Introduction
Hepatocellular carcinoma (HCC) is the fifth-most
common malignant tumor with poor prognosis globally in the previous
10 years, and also is the second most common cause of
cancer-associated mortalities in China in the previous 10 years
(1,2).
For patients with HCC who undergo surgery, radiotherapy and
chemotherapy treatment, the median survival rate is ~40% (range,
15–62%) after 5 years. If patients are diagnosed early and treated
appropriately, the 5-year survival rate may increase >70%
(3). To improve prognosis and
diagnosis, predictive biomarkers for HCC are required, allowing
screening of high risk patients in order to provide timely and
suitable treatment (4). To date,
certain oncofetal proteins are used in clinical settings as
diagnostic markers for a wide variety of tumors and to monitor
recurrence following treatment (5).
However, the accuracy, sensitivity and the detection thresholds for
these diagnostic markers are low. Therefore, novel biomarkers with
high specificities and sensitivities are necessary to ensure that
the optimal clinical decisions for patients with HCC are
available.
Over the previous decades, a number of
tissue-specific and circulating biomarkers of HCC have been
identified from retrospective studies, but the lack of clinical
validation has limited their use (6–8). The serum
levels of α-fetoprotein (AFP) and lens culinaris lectin-reactive
AFP, and imaging techniques, including ultrasonography, magnetic
resonance imaging or computer tomography are the gold standard to
identify suspected cases of HCC (7,8). However,
many early-stage HCC (<2 cm) patients have a low level of AFP,
and the detection of AFP can not effectively predict the HCC
leading to high false-negative rates (>30%) (9,10).
Therefore, novel diagnostic biomarkers with high specificity and
sensitivity are required.
Long noncoding RNAs (lncRNAs) are a type of
noncoding RNAs with >200 nucleotides, which act as the
regulatory molecules and may serve central roles in a variety of
diseases through complex mechanisms (11,12).
Therefore, identifying HCC-associated oncofetal lncRNAs may be
important for the diagnosis and treatment of HCC. Additionally,
previous studies have demonstrated that specific lncRNAs are
associated with cancer development, and are detectable in plasma,
highlighting the convenience and speed of detection of these
biomarkers (13,14). For example, the lncRNA SOCS2-antisense
RNA 1 is positively associated with castration-resistant prostate
cancer, whereas lncRNA neuroblastoma associated transcript 1 is
negatively associated with neuroblastoma in stage 2A (15–17).
Therefore, the levels of lncRNAs in plasma may act as potential
biomarkers for cancer detection, diagnosis, tumor grade screening
and recurrence monitoring during clinical treatment.
In the present study, the abnormal expression of
serum LRB1 was identified in patients with HCC. The potential value
of LRB1 as a biomarker for the detection and monitoring of
prognosis of patients with HCC was investigated, and the diagnostic
accuracy of LRB1 in sera was compared to des-γ-carboxy prothrombin
(DCP) and AFP, respectively and in different combinations, in
patients with HCC, consequently providing the optimal combination
of biomarker indices for HCC detection.
Materials and methods
Patients and serum samples
The present study was approved by Human Research
Ethics Committee of The First Hospital of Hebei Medical University
(Shijiazhuang, China), and all patients provided written informed
consent. The serum samples were collected from 326 patients with
HCC who underwent primary surgery treatment without radiotherapy or
preoperative chemotherapy treatment at The First Hospital of Hebei
Medical University between March 2011 and March 2015 (251 males and
75 females, 162 patients >60 years old and 164 patients ≤60
years old) and 73 healthy volunteers were recruited from the health
examination center of The First Hospital of Hebei Medical
University between March 2011 and March 2015 (43 males and 30
females, 32 volunteers >60 years old and 31 volunteers ≤60 years
old). The median expression level of LRB1 was used as the cut-off
value, and all of the patients with HCC were divided into two
groups according to the cut-off value of LRB1. Firstly, peripheral
blood was collected from each patient with HCC, then a second blood
sample was obtained 10 days after surgery using BD
Vacutainer® sodium heparin tubes (BD Biosciences,
Franklin Lakes, NJ, USA). The blood samples were centrifuged at 4°C
with 800 × g for 20 min, 2,000 × g for 10 min and then 5,000 × g
for 5 min for the prevention of contamination with nucleic acids.
Then, the serum samples were transferred into 1.5 ml tubes and
stored at −80°C until use.
HCC diagnosis and grade
determination
The HCC diagnoses were determined according to the
American Association for the Study of Liver Diseases practice
guidelines (18). The direct HCC
diagnosis gold standard is pathological detection, and the tumor
tissues are obtained through surgical resection or percutaneous
biopsy. Alternatively, blood biochemical detection and radiological
diagnosis are indirect methods of detection. For tumor stage
determination, Tumor Node Metastasis (TNM) stage was assessed
postoperatively according to the 7th edition of American Joint
Commission on Cancer (19), and the
clinical definition of HCC stage was determined based on the
Barcelona Clinic Liver Cancer (BCLC) classification (20).
Clinical chemistry and detection of
AFP and DCP
Blood samples were collected for detection of the
biomarker indices using the SpotChem EZ clinical chemistry analyzer
(ARKRAY Inc., Kyoto, Japan). The levels of serum AFP were analyzed
by Human α-Fetoprotein Quantikine® ELISA Kit (cat. no.
