Introduction
Lung cancer is one of the very few cancer types in
which a continuous increase in incidence has emerged over recent
years. Currently, ~1.6 million new cases of lung cancer are
diagnosed annually, worldwide (1). Of
these, >85% of lung cancer cases are classified as non-small
cell lung cancer (NSCLC) (2). Despite
advancements and improvements in surgical, and medical treatments
of this patient population, the 5-year survival rate of patients
with lung cancer remains at ~17.4% (2). This is largely due to the late stage at
which the majority of patients are diagnosed, and the lack of
effective treatments for this disease. In addition, local control
for early-stage NSCLC has dramatically improved over previous
decades for operable and inoperable patients; however, ~20% of
early-stage patients still develop distant metastasis (2,3). Thus,
understanding the molecular markers that regulate invasion and
disease spread is crucial in identifying novel and reliable
prognostic markers for NSCLC, and may be exploited to refine
patient selection for already existing therapies.
The FET (previously TET) gene family comprise fused
in sarcoma/translocated in liposarcoma (FUS/TLS), Ewing's sarcoma
and TATA-binding protein-associated factor 15. These proteins,
encoded by the FET gene family, are similar in structure and
function (4). FET proteins contain an
N-terminal domain, a G-rich domain, an RRM (RNA-binding domain), a
zinc finger motif and a C-terminal RGG-rich (arginine-glycine-rich)
domain (4,5). Multifunctional characteristics of the
FET protein family have been authenticated. The N-terminal domain,
rich in Gln, Gly, Ser and Tyr, has a transcriptional activation
function (6,7). The RRM, zinc-finger and RGG-rich domains
contribute to the RNA-binding ability of the FET-proteins (8–10). In
addition, FET proteins also bind single-stranded DNA and possibly
double-stranded DNA. The unique functions also include pre-mRNA
splicing and DNA repair and recombination (11). Notably, chromosomal rearrangements
result in the 5′ regions of FET genes being fused with different
transcription factor genes in sarcomas or other types of cancer
(12–14). FUS/TLS was identified as a fusion gene
with DNA damage inducible transcript 3 (CHOP) (also known as
GADD153, DDIT3) in human myxoid liposarcoma with chromosomal
translocation t(12; 16)(q13; p11) (15). In myxoid liposarcoma, >85% of cases
were associated with the translocation of FUS/TLS (4,12).
Subsequently, TLS/FUS-ERG ETS transcription factor (ERG) chimeric
transcripts were observed to contribute an important role in the
development of acute myeloid leukemia (16,17). In
addition, overexpression of FUS/TLS protein or mRNA has been
reported in liposarcoma cell lines (18), breast cancer cells (19) and sporadic colorectal cancer cells
(20). Thus, FUS/TLS serves an
important function in the development of cancer.
Epithelial-mesenchymal transition (EMT) is a crucial
event in cancer development and metastasis (21). One of the key processes of EMT is the
loss of E-cadherin. E-cadherin has been reported to be a hallmark
in the progression of NSCLC (22). A
previous study indicated that FUS/TLS protein was located in the
spreading initiation centers of adhering cells and was involved in
cell spreading (23), while
E-cadherin primarily expressed on the membrane of epithelial cells,
contributes an important role in cell-cell adhesion (24). This suggested that FUS may be involved
in regulating E-cadherin expression. The aim of the present study
was to examine FUS/TLS levels in NSCLC and matched paratumor
tissues, and evaluate the association between FUS/TLS expression
and the clinicopathological characteristics of patients with NSCLC.
In addition, the prognostic effect of FUS/TLS and E-cadherin
expression was determined by multivariate analysis, either as
FUS/TLS as an independent parameter or combined with
E-cadherin.
Materials and methods
Patients and specimens
From January 2005 to December 2005, archival
specimens were consecutively collected from 208 patients (male 148,
female 60; median age 65 years; age range 40–83 years) with NSCLC
who received curative resection at Zhongshan Hospital of Fudan
University (Shanghai, China). Patient characteristics: All the
patients' clinicopathological information were in agreement with
the description of the authors previous study (25). The collection and conservation of
patient samples, and details were permitted with the evidence of
written informed consent. The follow-up was terminated in July
2010. The median follow-up duration was 43 months (range, 1–66
months). The overall survival (OS) was calculated from the day of
surgery to the date of either mortality as a result of lung cancer
or the last follow-up. Ethical approval was obtained from the
Zhongshan Hospital Research Ethics Committee (Shanghai, China).
Cell culture
The 16HBE cell line was obtained from Xiangbio
(Shanghai, China). H460, 95-C, A549 and 95-D cell lines were
purchased from the Institute of the Biochemistry and Cell Biology
of the Chinese Academy of Sciences (Shanghai, China). 16HBE, A549
and H460 cell lines were maintained in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.) and antibiotics (100 IU/ml penicillin, and 100
µg/ml streptomycin sulfate). 95-C and 95-D cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS and antibiotics. All cell lines were incubated in a
humidified atmosphere of 5% CO2 at 37°C.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of tissues was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was
reverse-transcribed to cDNA using PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol. Amplification and detection were performed
using the ABI PRISM 7900 Sequence Detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) starting with 1 µl cDNA
and SYBR Green Real-Time PCR Master mix (Takara Biotechnology Co.,
Ltd.). Primers were designed as follows: FUS/TLS forward,
5′-AGCTGAAGGGAGAGGCAAC-3′ and reverse, 5′-GGCGAGTAGCAAATGAGACC-3′;
GADPH forward, 5′-GGTATGACAACGAATTTGGC-3′ and reverse,
5′-GAGCACAGGGTACTTTATTG-3′. Relative expression levels of the gene
of interest were calculated using the 2−ΔΔCq method
(26). GADPH was used as an internal
standard and triplicate RT-qPCR samples were performed in each
assay. GraphPad Prism 5.0 software was applied to the
quantification of relative mRNA expression (GraphPad Software,
Inc., La Jolla, CA, USA).
