15 sec, 60°C for 60 sec, and 72°C
for 30 sec. With let-7a as endogenous control (12), the relative expression of miRNA was
calculated automatically by RT-PCR instrument.
Analysis of the specificity of miR-222
stem-loop RT-PCR
A total of 5 µg total RNA was used to isolate low
molecular weight RNAs. After denaturing by 15% urea solution, miRNA
precursor and mature miRNA were then separated by 8% PAGE.
Stem-loop RT-PCR was then performed to detect miR-222. Reaction
volume was 20 µl. First, 2 µg of total RNA, 1 µl of RT primer
(stem-loop primer) and 1 µl of dNTPs were mixed, and DEPC water was
added to make a volume of 14 µl. After denaturation at 65°C for 5
min, in the produced was kept on ice. Then 1 µl of reverse
transcriptase AMV, 4 µl of buffer and 1 µl of RNase inhibitor were
added. Reaction conditions were: 16°C for 30 min, followed by 60
cycles of 20°C for 30 sec, 42°C for 30 sec and 50°C for 1 sec, and
85°C for 10 min.
Statistical analysis
Data were analyzed using SPSS 19.0 software (Beijing
Xinmei Jiahong Technology Co., Ltd., Beijing, China). Measurement
data were expressed as mean ± standard deviation, ANOVA was used
for multiple group comparisons and the post hoc test was Dunnetts
test. Survival analysis was performed using GraphPad Prism 5 with
α=0.05 as standard.
Results
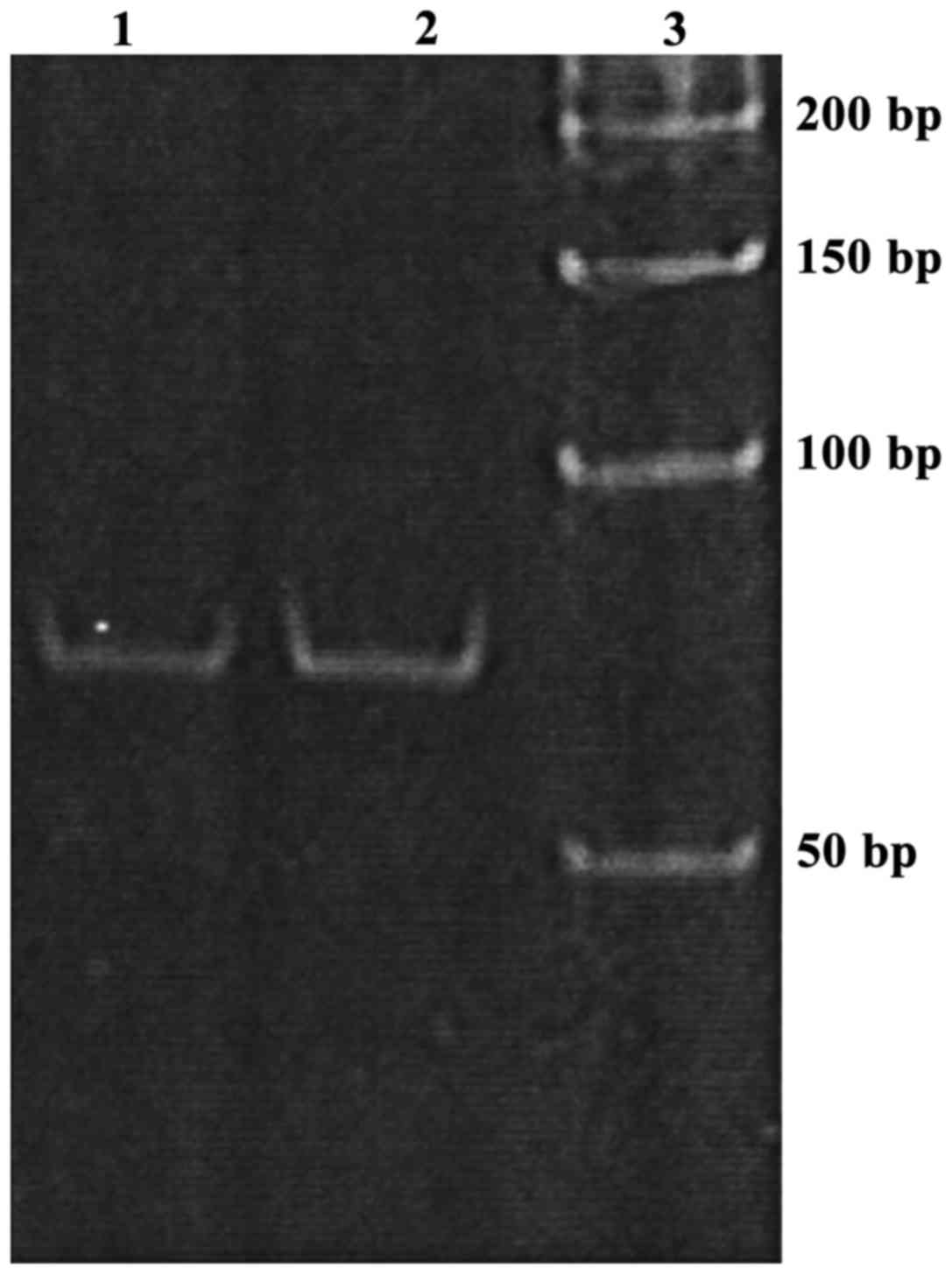
Establishment of stem-loop RT-PCR
detection method
The product of miRNA amplification by stem-loop
RT-PCR showed a single band after 8% PAGE. So stem-loop RT-PCR
method can specifically amplify miR-222 (Fig. 1).
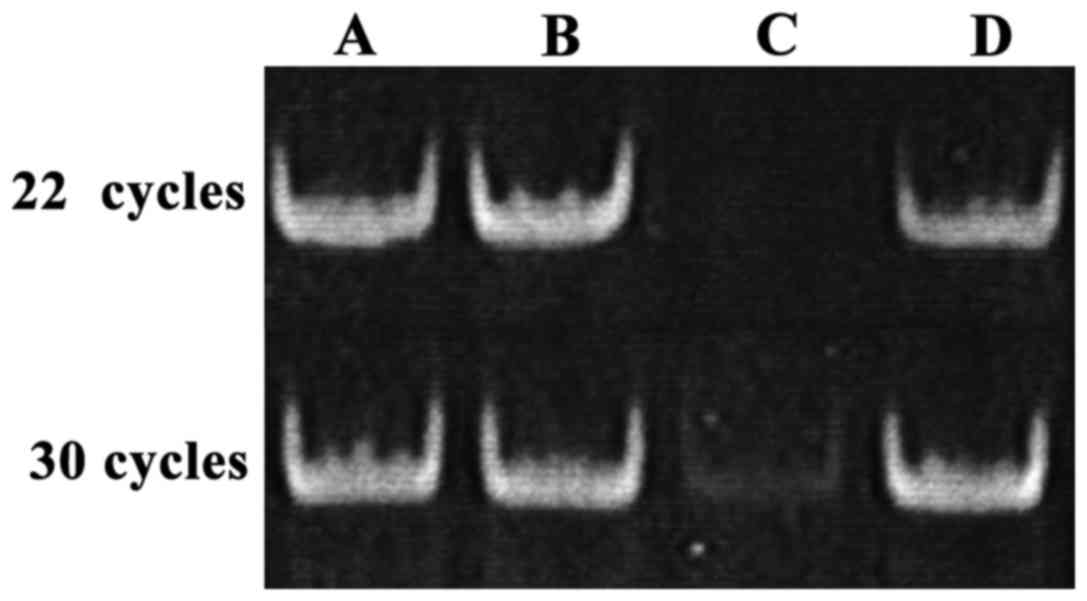
As shown in Fig. 2,
after 8% PAGE, gray values of the bands derived from PCR reaction
with 22 or 30 cycles were similar. PCR with 30 cycles provided a
faint band, while PCR with 22 cycles failed to show this band. We
speculated that low RNA recovery rate is the reason why
amplification efficiency of mature miRNA was lower than total RNA
amplification efficiency. Small RNA recovery efficiency was only
50%.