DAFP00; R&D Systems, Minneapolis, MN, USA) according to the
manufacturer's protocol. The serum DCP concentrations were detected
by Lumipulse G1200 auto-analyzer (FUJIREBIO Inc., Tokyo, Japan)
according to the manufacturer's protocol. All the experiments were
repeated three times, and the average values were calculated.
RNA extraction and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from serum samples using the
RNA Isolation kit (Axygen Scientific, Inc., Union City, CA, USA)
according to the manufacturer's protocol. Triplicates of each gene
and each specimen were used, with GAPDH as an internal standard.
The single-strand cDNA for PCR template was synthesized from 10 µg
of total RNA by ReverTra Ace qPCR RT kit (cat. no. FSQ-101; Toyobo
Life Science, Osaka, Japan) from the extracted total RNA. StepOne™
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used in the RT-PCR assay. The RT-PCR was performed with a
total reaction volume of 20 µl, including 10 µl Power SYBR Green
PCR Master mix (Roche Diagnostics, Indianapolis, IN, USA), 5 pmol
of forward and reverse primer respectively and 2 µl of cDNA.
Quantification cycle (Cq) was observed in the amplification with 35
cycles of 1 min at 95°C, 1 min at 58°C, and 1 min at 72°C. The
results were normalized to GAPDH, which served as the endogenous
control, and the relative expression of LRB1 was quantified using
the 2−ΔΔCq method (21).
The PCR primer sequences for LRB1 and GAPDH were as follows: LRB1
forward, 5′-TCATGCGATAGCTGAACGCTA-3′ and reverse,
5′-GAGGCCGGTAGTCGTAACT-3′; GAPDH forward,
5′-ATTCCACCCATGGCAAATTC-3′ and reverse,
5′-TGGGATTTCCATTGATGACAAG-3′.
Microarray analysis of lncRNAs
Total RNA was extracted from serum samples using an
RNA Isolation kit (Axygen Scientific, Inc.) according to the
manufacturer's protocol., the extracted RNA was amplified and
transcribed into fluorescence-labeled cDNA using Quick Amp Labeling
kit (Agilent Technologies, Inc., Santa Clara, CA, USA), the labeled
cDNA was hybridized onto a lncRNA Array 2.0 (8×60 K array;
ArrayStar, Inc., Rockville, MD, USA) for lncRNA expression
detection. The array was scanned using Agilent Scanner G2505C
(Agilent Technologies, Inc.), and the array images were obtained by
Agilent Feature Extraction software (version 11.0.1.1; Agilent
Technologies, Inc.). Accessible raw and normalization data was
processed using GeneSpring software (version GX v12.1, Agilent
Technologies, Inc.). Subsequently, functional annotation was
performed on the samples using the gene set enrichment analysis
(GSEA) method (22), GSEA was
supported by the Broad Institute website (http://www.broadinstitute.org/gsea/index.jsp),
and was performed using the GSEA software (version 2.2.2; Broad
Institute, Inc., Massachusetts Institute of Technology, and Regents
of the University of California).
Statistical analysis
All data were presented as the mean ± standard
deviation, and were analyzed using SPSS (version 20.0; IBM Corp.,
Armonk, NY, USA). The differences between two groups were analyzed
using Pearson's χ2 test, paired Student's t-test,
Wilcoxon test or Fisher's exact test. For comparisons between more
than two groups, the differences were estimated using
Kruskal-Wallis test, followed by Bonferroni post hoc testing.
Pearson's correlation coefficient was used to analyze the
correlation between LRB1 and AFP as well DCP respectively in
diagnosis of HCC. Receiver operator characteristic (ROC) curve and
area under the curve (AUC) analyses were used to detect the
accuracy of markers with a 95% confidence interval (CI). The
sensitivity represents the true positive rate and was used as the
y-axis, (1-specificity) represents the false positive rate and was
used as the x-axis. The value of AUC is the size of the area under
the ROC curve and is between 0.5 and 1, and the closer the AUC is
to 1, the better the diagnosis. The Kaplan-Meier method was used to
analyze the postoperative survival rate, and the log-rank test was
used to assess the difference of survival rate in different groups.