Western blot analysis
Total protein was prepared using
radioimmunoprecipitation assay lysis buffer (Beyotime; Shanghai,
China) protein concentration was determined using an Enhanced BCA
Protein Assay Kit (Beyotime Institute of Biotechnology, Haimen,
China). Equal amounts of protein (30 µg) were separated via 10%
SDS-PAGE and transferred onto polyvinylidene difluoride membranes
(Merck KGaA, Darmstadt, Germany). Membranes were blocked in 5%
fat-free milk at room temperature for 1 h, and subsequently
incubated with primary antibodies rabbit anti-FUS/TLS (dilution
1:4,000; cat. no. 11570-1-ap; ProteinTech Group, Inc., Chicago, IL,
USA) and mouse anti-β-actin (dilution 1:1,000; cat. no. AA128;
Beyotime Institute of Biotechnology), which was used as an internal
control. The primary antibodies were incubated at 4°C overnight.
Subsequent to washing with Tris-buffered saline containing 0.1%
Tween-20 (TBST), the membranes were incubated with a horseradish
peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary
antibody (catalog no. A0208 or A0216; dilution, 1:1,000; Beyotime
Institute of Biotechnology) at room temperature for 1 h. Finally,
the membranes were washed with TBST and visualized using ECL
solution (EMD Millipore, Billerica, MA, USA) using an automatic
chemiluminescence Image analysis system (Tanon Technology Co.,
Ltd., Shanghai, China), Image-Pro Plus software version 6.0 (Media
Cybernetics, Inc., Rockville, USA) was used to analyze relative
protein expression and exhibited as the density ratio vs. β-actin.
All experiments were performed in triplicate.
Immunofluorescence
The localization of FUS/TLS protein in 16HBE, A549,
H460, 95C and 95D cells was examined by immunofluorescence. Cells
were fixed with 4% paraformaldehyde (Yesen, Shanghai, China) for 30
min at room temperature and then permeated with 0.3% Triton X
(Beyotime Institute of Biotechnology) for 10 min, washed three
times with PBS. Then the cells were blocked with 5% BSA/PBS for 30
min at room temperature. Subsequently, cells were incubated at 4°C
overnight with rabbit anti-FUS/TLS. (1:400 dilution; catalog no.
11570-1-ap; ProteinTech Group, Inc.; Chicago, IL, USA). Following
washing, the cells were incubated with secondary anti-rabbit IgG
(1:400 dilution; catalog no. A0468; Beyotime; Shanghai, China) at
room temperature for 2 h. Nuclei were counterstained with DAPI.
(catalog no. C1006; Beyotime; Shanghai, China) for 5 min at room
temperature and washed with PBS. The locations of FUS/TLS were
detected by fluorescence microscopy (Olympus Corporation, Tokyo,
Japan) used a magnification of ×200.
Tissue microarray (TMA) and
immunohistochemistry (IHC)
TMAs were constructed by Shanghai Biochip Co., Ltd.
(Shanghai, China). Pathological types of all samples were reviewed
by hematoxylin and eosin staining, in accordance with protocols
from a previous study (25). IHC of
paraffin sections was performed according to a protocol from a
previous study (27). In brief,
paraffin sections were deparaffinized by heating at 65°C for 2 h,
subsequently washed with xylene and rehydrated in ethanol. After
antigen retrieval was performed by incubating in 10 mmol/l Citrate
Sodium Buffer (pH 6.0; Yesen) and the slides boiled in a microwave,
slides were incubated in 0.3% H2O2 for 15 min
to block endogenous peroxidase activity at room temperature. Slides
were rinsed in PBS and immersed in 5% BSA for 1 h at room
temperature to block nonspecific binding sites. Subsequently, the
sections were incubated with primary antibody, rabbit anti-human
FUS/TLS (dilution, 1:5,000; catalog no. 11570-1-ap; ProteinTech
Group, Inc.; Chicago, IL, USA), overnight at 4°C. The following
day, samples were washed three times with PBS. After 30 min of
incubating slides in horseradish peroxidase labeled secondary
antibody (GTVision III immunohistochemical kit; cat. no. GK500705;
Gene Tech, Shanghai, China) at room temperature and washing in PBS
buffer, the slides were stained with 3,3′-diaminobenzidine
(DAB)-H2O2 under a microscope for 2 min at
room temperature, and hematoxylin was used to counterstain the
nuclei at room temperature for 40 sec. Finally, the sections were
dehydrated and covered with glass microscope glass using neutral
resins.
Evaluation of immunostaining intensity
of TMAs
The staining results scored were as follows. The
proportion of immunoreactive cells in total cell number of every
point: 0 (0%), 1 (>0% to 25%), 2 (>25% to 50%), 3 (>50% to
75%) and 4 (>75%). The cellular staining intensity was
determined by the degree of color: 0 (none), 1 (weak), 2
(moderate), 3 (strong) (28). The
final score for FUS/TLS expression was the summation of both
scores. The combined scores were divided into negative (−, 0–1),
weak positive (+, 2–3), moderate positive (++, 4–5) and strong
positive (+++, 6–7), respectively. For statistical analysis, the
negative and weak positive staining were considered the FUS/TLS low
level group, and the moderate and strong positive staining were
deemed the FUS high level group.
Statistical analysis
SPSS 21.0 software package (SPSS, Chicago, IL, USA)
was utilized for statistical analyses. OS was plotted using the
Kaplan-Meier method (log-rank test). Spearman's rank correlation
analysis identified a correlation between FUS/TLS and E-cadherin
expression. Significant factors in univariate analysis were further
incorporated into multivariate Cox proportional hazards regression
model to selection of independent prognostic factors. The
differences between categorical variables were analyzed by
χ2 test. P<0.05 was considered to indicate a
statistically significant difference (two-tailed).