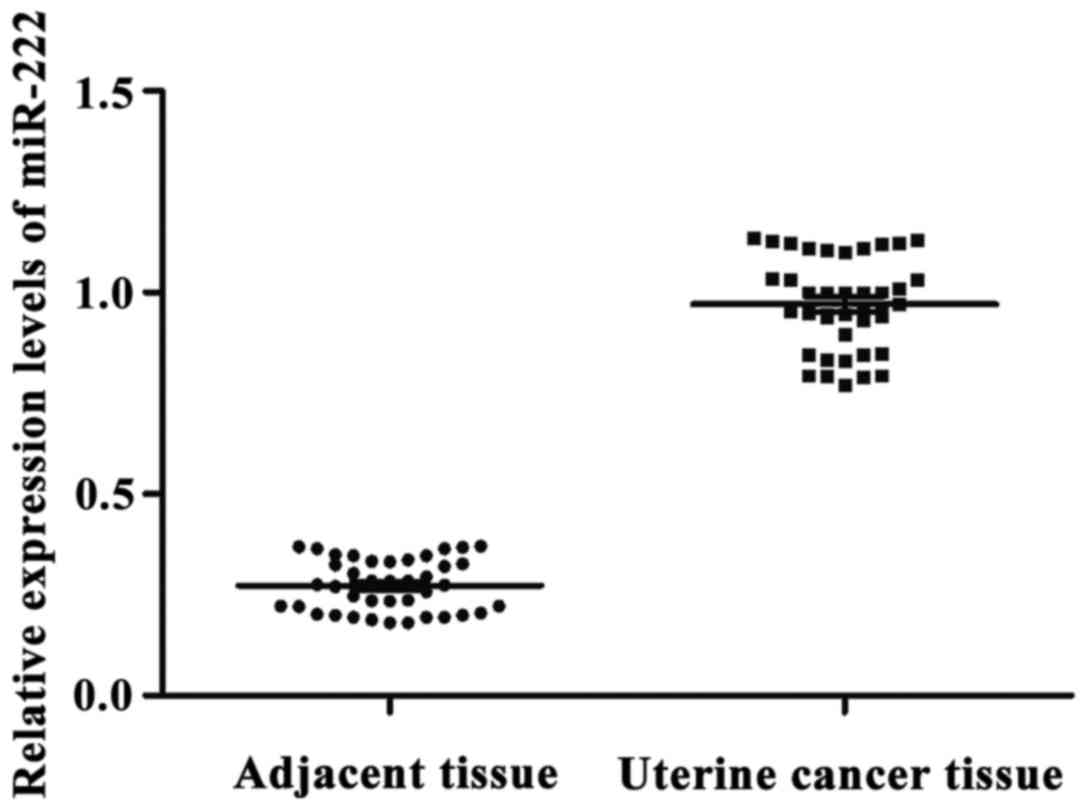
miR-222 expression in uterine cancer
and adjacent tissues
With let-7a as endogenous control, the relative
expression levels of miR-222 in adjacent and uterine cancer tissues
were 0.275±0.094 and 0.953±0.183, respectively. The expression
level of miR-222 in adjacent tissues was significantly lower than
that in uterine cancer tissues. The difference was statistically
significant (p<0.05) (Fig. 3).
Correlation between miR-222 expression
and a variety of clinicopathological features of uterine
cancer
The expression level of miR-222 was different in
uterus cancer tissues with the different differentiation degrees.