The correlation between serum LRB1 level with pathological and
clinical characteristics was analyzed with the Spearman rank
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Evaluation of serum LRB1 levels in
patients with HCC
In order to investigate the hypothesis that the
serum level of LRB1 is a potential biomarker for the diagnosis of
HCC, an lncRNA expression microarray and RT-qPCR for the lncRNA
expression detection was performed with samples from 326 patients
with HCC and 73 healthy volunteers. The results of the lncRNA
expression microarray indicated that the serum LRB1 expression was
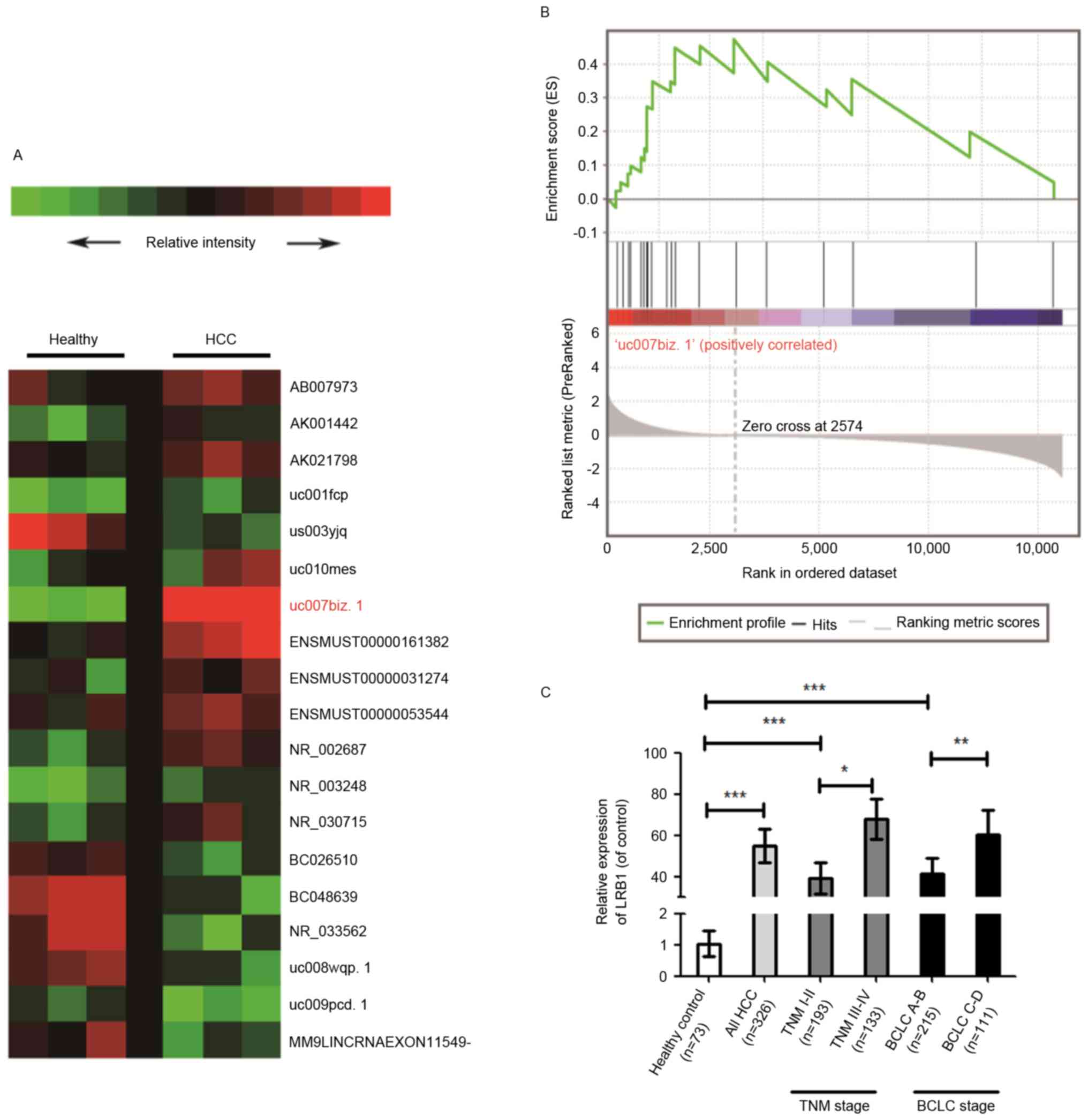
significantly increased (P<0.001) in patients with HCC compared
with healthy volunteers (Fig. 1A),
and the results of GSEA suggested that the expression of LRB1 was
associated with HCC (Fig. 1B). The
RT-qPCR assay also confirmed that the expression of serum LRB1 in
patients with HCC was significantly increased compared with healthy
volunteers, and that the expression of LRB1 exhibited a positive
association with HCC stage (Fig. 1C).
Concurrently, the expression of AFP and DCP was analyzed, and the
results were similar to those of LRB1. The expression levels of AFP
and DCP in patients with HCC were significantly increased compared
with healthy volunteers (Table
I).
 | Table I.Levels of LRB1, AFP and DCP
expression. |
Table I.
Levels of LRB1, AFP and DCP
expression.
| Parameters | LRB1 (ng/ml) | AFP (ng/ml) | DCP (mAU/ml) |
|---|
| Control (n=73) | 1.05±0.40 | 13.96±0.83 | 30.27±4.22 |
| HCC (n=326) | 54.83±8.21 | 1,046.72±135.18 | 5,934.72±416.73 |
| P-value (control vs.
HCC) | <0.0001 | <0.0001 | <0.0001 |
| TNM stage |
|
|
|
| TNM I–II
(n=193) | 39.25±7.76 | 946.17±159.33 |
4,352.33±496.04 |
| TNM
III–IV (n=133) | 68.03±9.66 |
1,191.25±218.04 |
7,130.25±569.23 |
| P-value
(control vs. TNM I–II) | <0.001 | <0.001 | <0.001 |
| P-value
(TNM I–II vs. TNM III–IV) | 0.027 | 0.074 | 0.037 |
| BCLC stage |
|
|
|
| BCLC
A-B (n=215) | 41.32±7.64 |
1,002.33±132.57 |
4,342.86±672.03 |
| BCLC
C-D (n=111) | 60.31±11.77 |
1,160.27±194.28 |
7,361.07±548.28 |
| P-value
(control vs. BCLC A-B) | <0.001 | <0.001 | <0.001 |
| P-value
(BCLC A-B vs. BCLC C-D) | 0.007 | 0.088 | 0.031 |
Association between serum LRB1
expression levels and clinicopathological factors in patients with
HCC
In order to detect whether the serum level of LRB1
was associated with the clinicopathological factors in patients
with HCC, 326 patients with HCC were enrolled in the present study.