Results
FUS/TLS is highly expressed in NSCLC
tissues
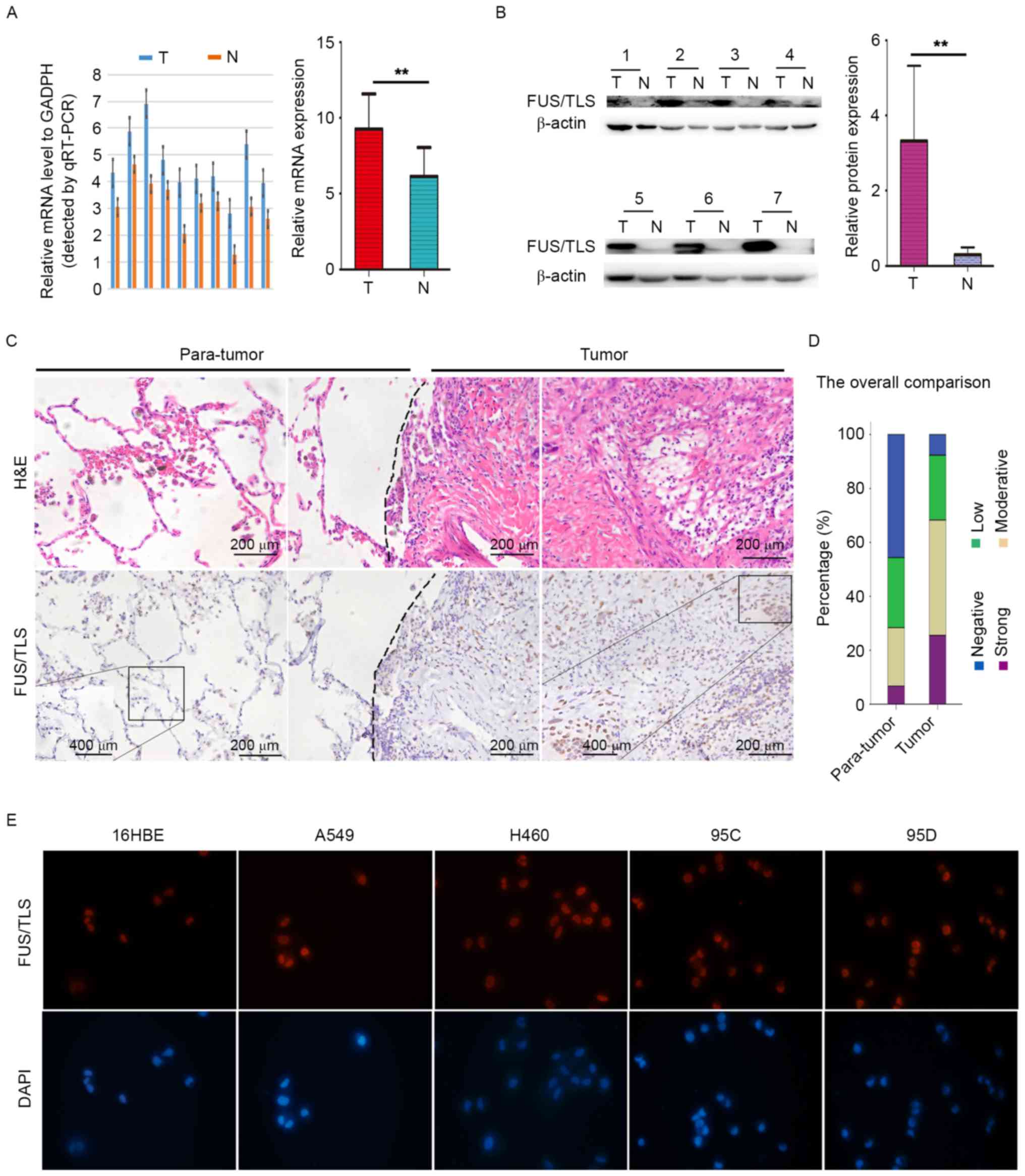
RT-qPCR, western blotting and immunohistochemistry
were performed to detect mRNA, and protein levels of FUS/TLS in
NSCLC tissues and matched adjacent nontumorous tissues. FUS/TLS
expression was significantly increased in NSCLC compared with that
in corresponding adjacent normal lung tissues at the mRNA
(9.27±0.73 vs. 6.15±0.60; Fig. 1A)
and protein (3.32±0.75 vs. 0.30±0.07; Fig. 1B) levels. The pathological type of
NSCLC and normal lung tissues were confirmed by hematoxylin and
eosin staining (Fig. 1C).
Immunohistochemical analysis demonstrated that strong intensity
FUS/TLS staining in NSCLC tissues was markedly higher than that in
paratumor tissues (Fig. 1C). Overall,
FUS/TLS-positive immunostaining was more frequent in tumor tissues
(49.5%, 103/208) compared with matched nontumor tissues (28.4%,
59/208; Fig. 1D).
Immunohistochemically, FUS/TLS demonstrated primarily nuclear and
to a lesser degree, cytoplasmic localization in NSCLC tissues. The
expression of FUS/TLS in normal lung endothelial cells (16HBE) and
NSCLCs cell lines (A549, H460, 95C and 95D) was further examined by
immunofluorescence. The results presented in Fig. 1E demonstrate a predominant nuclear
localization of FUS/TLS expression and high level of FUS/TLS was
observed in NSCLC cells. The aforementioned results indicate that
FUS/TLS contributes to the onset and progression of NSCLC.
Association between FUS/TLS and
clinicopathological features of NSCLCs
The association between the clinicopathological
features of the patient cohort and FUS/TLS was analyzed. Detailed
clinical and pathological information was presented in a previous
study (29). Briefly, analysis
included a total of 208 cases of primary NSCLC, and the cohort
consisted of 60 (28.8%) women and 148 (71.2%) men. The number of
patients with squamous cell carcinoma, adenocarcinoma and other
pathologic subtypes of NSCLC was 85, 110 and 13, respectively.
There were 144 (69.2%) tumors in tumor node metastasis (TNM) stages
I–II and 64 (30.8%) tumors in stages III–IV. In addition, tumors
with low and high differentiation were 93 and 115 respectively.