The expression level of miR-222 in grade III uterine cancer is
obviously higher than that in grade I and II. The expression level
of miR-222 was increased with the increased degree of malignancy.
There was no significant correlation between miR-222 expression and
age, tumor size, gross type, pathological type and FIGO stage
(p>0.05) (Table II).
 | Table II.Correlation between miR-222 expression
and a variety of clinicopathological features of uterine
cancer. |
Table II.
Correlation between miR-222 expression
and a variety of clinicopathological features of uterine
cancer.
| Clinicopathological
features | Cases | miR-222
expression | P-value |
|---|
| Age (years) |
|
|
|
|
<40 | 17 | 0.394±0.074 | 0.916 |
|
40–60 | 21 | 0.371±0.068 |
|
|
>60 | 28 | 0.474±0.085 |
|
| Tumor size (cm) |
|
|
|
|
<4 | 27 | 0.527±0.093 | 0.617 |
| ≥4 | 39 | 0.416±0.078 |
|
| Gross type |
|
|
|
| Exogenous
type | 32 | 0.371±0.064 | 0.563 |
|
Endogenous type | 19 | 0.398±0.069 |
|
| Ulcer
type | 11 | 0.474±0.086 |
|
| Neck
type | 4 | 0.973±0.184 |
|
| Tissue
differentiation |
|
|
|
| Large
cell keratinizing (I) | 9 | 0.214±0.047 | 0.001 |
|
Non-Keratinizing type
(II) | 19 | 0.289±0.051 |
|
| Small
cell type (III) | 38 | 0.656±0.117 |
|
| Pathological
type |
|
|
|
| Squamous
cell carcinoma | 53 | 0.427±0.082 | 0.628 |
|
Adenocarcinoma | 9 | 0.416±0.079 |
|
|
Adenosquamous carcinoma | 4 | 0.398±0.077 |
|
| FIGO stage |
|
|
|
| Stage
I | 6 | 0.234±0.049 | 0.284 |
| Stage
II | 25 | 0.425±0.082 |
|
| Stage
III | 18 | 0.419±0.084 |
|
| Stage
IV | 17 | 0.427±0.087 |
|
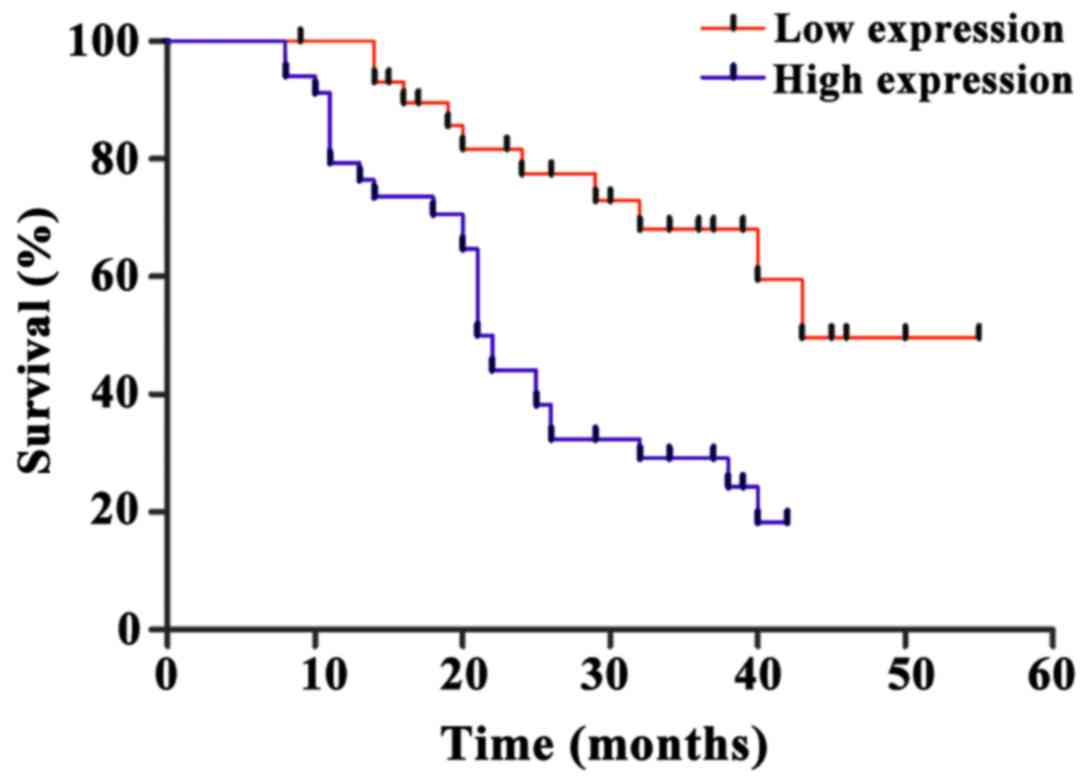
Correlation between miR-222 expression
and prognosis
Follow-up study of 66 uterine cancer patients showed
that the median survival time of miR-222 high expression uterine
cancer patients was 21.4±0.8 months, while the median survival time
of miR-222 low expression uterine cancer patients was 44.9±1.7
months. The expression level of miR-222 was significantly
negatively correlated with the survival of uterine cancer patients
(p<0.05) (Fig. 4).
Discussion
Uterine cancer is the second most common malignancy
in women, and its incidence is only lower than that of breast
cancer (13). However, pathogenesis
of uterine cancer has not been fully understood, leading to poor
treatment outcomes as well as high recurrence and mortality rate
(14). As a class of highly conserved
endogenous regulatory RNAs, miRNAs bind to mRNAs transcribed from
target genes to inhibit further translation, thereby regulating the
expression of target genes and participating in a series of