The serum samples were divided into two independent groups
according to the median expression level (47.24 ng/ml) of LRB1 in
HCC patients. The results indicated that the serum LRB1 expression
level was associated with AFP expression, larger tumor size, tumor
stage and venous invasion (Table
II). However, there was no significant association with sex,
age and hepatitis B virus infection (Table II). These results suggest that LRB1
may serve an important role in hepatocarcinogenesis and tumor
progression.
 | Table II.Clinicopathological association of
LRB1 expression levels in patients with HCC. |
Table II.
Clinicopathological association of
LRB1 expression levels in patients with HCC.
|
| LRB1 |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low | Higha | χ2 |
P-valueb |
|---|
| All cases | 159 | 167 |
|
|
| Age |
|
| 0.361 | 0.537 |
|
>60 | 77 | 85 |
|
|
|
≤60 | 82 | 82 |
|
|
| Sex |
|
| 0.243 | 0.674 |
|
Male | 121 | 130 |
|
|
|
Female | 38 | 37 |
|
|
| HBs antigen |
|
| 2.094 | 0.157 |
|
Present | 83 | 90 |
|
|
|
Absent | 76 | 77 |
|
|
| HBe antigen |
|
| 2.993 | 0.131 |
|
Present | 73 | 79 |
|
|
|
Absent | 86 | 88 |
|
|
| Liver
cirrhosis |
|
| 0.405 | 0.339 |
|
Present | 79 | 86 |
|
|
|
Absent | 80 | 81 |
|
|
| AFP, ng/ml |
|
| 10.329 | 0.002 |
|
>20 | 92 | 155 |
|
|
|
≤20 | 67 | 12 |
|
|
| Tumor size, cm |
|
| 11.905 | 0.001 |
|
>5 | 33 | 129 |
|
|
| ≤5 | 126 | 38 |
|
|
| TNM stage |
|
| 14.551 | 0.002 |
| I | 61 | 7 |
|
|
| II | 62 | 53 |
|
|
|
III | 32 | 64 |
|
|
| IV | 4 | 43 |
|
|
| BCLC stage |
|
| 14.229 | 0.001 |
| A | 62 | 4 |
|
|
| B | 73 | 59 |
|
|
| C | 21 | 66 |
|
|
| D | 3 | 38 |
|
|
| Venous
invasion |
|
| 6.327 | 0.018 |
|
Present | 26 | 108 |
|
|
|
Absent | 133 | 59 |
|
|
| Tumor
microsatellite |
|
| 0.975 | 0.328 |
|
Present | 83 | 89 |
|
|
|
Absent | 76 | 78 |
|
|
| Tumor
encapsulation |
|
| 1.704 | 0.115 |
|
Present | 93 | 98 |
|
|
|
Absent | 66 | 69 |
|
|
Serum LRB1 levels in HCC tumors may be
used for prediction of tumor prognosis according to tumor
stage
In order to determine the association between serum
LRB1 level and HCC progression, patients with HCC were divided into
several groups according to TNM or BCLC stages, and the serum LRB1
levels were presented in Fig. 1C and
Table I. The results suggested that
the LRB1 levels in patients with HCC were significantly increased
compared with the healthy controls. In patients with TNM stages
I–II, the levels of LRB1 were significantly decreased compared with
patients with TNM stages III–IV. Similarity, the levels of LRB1 in
patients with BCLC stages A-B were significantly decreased compared
with patients with BCLC stages C-D, where BCLC A-B and BCLC C-D
represent early-stage and represents late-stage disease,
respectively. Concurrently, the DCP and AFP levels were also
detected in patients with HCC. When stratified by TNM or BCLC
stages, the levels of DCP in patients with TNM I–II or BCLC A-B
were significantly decreased compared with patients with TNM III–IV
or BCLC C-D (Table I). However, no
statistically significant differences in AFP levels between
patients with different TNM or BCLC stages were observed (Table I). Compared with the healthy control
group, LRB1, DCP and AFP levels in patients with early stage HCC
were significantly increased compared with the healthy group
(Table I). Subsequently, survival
analysis of the follow-up data of patients with HCC was performed.