Lymph node metastasis was identified in 90 (43.3%) of samples.
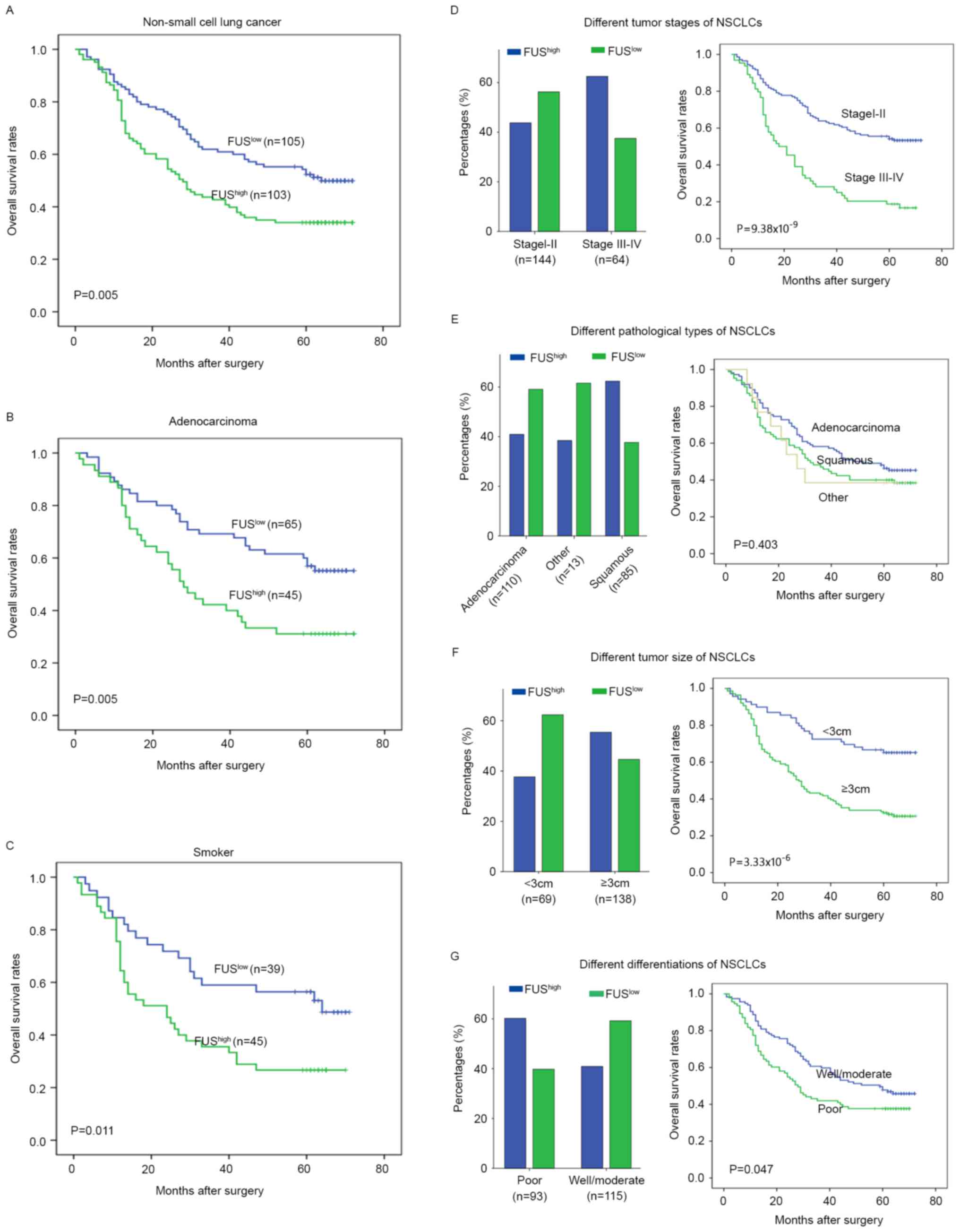
The association between FUS/TLS expression with
clinicopathological parameters was demonstrated in Table I and Fig.
2. FUS/TLS expression was significantly associated with
histological type (P=0.009), high TNM stage (P=0.016), poor tumor
differentiation (P=0.008) and larger tumor size (P=0.019). The
proportion of patients with high FUS/TLS levels in TNM stage III–IV
(62.5%, 40/64) was greater than patients in TNM stage I–II (43.75%,
63/144; Fig. 2D). Compared with
patients with adenocarcinoma (40.91%, 45/110), high levels of
FUS/TLS were more common in patients with squamous cell carcinoma
(62.35%, 53/85; Fig. 2E). Notably,
high FUS level patients with larger tumor size (≥3 cm, 55.4%,
77/139) were more frequent than patients with smaller tumor size
(<3 cm, 37.68%, 26/69; Fig. 2F).
Furthermore, a high proportion of elevated FUS/TLS level was also
present in tissues with poorer differentiation (60.22%, 56/93) than
well/moderate differentiation (47/115; Fig. 2G). Different subtype of NSCLCs
revealed different OS rates. The patients with higher TNM stage,
larger tumor size, and poorer differentiation exhibited a more
unfavorable OS rate. These clinicopathological features were all
significantly associated with high FUS/TLS expression. However,
other clinicopathological features, including age and smoking
status, were not directly associated with the level of FUS/TLS.
 | Table I.Association of expression of FUS/TLS
and E-cadherin with clinicopathological features of NSCLC
tissues. |
Table I.