physiological and pathological processes (15). miRNA can play a role as oncogene or
tumor suppressor gene in the occurrence and development of a tumor
(16).
Analysis of the expression pattern of miRNAs in
invasive cervical squamous cell carcinoma showed that most miRNAs
were upregulated and only two were downregulated (16). The expression level of miR-127 is
significantly correlated with lymph node metastasis (17). Some scholars found that in colon
cancer tissues, miR-143 and miR-145 expression were significantly
downregulated (18,19). In addition, abnormal expression of a
large number of miRNAs in solid tumors was observed, and most
miRNAs, such as miR-21, miR-92 and miR-155, were usually
upregulated (20,21).
In recent years, studies have shown that the
expression level of miR-222 is increased to varying degrees in
tumor tissue or tumor cell lines of different types of cancer
(22,23). miR-222 is involved in the occurrence
and development of a variety of cancers, such as bladder cancer
(24), prostate cancer (25), and melanoma (26). Among these cancers, target genes are
usually tumor suppressor genes or pre-apoptotic factors. miR-222
inhibits cell apoptosis and participates in the occurrence and
development of tumor through the regulation of expression of its
target genes (23). Results of this
study showed that the stem-loop RT-PCR can specifically amplify
miRNA. miR-222 was significantly upregulated in uterine cancer
tissue, suggesting that miR-222 may be involved in the occurrence
and development of uterine cancer. It has been reported that
miR-222 can regulate the expression of PTEN to affect the
radiosensitivity, cell growth and invasion of gastric cancer
SGC7901 cells. When miR-222 was downregulated, the malignant
behavior of SGC7901 cells was significantly inhibited, suggesting
that miR-222 may play a role as oncogene in gastric cancer
(27). Different expression levels of
miRNA were observed in uterus cancer tissues with the different
differentiation degrees. The expression level of miR-222 in grade
III uterine cancer was obviously higher than that in grade I and
II. The expression level of miR-222 was increased with the
increased degree of malignancy. Therefore, we speculate that
miR-222 expression upregulation in uterine cancer may silence
downstream tumor suppressor genes such as p27 (28), p57 (29)
and Bmf (30). In addition, the
expression level of miR-222 was negatively correlated with the
survival time of patients with uterine cancer, suggesting that the
expression of miR-222 in uterine cancer may serve as an indicator
for the prognosis of this disease.