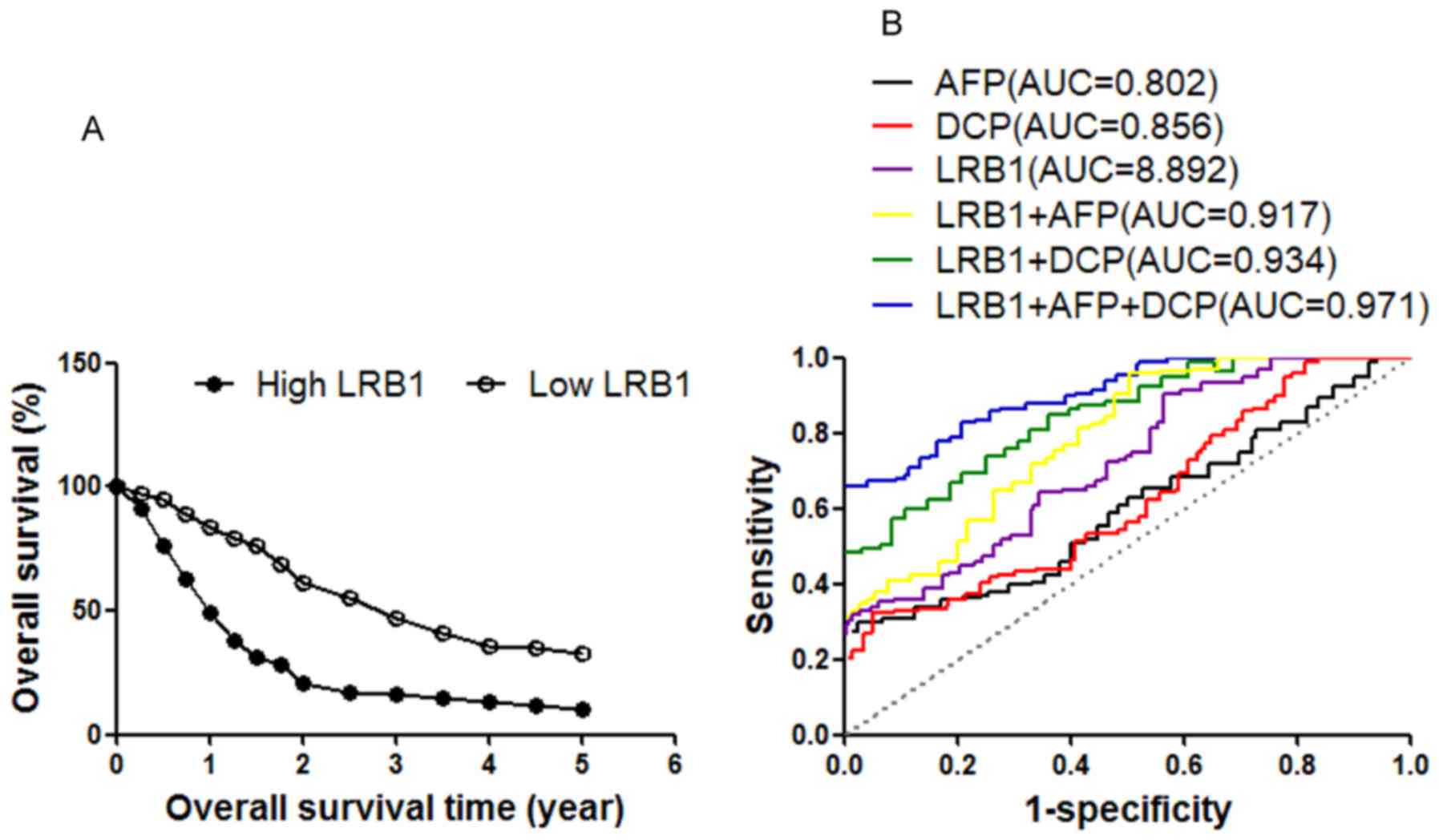
As indicated by Kaplan-Meier survival analysis, patients with HCC
with low serum LRB1 levels exhibited improved overall survival
compared with patients with high serum LRB1 levels (Fig. 2A). These results indicate that the
serum level of LRB1 in patients with HCC was associated with tumor
prognosis, and may be used as an indicator of tumor stage
detection.
Accuracy of LRB1, AFP and DCP markers
for the diagnosis of HCC
Based on the ROC curve analysis (Fig. 2B), the optimum cut-off values of LRB1,
AFP and DCP were calculated as 3.481, 9.36 ng/ml and 41.26 mAU/ml,
respectively. The results for AUC, 95% CI, sensitivity and
specificity values for all biomarkers were calculated (Table III). In present clinical detection,
the threshold values of AFP and DCP were 10 ng/ml and 35 mAU/ml,
respectively, and the threshold values of AFP and DCP in our
research were 9.36 ng/ml and 41.26 mAU/ml, respectively; therefore,
the clinical examination threshold is notably similar to the
threshold in our experiment. ROC analysis was performed to assess
the ability of the markers in distinguishing between patients with
HCC and healthy controls. The results indicated that the AUC value
for LRB1 marker was higher compared with the values for AFP or DCP
(Fig. 2B; Table III). According to Pearson's
correlation analysis, the results indicated that there were no
statistically significant correlations between LRB1 and AFP or
DCP.
 | Table III.Accuracy of LRB1, AFP and DCP markers
for the diagnosis of HCC. |
Table III.