Association of expression of FUS/TLS
and E-cadherin with clinicopathological features of NSCLC
tissues.
|
|
| FUS/TLS |
| E-cadherin |
|
|---|
|
|
|
|
|
|
|
|---|
| Variables | No. | Low | High |
P-valuea | Low | High |
P-valuea |
|---|
| Age, years |
|
|
| 0.074 |
|
| 0.092 |
|
<60 | 102 | 58 | 44 |
| 66 | 36 |
|
|
≥60 | 106 | 47 | 59 |
| 56 | 50 |
|
| Sex |
|
|
| 0.022 |
|
| 0.877 |
|
Male | 148 | 67 | 81 |
| 86 | 62 |
|
|
Female | 60 | 38 | 21 |
| 36 | 24 |
|
| Smoking status |
|
|
| 0.397 |
|
| 0.567 |
|
Smokers | 84 | 39 | 45 |
| 47 | 37 |
|
|
Non-smokers | 124 | 66 | 58 |
| 75 | 49 |
|
| Histologic
type |
|
|
| 0.009a |
|
| 0.585 |
|
Squamous carcinoma | 85 | 32 | 53 |
| 52 | 33 |
|
|
Adenocarcinoma | 110 | 65 | 45 |
| 64 | 46 |
|
|
Otherb | 13 | 8 | 5 |
| 6 | 7 |
|
|
Differentiation |
|
|
| 0.008a |
|
| 0.571 |
|
Well/moderate | 115 | 68 | 47 |
| 65 | 50 |
|
|
Poor | 93 | 37 | 56 |
| 57 | 36 |
|
| Tumor stage |
|
|
| 0.016a |
|
| 0.222 |
|
I+II | 144 | 81 | 63 |
| 80 | 64 |
|
|
III+IV | 64 | 24 | 40 |
| 42 | 22 |
|
| Lymph node
metastasis |
|
|
| 0.263 |
|
| 0.011a |
|
Yes | 90 | 41 | 49 |
| 62 | 28 |
|
| No | 118 | 64 | 54 |
| 60 | 58 |
|
| Tumor size, cm |
|
|
| 0.019a |
|
| 0.072 |
|
<3 | 69 | 43 | 26 |
| 34 | 35 |
|
| ≥3 | 139 | 62 | 77 |
| 88 | 51 |
|
High level of FUS was associated with
poor prognosis of patients with NSCLC
The 5-year OS rate after surgery was 47.6% for the
entire cohort. Patients with high FUS/TLS levels exhibited a worse
prognosis compared with those with low FUS/TLS expression (P=0.005;
Fig. 2A). The relationship between
FUS/TLS expression and prognosis by various subset analyses was
examined. High FUS/TLS level was associated with a poorer outcome
in patients with adenocarcinoma (P=0.005; Fig. 2A), whereas no distinct statistical
difference was observed in patients with squamous cell carcinoma
(P=0.314). It was of interest that smokers with high FUS/TLS levels
underwent a worse prognosis compared with smokers with low FUS/TLS
levels (P=0.011; Fig. 2C). In
addition, the level of FUS/TLS had an indistinctive effect on
non-smokers within this cohort (P=0.150). These data further
revealed that high FUS levels may predict a poor prognosis for
patients with NSCLC.
The univariate analysis indicated that FUS/TLS
expression, tumor size, TNM stage, lymph node metastasis and tumor
differentiation demonstrated a distinct impact on OS. In
multivariate analysis, tumor size and lymph node metastasis
remained associated with OS; however, FUS/TLS levels lost value as
an independent predictor of OS (P=0.093). Owing to the association
observed with TNM stage (P=0.016) and tumor size (P=0.019), FUS/TLS
level significantly influences the OS rate of patients (Table II).
 | Table II.Univariate and multivariate analysis
of factors associated with OS. |
Table II.
Univariate and multivariate analysis
of factors associated with OS.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender |
|
|
|
|
|
|
| (Male
vs. female) | 0.789 | 0.526–1.183 | 0.251 |
|
|
|
| Smoking status |
|
|
|
|
|
|
|
(Non-smokers vs. smokers) | 1.284 | 0.895–1.843 | 0.175 |
|
|
|
| Tumor size |
|
|
|
|
|
|
| (≥3 cm
vs. <3 cm) | 2.755 | 1.758–4.318 |
9.76×10−6 | 2.059 | 1.296–3.271 | 0.002a |
| Tumor stage |
|
|
|
|
|
|
| (III–IV
vs. I–II) | 2.771 | 1.922–3.993 |
4.65×10−8 | 1.430 | 0.904–2.262 | 0.127 |
| Lymph node
metastasis |
|
|
|
|
|
|
| (Yes
vs. no) | 3.042 | 2.103–4.399 |
3.47×10−9 | 2.172 | 1.371–3.443 | 0.001a |
|
Differentiation |
|
|
|
|
|
|
|
(Well/moderate vs. poor) | 1.431 | 1.000–2.049 | 0.050a | 1.122 | 0.772–1.629 | 0.546 |
| FUS/TLS level |
|
|
|
|
|
|
| (Low
vs. high) | 1.667 | 1.160–2.394 | 0.006a | 1.387 | 0.947–2.030 | 0.093 |
| E-cadherin
expression |
|
|
|
|
|
|
| (Low
vs. high) | 0.655 | 0.450–0.954 | 0.027a | 0.785 | 0.538–1.147 | 0.211 |
| FUS(TLS)/E-cadherin
expression |
|
| 0.001a |
|
| 0.044a |
| 1 vs.
3 | 1.461 | 1.144–1.866 | 0.002a | 1.255 | 0.968–1.628 | 0.086 |
| 2 vs.
3 | 1.959 | 1.310–2.930 | 0.001a | 1.683 | 1.115–2.539 | 0.013a |
| 1+2 vs.