In conclusion, upregulated expression of miR-222 in
uterine cancer may play an important role in the occurrence and
development of uterine cancer. Our future studies will focus on the
function of target genes of miR-222 to identify new targets for the
treatment of uterine cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XN conceived and designed the study, and drafted the
report. XN and HT collected, analyzed and interpreted the
experimental data, and revised the manuscript critically for
important intellectual content. Both authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Dongying People's Hospital (Dongying, China). Signed written
informed consents were obtained from the patients or the
guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kowalewska M, Bakula-Zalewska E,
Chechlinska M, Goryca K, Nasierowska-Guttmejer A, Danska-Bidzinska
A and Bidzinski M: MicroRNAs in uterine sarcomas and mixed
epithelial-mesenchymal uterine tumors: A preliminary report. Tumour
Biol. 34:2153–2160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peralta-Zaragoza O, Deas J, Meneses-Acosta
A, de la O-Gómez F, Fernández-Tilapa G, Gómez-Cerón C,
Benítez-Boijseauneau O, Burguete-García A, Torres-Poveda K,
Bermúdez-Morales VH, et al: Relevance of miR-21 in regulation of
tumor suppressor gene PTEN in human cervical cancer cells. BMC
Cancer. 16:2152016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang H, Shuang D, Yi Z, Sheng H and Liu Y:
Up-regulated microRNA-155 expression is associated with poor
prognosis in cervical cancer patients. Biomed Pharmacother.
83:64–69. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pang H and Yue X: MiR-205 serves as a
prognostic factor and suppresses proliferation and invasion by
targeting insulin-like growth factor receptor 1 in human cervical
cancer. Tumour Biol. 39:10104283177013082017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun Y, Zhang B, Cheng J, Wu Y, Xing F,
Wang Y, Wang Q and Qiu J: MicroRNA-222 promotes the proliferation
and migration of cervical cancer cells. Clin Invest Med.
37:E1312014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu P, Sun M, Jiang W, Zhao J, Liang C and
Zhang H: Identification of targets of miRNA-221 and miRNA-222 in
fulvestrant-resistant breast cancer. Oncol Lett. 12:3882–3888.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang DQ, Zhou CK, Jiang XW, Chen J and
Shi BK: Increased expression of miR-222 is associated with poor
prognosis in bladder cancer. World J Surg Oncol. 12:2412014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ahmed FE, Amed NC, Vos PW, Bonnerup C,
Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE and Allison RR:
Diagnostic microRNA markers to screen for sporadic human colon
cancer in blood. Cancer Genomics Proteomics. 9:179–192.
2012.PubMed/NCBI
|
|
10
|
Lorimer IA: Regulation of p27Kip1 by miRNA
221/222 in glioblastoma. Cell Cycle. 8:26852009.PubMed/NCBI
|
|
11
|
Zhao Y, Wang Y, Yang Y, Liu J, Song Y, Cao
Y, Chen X, Yang W, Wang F, Gao J, et al: MicroRNA-222 controls
human pancreatic cancer cell line capan-2 proliferation by p57
targeting. J Cancer. 6:1230–1235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong Y, Si JW, Li WT, Liang L, Zhao J,
Zhou M, Li D and Li T: miR-200a/miR-141 and miR-205 upregulation
might be associated with hormone receptor status and prognosis in
endometrial carcinomas. Int J Clin Exp Pathol. 8:2864–2875.
2015.PubMed/NCBI
|
|
14
|
Tseng JH, Bisogna M, Hoang LN, Olvera N,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Levine DA and
Jelinic P: miR-200c-driven mesenchymal-to-epithelial transition is
a therapeutic target in uterine carcinosarcomas. Sci Rep.