Accuracy of LRB1, AFP and DCP markers
for the diagnosis of HCC.
| HCC vs.
control | AUC | 95% CI | Sensitivity, % | Specificity, % | PPV, % | NPV, % | +LR | -LR |
|---|
| LRB1 | 0.892 | 0.843–0.922 | 92.43 | 71.85 | 81.37 | 82.19 | 3.97 | 0.29 |
| AFP | 0.802 | 0.769–0.834 | 61.72 | 83.63 | 87.02 | 76.32 | 7.42 | 0.37 |
| DCP | 0.856 | 0.773–0.879 | 63.08 | 89.41 | 94.26 | 77.22 | 10.83 | 0.41 |
| LRB1+AFP | 0.917 | 0.869–0.938 | 79.32 | 79.38 | 84.52 | 84.91 | 4.62 | 0.21 |
| LRB1+DCP | 0.934 | 0.893–0.953 | 81.69 | 83.25 | 92.19 | 87.36 | 8.44 | 0.17 |
| LRB1+AFP+DCP | 0.971 | 0.942–0.988 | 86.33 | 87.64 | 93.04 | 89.08 | 9.01 | 0.11 |
Therefore, it was subsequently evaluated if the
combination of these three HCC markers may improve the accuracy of
diagnosis. The results indicated that the diagnostic accuracy of
LRB1 combined with AFP or DCP were markedly increased compared with
the accuracy of using LRB1 alone (Fig.
2B; Table III). In addition,
the diagnostic accuracy may be optimized when a combination of all
three markers, LRB1, AFP and DCP, was employed. (Fig. 2B; Table
III).
The accuracy of using each of the markers (LRB1, AFP
and DCP) alone and a combination of these markers for the diagnosis
of early-stage HCC (TNM stages I–II or BCLC stages A-B) was also
assessed. The results indicated that the AUC value of the LRB1
marker was higher compared with AFP and DCP for TNM stages I–II
(Fig. 3A; Table IV) or BCLC A-B (Fig. 3B; Table
V). In addition, when a combination of the three markers was
employed for the diagnosis of TNM stages I–II vs. control (Fig. 3A; Table
IV) or BCLC stages A-B vs. control (Fig. 3B; Table
V), the AUC value was the highest. These results suggested that
serum LRB1 has a high accuracy for the diagnosis of HCC and
early-stage of the disease, and the combination of LRB1, AFP and
DCP may increase the accuracy of diagnosis of HCC and early-stage
of the disease.
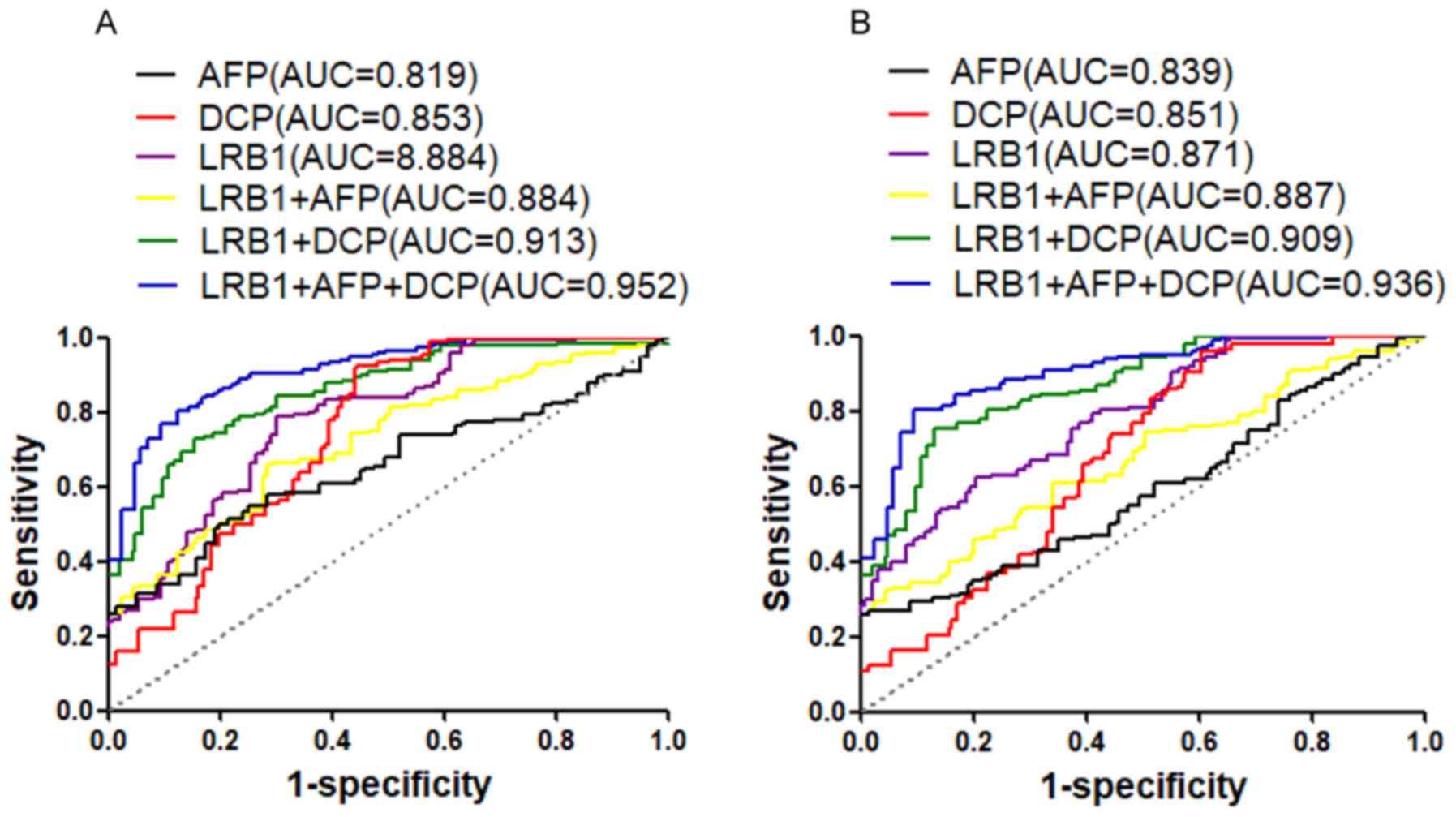 | Figure 3.ROC curves of LRB1, AFP and DCP and
combinations of these markers for the diagnosis of early-stage HCC.