3 | 2.037 | 1.414–2.934 |
2.03×10−4 | 1.634 | 1.122–2.381 | 0.010a |
High level of FUS/TLS impaired
E-cadherin expression and the combination of the two markers
defined a subset of NSCLC patients with worse prognosis
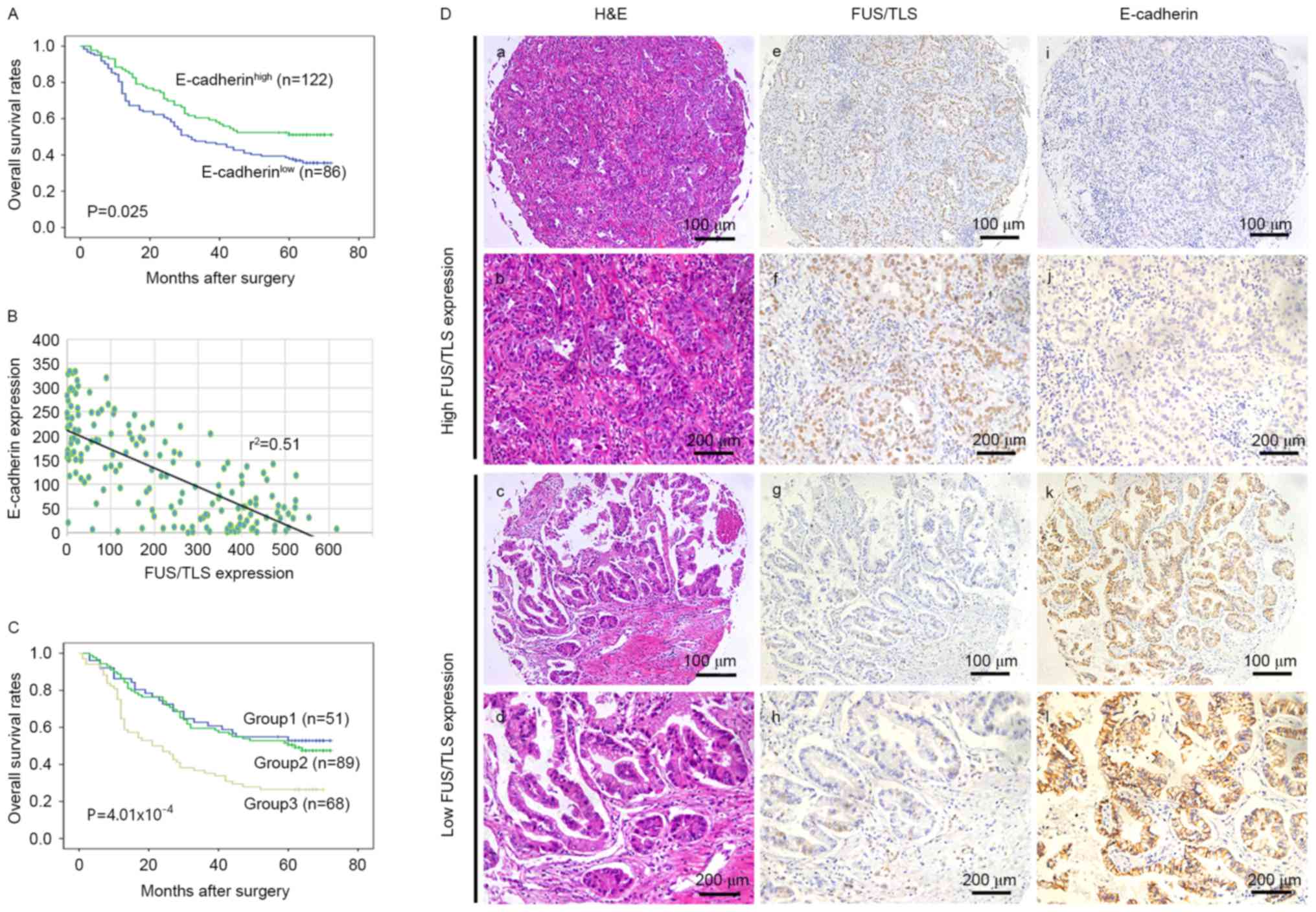
The association between E-cadherin expression with
clinicopathological characteristics are exhibited in Table I. No significant associations were
observed between impaired E-cadherin expression and any of the
clinicopathological variables, except lymph nodal involvement
(P=0.011). Patients with low E-cadherin expression exhibited a
shorter OS compared to those with high E-cadherin expression
(P=0.025; Fig. 3A). In univariate
analysis, E-cadherin expression demonstrated a significant
influence on OS (P=0.027). However, in the multivariate analysis,
E-cadherin was not significant as an independent predictor of
survival (P=0.211; Table II).
The association between FUS/TLS and E-cadherin
expression was also examined (Fig.
3). A significant negative association was observed between
FUS/TLS and E-cadherin expression (r2=0.51 and P=0.036;
Fig. 3B and Table III). High FUS/TLS expression was
associated with impaired E-cadherin expression and low FUS/TLS
expression was associated with increased E-cadherin expression,
representative images are shown in Fig.
3D. To investigate the combined influence of FUS/TLS and
E-cadherin expression on the prognosis of patients with NSCLC,
patients were divided into three groups: Group 1, low FUS/TLS and
preserved E-cadherin (n=51); Group 2, FUS/TLS and E-cadherin high
or low (n=89); Group 3, high FUS/TLS and impaired E-Cadherin
(n=68). Significant differences of 5-year OS rates were observed
among the three groups (Fig. 3C).
Group 1 and 2 exhibited a more favorable 5-year OS rates (52.9 and
48.3%, respectively) as compared with Group 3 (26.5%). In addition,
the multivariate analysis demonstrated the index that combined the
two markers (FUS/TLS and E-cadherin) was an independent prognostic
factor in patients' overall survival (P=0.044; Table II).
 | Table III.Association between FUS/TLS and E-cad
expression in NSCLC tissues. |
Table III.
Association between FUS/TLS and E-cad
expression in NSCLC tissues.
|
| FUS/TLS |
|
|
|---|
|
|
|
|
|
|---|
| E-cadherin | Low | High | Total | P-value |
|---|
| Low | 54 | 68 | 122 | 0.036a |
| High | 51 | 35 | 86 |
|
| Total | 105 | 103 | 208 |
|
Discussion
In the present study, it was demonstrated that the
mRNA and protein levels of FUS/TLS were upregulated in NSCLC
tissues compared with corresponding normal tissues. Increased
expression of FUS/TLS was associated with higher TNM stage, poor
tumor differentiation and large tumor size, and may be beneficial
in predicting poor prognosis in patients with NSCLC. Furthermore,
it was revealed that there was a significant negative association
between FUS/TLS and E-cadherin expression. Combined evaluation of
FUS/TLS and E-cadherin expression was an independent prognostic
factor that defined a new subgroup of patients with NSCLC with
shortened survival. The results from the present study indicated
that high FUS/TLS levels contributed to the progression of
NSCLC.