7:36142017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun H, Xin J, Lu Z, Wang N, Liu N and Guo
Q: Potential molecular mechanisms for improved prognosis and
outcome with neoadjuvant chemotherapy prior to laparoscopical
radical hysterectomy for patients with cervical cancer. Cell
Physiol Biochem. 32:1528–1540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiong X, Cheng J, Liu X, Tang S and Luo X:
Correlation analysis between miR-124 rs531564 polymorphisms and
susceptibility to cervical cancer. Nan Fang Yi Ke Da Xue Xue Bao.
34:210–213. 2014.(In Chinese). PubMed/NCBI
|
|
17
|
He Z, Xu H, Meng Y and Kuang Y: miR-944
acts as a prognostic marker and promotes the tumor progression in
endometrial cancer. Biomed Pharmacother. 88:902–910. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu B, Che Q, Qiu H, Bao W, Chen X, Lu W,
Li B and Wan X: Elevated MiR-222-3p promotes proliferation and
invasion of endometrial carcinoma via targeting ERα. PLoS One.
9:e875632014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yue Z, Yun-Shan Z and Feng-Xia X: miR-205
mediates the inhibition of cervical cancer cell proliferation using
olmesartan. J Renin Angiotensin Aldosterone Syst.
17:14703203166633272016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang T, Zou P, Wang T, Xiang J, Cheng J,
Chen D and Zhou J: Down-regulation of miR-320 associated with
cancer progression and cell apoptosis via targeting Mcl-1 in
cervical cancer. Tumour Biol. 37:8931–8940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin ZL, Wang YL, Ge SF, Guo TT, Wang L,
Zheng XM and Liu J: Reduced expression of miR-503 is associated
with poor prognosis in cervical cancer. Eur Rev Med Pharmacol Sci.
19:4081–4085. 2015.PubMed/NCBI
|
|
22
|
Li W, Liang J, Zhang Z, Lou H, Zhao L, Xu
Y and Ou R: MicroRNA-329-3p targets MAPK1 to suppress cell
proliferation, migration and invasion in cervical cancer. Oncol
Rep. 37:2743–2750. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Dong Y, Ti H, Zhao J, Wang Y, Li
T and Zhang B: Down-regulation of miR-145 and miR-143 might be
associated with DNA methyltransferase 3B overexpression and worse
prognosis in endometrioid carcinomas. Hum Pathol. 44:2571–2580.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun J, Chu H, Ji J, Huo G, Song Q and
Zhang X: Long non-coding RNA HOTAIR modulates HLA-G expression by
absorbing miR-148a in human cervical cancer. Int J Oncol.
49:943–952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen SN, Wang LF, Jia YF, Hao YQ, Zhang L
and Wang H: Upregulation of microRNA-224 is associated with
aggressive progression and poor prognosis in human cervical cancer.
Diagn Pathol. 8:692013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Azizmohammadi S, Safari A, Azizmohammadi
S, Kaghazian M, Sadrkhanlo M, Yahaghi E, Farshgar R and
Seifoleslami M: Molecular identification of miR-145 and miR-9
expression level as prognostic biomarkers for early-stage cervical
cancer detection. QJM. 110:11–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Zeng J, Pan J, Geng X, Liu Y, Wu
J, Song P, Wang Y, Jia J and Wang L: MicroRNA-200c is involved in
proliferation of gastric cancer by directly repressing p27Kip1.
Biochem Biophys Rep. 8:227–233. 2016.PubMed/NCBI
|
|
29
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu L, Cheng X, Shi J, Jiacheng L, Chen G,
Jin H, Liu AB, Pyo H, Ye J, Zhu Y, et al: Crosstalk between bone
marrow-derived myofibroblasts and gastric cancer cells regulates
cancer stemness and promotes tumorigenesis. Oncogene. 35:5388–5399.
2016. View Article : Google Scholar : PubMed/NCBI
|