(A) ROC curve analysis to distinguish HCC patients with TNM stages
I–II from healthy controls (B) and to distinguish HCC patients with
BCLC stages A-B from healthy controls. LRB1, lncRNA uc007biz.1;
HCC, hepatocellular carcinoma; ROC, receiver operator
characteristic; AFP, α-fetoprotein; DCP, des-γ-carboxy prothrombin;
AUC, areas under the curve; TNM, tumor-node metastasis; BCLC,
Barcelona Clinic Liver Cancer; HCC, hepatocellular carcinoma. |
 | Table IV.Accuracy of LRB1, AFP and DCP markers
for the diagnosis of early-stage (TNM I–II) HCC. |
Table IV.
Accuracy of LRB1, AFP and DCP markers
for the diagnosis of early-stage (TNM I–II) HCC.
| Early stage of HCC
(TNM I–II) vs. control | AUC | 95% CI | Sensitivity, % | Specificity, % | PPV, % | NPV, % | +LR | -LR |
|---|
| LRB1 | 0.884 | 0.775–0.892 | 89.76 | 73.66 | 71.26 | 83.26 | 2.57 | 0.17 |
| AFP | 0.819 | 0.761–0.887 | 67.54 | 87.93 | 86.88 | 77.46 | 6.35 | 0.41 |
| DCP | 0.853 | 0.796–0.894 | 68.37 | 91.65 | 92.35 | 73.49 | 9.35 | 0.45 |
| LRB1+AFP | 0.884 | 0.825–0.916 | 84.29 | 77.44 | 79.43 | 80.68 | 4.11 | 0.19 |
| LRB1+DCP | 0.913 | 0.881–0.947 | 79.05 | 89.47 | 89.68 | 79.49 | 8.95 | 0.25 |
| LRB1+AFP+DCP | 0.952 | 0.919–0.978 | 86.33 | 90.28 | 92.33 | 82.18 | 9.07 | 0.18 |
 | Table V.Accuracy of LRB1, AFP and DCP markers
for the diagnosis of early-stage (BCLC A-B) HCC. |
Table V.
Accuracy of LRB1, AFP and DCP markers
for the diagnosis of early-stage (BCLC A-B) HCC.
| Early stage of HCC
(BCLC A-B) vs. control | AUC | 95% CI | Sensitivity, % | Specificity, % | PPV, % | NPV, % | +LR | -LR |
|---|
| LRB1 | 0.871 | 0.847–0.896 | 88.38 | 76.79 | 73.42 | 85.47 | 2.13 | 0.23 |
| AFP | 0.839 | 0.819–0.895 | 68.47 | 88.26 | 87.39 | 79.33 | 5.61 | 0.42 |
| DCP | 0.851 | 0.824–0.879 | 69.35 | 92.47 | 93.15 | 75.79 | 9.85 | 0.38 |
| LRB1+AFP | 0.887 | 0.837–0.926 | 82.83 | 81.16 | 82.16 | 83.41 | 3.64 | 0.18 |
| LRB1+DCP | 0.909 | 0.874–0.933 | 81.49 | 88.35 | 90.43 | 81.26 | 7.06 | 0.24 |
| LRB1+AFP+DCP | 0.936 | 0.908–0.954 | 87.06 | 91.56 | 92.75 | 84.63 | 9.18 | 0.16 |
Discussion
HCC is one of the common types of malignant tumor
that is associated with a poor prognosis, and the second most
common cause of cancer-associated mortality in China (1–3). HCC
frequently occurs in patients with liver cirrhosis, and the low
rates of early diagnosis and high recurrence rates result in a poor
prognosis (23). Detection of
early-stage HCC increases the availability of appropriate
treatment, including local ablative therapy, resection or liver
transplantation. These treatments may prolong survival (24,25).
Although the markers DCP and AFP are widely used for the detection
of HCC, the sensitivity and specificity values are not optimum
(9,26,27).
Consequently, novel molecular biomarkers for the diagnosis of HCC
and monitoring of therapy are urgently required, particularly in
early-stage cancer screening protocols.