FUS/TLS, a ubiquitous and multifunctional DNA and
RNA-binding protein, participates in a wide range of cellular
processes, including RNA transcription, microRNA biogenesis,
splicing, and nucleo-cytoplasmic shuttling (30–32). In
addition, loss of FUS/TLS function significantly restricts cell
proliferation (33). The functions of
the fusion gene FUS-CHOP and TLS/FUS-ERG in solid tumor, and acute
myeloid leukemia have been extensively studied (16,17,34). For
example, FUS/TLS combined with the androgen receptor was reported
to promote prostate cancer cell proliferation (35). Furthermore, FUS/TLS physically binds
with nuclear paraspeckle assembly transcript 1 and contributes an
important role in the survival of breast cancer cells (19). FUS/TLS has been demonstrated to
participate in various types of cancer and neurodegeneration.
However, little attention has been given to the investigation of
the association between FUS/TLS and lung cancer. The results from
the present study indicated that elevated FUS/TLS expression
significantly associated with shortened survival in patients with
NSCLC. Clinically, FUS/TLS expression in lung adenocarcinoma and
squamous cell carcinoma was present heterogeneously. Upregulated
FUS/TLS expression was observed in a higher proportion in squamous
cell carcinoma tissues (62.35%) than in adenocarcinoma tissues
(40.91%). Although the gene expression profiles of the two
histological subtypes vary, the biological reason remains unclear.
Furthermore, higher tumor TNM stage, larger tumor size and poor
differentiation, the three factors that predict a poor survival,
were all significantly associated with high FUS/TLS expression.
Subgroup analyses revealed that the proportion of elevated FUS/TLS
level in patients with NSCLC with TNM stage III–IV was
significantly higher than patients with TNM stage I–II. The same
phenomenon was demonstrated in patients with tumor size ≥3 cm and
poor differentiation compared with tumor size <3 cm and well or
moderate differentiation, respectively.
E-cadherin, an important cell-to-cell adhesion
molecule serves a critical role in cancer development and
metastasis. Loss of E-cadherin expression in epithelial cancer
cells lead to loss of cell polarity and epithelial markers, and
increased motility due to acquired mesenchymal markers (3). E-cadherin is also considered to be a
hallmark of NSCLC, associated with invasiveness, metastasis and
prognosis (4). The results from the
present study indicated that FUS/TLS and E-cadherin had a
significant influence on the prognosis of patients with NSCLCs.
However, in multivariate analysis, the significance value was lost
as an independent predictor of OS. It was also demonstrated in the
present study that there were potential associations between
FUS/TLS and E-cadherin expression in NSCLC. An important finding is
the statistically significant inverse association between FUS/TLS
and E-cadherin expression in NSCLC. Furthermore, in the
multivariate analysis, high FUS/TLS expression in combination with
low E-cadherin expression was an independent prognostic factor for
shortened OS.
In summary, FUS/TLS may represent a potential
prognostic biomarker of NSCLC. This combination may provide a novel
effective risk stratification scheme for NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation (grant no. 81560401).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DX and YBW designed the experiment and wrote the
paper. CJ and JJL performed the assays. JG, HBW and SQZ
participated in clinical data collection. YFL and XL supervised the
research program and performed the data analysis. JYD and JJX had
significant roles in the study design and manuscript review. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Zhongshan
Hospital Research Ethics Committee (Shanghai, China). The
collection and conservation of patient samples, and details were
permitted with the evidence of written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ettinger DS, Wood DE, Akerley W, Bazhenova
LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA,
Dilling TJ, et al: NCCN guidelines insights: non-small cell lung
cancer, version 4.2016. J Natl Compr Canc Netw. 14:255–264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zappa C and Mousa SA: Non-small cell lung
cancer: Current treatment and future advances. Transl Lung Cancer
Res. 5:288–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Trautmann M, Menzel J, Bertling C, Cyra M,
Isfort I, Steinestel K, Elges S, Grünewald I, Altvater B, Rossig C,
et al: FUS-DDIT3 fusion protein driven IGF-IR signaling is a
therapeutic target in myxoid liposarcoma. Clin Cancer Res.
23:6227–6238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morohoshi F, Ootsuka Y, Arai K, Ichikawa
H, Mitani S, Munakata N and Ohki M: Genomic structure of the human
RBP56/hTAFII68 and FUS/TLS genes. Gene. 221:191–198. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bertolotti A, Bell B and Tora L: The
N-terminal domain of human TAFII68 displays transactivation and
oncogenic properties. Oncogene. 18:8000–8010. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zinszner H, Albalat R and Ron D: A novel
effector domain from the RNA-binding protein TLS or EWS is required
for oncogenic transformation by CHOP. Genes Dev. 8:2513–2526. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lerga A, Hallier M, Delva L, Orvain C,
Gallais I, Marie J and Moreau-Gachelin F: Identification of an RNA
binding specificity for the potential splicing factor TLS. J Biol
Chem. 276:6807–6816. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nguyen CD, Mansfield RE, Leung W, Vaz PM,
Loughlin FE, Grant RP and Mackay JP: Characterization of a family
of RanBP2-type zinc fingers that can recognize single-stranded RNA.
J Mol Biol. 407:273–283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ohno T, Ouchida M, Lee L, Gatalica Z, Rao
VN and Reddy ES: The EWS gene, involved in Ewing family of tumors,
malignant melanoma of soft parts and desmoplastic small round cell
tumors, codes for an RNA binding protein with novel regulatory
domains. Oncogene. 9:3087–3097. 1994.PubMed/NCBI
|
|
11
|
Cassiday LA and Maher LJ III: Having it
both ways: Transcription factors that bind DNA and RNA. Nucleic
Acids Res. 30:4118–4126. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Panagopoulos I, Aman P, Mertens F, Mandahl
N, Rydholm A, Bauer HF and Mitelman F: Genomic PCR detects tumor
cells in peripheral blood from patients with myxoid liposarcoma.
Genes Chromosomes Cancer. 17:1021996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Campos-Melo D, Droppelmann CA, Volkening K
and Strong MJ: RNA-binding proteins as molecular links between
cancer and neurodegeneration. Biogerontology. 15:587–610. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Riggi N, Cironi L, Suva ML and Stamenkovic
I: Sarcomas: Genetics, signalling, and cellular origins. Part 1:
The fellowship of TET. J Pathol. 213:4–20. 2007.
|
|
15
|
Crozat A, Aman P, Mandahl N and Ron D:
Fusion of CHOP to a novel RNA-binding protein in human myxoid
liposarcoma. Nature. 363:640–644. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ichikawa H, Shimizu K, Hayashi Y and Ohki
M: An RNA-binding protein gene, TLS/FUS, is fused to ERG in human
myeloid leukemia with t(16;21) chromosomal translocation. Cancer
Res. 54:2865–2868. 1994.PubMed/NCBI
|
|
17
|
Kong XT, Ida K, Ichikawa H, Shimizu K,
Ohki M, Maseki N, Kaneko Y, Sako M, Kobayashi Y, Tojou A, et al:
Consistent detection of TLS/FUS-ERG chimeric transcripts in acute
myeloid leukemia with t(16;21)(p11;q22) and identification of a
novel transcript. Blood. 90:1192–1199. 1997.PubMed/NCBI
|
|
18
|
Spitzer JI, Ugras S, Runge S, Decarolis P,
Antonescu C, Tuschl T and Singer S: mRNA and protein levels of FUS,
EWSR1, and TAF15 are upregulated in liposarcoma. Genes Chromosomes
Cancer. 50:338–347. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q,
Su X, Peng L and Jiao B: NEAT1 is required for survival of breast
cancer cells through FUS and miR-548. Gene Regul Syst Bio.
10:11–17. 2016.PubMed/NCBI
|
|
20
|
Lepourcelet M, Tou L, Cai L, Sawada J,
Lazar AJ, Glickman JN, Williamson JA, Everett AD, Redston M, Fox
EA, et al: Insights into developmental mechanisms and cancers in
the mammalian intestine derived from serial analysis of gene
expression and study of the hepatoma-derived growth factor (HDGF).
Development. 132:415–427. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bremnes RM, Veve R, Gabrielson E, Hirsch
FR, Baron A, Bemis L, Gemmill RM, Drabkin HA and Franklin WA:
High-throughput tissue microarray analysis used to evaluate biology
and prognostic significance of the E-cadherin pathway in
non-small-cell lung cancer. J Clin Oncol. 20:2417–2428. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andersson MK, Stahlberg A, Arvidsson Y,
Olofsson A, Semb H, Stenman G, Nilsson O and Aman P: The
multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell
type-specific expression patterns and involvement in cell spreading
and stress response. BMC Cell Biol. 9:372008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirohashi S: Inactivation of the
E-cadherin-mediated cell adhesion system in human cancers. Am J
Pathol. 153:333–339. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gu J, Ding JY, Lu CL, Lin ZW, Chu YW, Zhao
GY, Guo J and Ge D: Overexpression of CD88 predicts poor prognosis
in non-small-cell lung cancer. Lung Cancer. 81:259–265. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao YF, Wu YB, Long X, Zhu SQ, Jin C, Xu
JJ and Ding JY: High level of BRD4 promotes non-small cell lung
cancer progression. Oncotarget. 7:9491–9500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liao Y, Gu J, Wu Y, Long X, Ge DI, Xu J
and Ding J: Low level of 5-Hydroxymethylcytosine predicts poor
prognosis in non-small cell lung cancer. Oncol Lett. 11:3753–3760.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao GY, Ding JY, Lu CL, Lin ZW and Guo J:
The overexpression of 14-3-3ζ and Hsp27 promotes non-small cell
lung cancer progression. Cancer. 120:652–663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Uranishi H, Tetsuka T, Yamashita M,
Asamitsu K, Shimizu M, Itoh M and Okamoto T: Involvement of the
pro-oncoprotein TLS (translocated in liposarcoma) in nuclear
factor-kappa B p65-mediated transcription as a coactivator. J Biol
Chem. 276:13395–13401. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zinszner H, Sok J, Immanuel D, Yin Y and
Ron D: TLS (FUS) binds RNA in vivo and engages in
nucleo-cytoplasmic shuttling. J Cell Sci. 110:1741–1750.
1997.PubMed/NCBI
|
|
32
|
Mastrocola AS, Kim SH, Trinh AT,
Rodenkirch LA and Tibbetts RS: The RNA-binding protein fused in
sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase
(PARP) in response to DNA damage. J Biol Chem. 288:24731–24741.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ward CL, Boggio KJ, Johnson BN, Boyd JB,
Douthwright S, Shaffer SA, Landers JE, Glicksman MA and Bosco DA: A
loss of FUS/TLS function leads to impaired cellular proliferation.
Cell Death Dis. 5:e15722014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Panagopoulos I, Aman P, Fioretos T,
Höglund M, Johansson B, Mandahl N, Heim S, Behrendtz M and Mitelman
F: Fusion of the FUS gene with ERG in acute myeloid leukemia with
t(16;21)(p11;q22). Genes Chromosomes Cancer. 11:256–262. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Haile S, Lal A, Myung JK and Sadar MD:
FUS/TLS is a co-activator of androgen receptor in prostate cancer
cells. PLoS One. 6:e241972011. View Article : Google Scholar : PubMed/NCBI
|