Previous studies investigating the human genome and
transcriptome sequencing revealed that only a minority of gene
transcripts encodes protein. The majority of the genome that is
transcribed does not encode protein (12). lncRNAs are a type of noncoding RNA
molecules with >200 nucleotides (12). Previous studies have demonstrated that
dysregulation of lncRNAs may alter epigenetic information and
promote cell growth, resulting in tumor growth and progression
(28,29). The survival of cancer cells and
proliferation is associated with the expression level of lncRNAs,
which is associated with cancer diagnosis and prediction of tumor
prognosis (30,31). Therefore, the identification of
cancer-associated lncRNAs and their clinical functions is
important, and may contribute to the identification of novel cancer
biomarkers, and clarify oncogenic and tumor networks. In previous
studies, serum lncRNAs have been used as biomarkers to identify
various types of cancer, including lung, gastric and breast cancer
(32–34). However, a small number of studies
demonstrated the potential use of serum lncRNAs in the diagnosis
and prediction of prognosis of HCC (35). Therefore, there may be a novel lncRNA
biomarker for the diagnosis of early-stage HCC.
In the present study, a lncRNA expression
microarray, GSEA and RT-qPCR analyses were conducted in 326
patients with HCC and 73 healthy volunteers, the results of which
suggested that LRB1 is significantly associated with a risk of HCC,
and that the serum LRB1 level is associated with HCC early
diagnosis and tumor prognosis. The serum LRB1 levels in patients
with HCC were significantly increased compared with the healthy
volunteers. In current clinical HCC detection, AFP and DCP are
widely used as biomarkers, but the associated high false-positive
rates limit the use of these markers.
In the present study, the results indicated that
LRB1 level was positively associated with AFP expression, large
tumor size, tumor stage (TNM or BCLC stage) and venous invasion.
Additionally, the association between LRB1 level and clinical
outcomes were detected. The Kaplan-Meier analysis suggested that
LRB1 level was significantly associated with overall survival of
patients with HCC, and higher LRB1 levels were associated with
poorer recurrence-free survival rates. In addition, in the present
study, the AUC and sensitivity values for LRB1 for distinguishing
between patients with HCC and healthy controls were higher compared
with DCP or AFP, but the specificity value was lower for LRB1
compared with DCP or AFP. When a combination of all three markers
(LRB1, AFP and DCP) was employed for the diagnosis of HCC, the AUC
and specificity values were markedly higher compared with using
LRB1 alone. However, the sensitivity value of using a combination
of all three markers was lower compared with using LRB1 alone.
These results indicated that there is a high
expression of LRB1 in HCC and therefore this may be used to may
predict the prognosis and overall survival of patients with HCC.
Consequently, serum LRB1 may be a novel biomarker for HCC diagnosis
and prediction of prognosis.
Previous studies have demonstrated that circulating
lncRNAs are associated with tumor dynamics (11–15). In
the present study, serum LRB1 levels demonstrated clinical
significance in the diagnosis of HCC and prediction of tumor
prognosis, as high serum LRB1 levels were associated with higher
degrees of malignancy of HCC. These findings suggested that higher
serum LRB1 level may be an independent prognostic factor of poor
prognosis of patients with HCC, and that serum LRB1 level was
associated with the early stages of HCC. Taken together, serum LRB1
level may be used for monitoring, diagnosis and prediction of
prognoses of HCC, and employing a combination of LRB1, AFP and DCP
may increase diagnostic accuracy.
In summary, to the best of our knowledge, the
present study was the first to reveal the use of serum LRB1 level
in cancer diagnosis and prognosis. LRB1 may act as an important key
regulator in HCC progression and be a potential therapeutic target
for treatment of HCC. Although the sample size of the present study
was small, the data suggested that serum LRB1 may serve as a useful
biomarker for the diagnosis of HCC and prediction of prognosis.
When a combination of LRB1, DCP and AFP was employed, the
diagnostic accuracy was markedly increased. Large-scale prospective
studies are required to validate the accuracy and effectiveness of
LRB1 as a biomarker of HCC, including the use of a combination of
LRB1, DCP and AFP markers.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trépo C, Chan HL and Lok A: Hepatitis B
virus infection. Lancet. 384:2053–2063. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu LC, Cheng AL and Poon RT: Recent
advances in the prevention of hepatocellular carcinoma recurrence.
Semin Liver Dis. 34:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Zhang A and Sun H: Power of
metabolomics in diagnosis and biomarker discovery of hepatocellular
carcinoma. Hepatology. 57:2072–2077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zucman-Rossi J, Villanueva A, Nault JC and
Llovet JM: Genetic landscape and biomarkers of hepatocellular
carcinoma. Gastroenterology. 149:1226–1239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bruix J, Reig M and Sherman M:
Evidence-based diagnosis, staging, and treatment of patients with
hepatocellular carcinoma. Gastroenterology. 150:835–853. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frau M, Biasi F, Feo F and Pascale RM:
Prognostic markers and putative therapeutic targets for
hepatocellular carcinoma. Mol Aspects Med. 31:179–193. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sauzay C, Petit A, Bourgeois AM, Barbare
JC, Chauffert B, Galmiche A and Houessinon A: Alpha-foetoprotein
(AFP): A multi-purpose marker in hepatocellular carcinoma. Clin
Chim Acta. 463:39–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng J, Wang W, Zhang Y, Liu X, Li M, Wu
Z, Liu Z, Lv Y and Wang B: Prognostic role of pre-treatment serum
AFP-L3% in hepatocellular carcinoma: Systematic review and
meta-analysis. PLoS One. 9:e870112014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schmitt AM and Chang HY: Long noncoding
rnas in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mohankumar S and Patel T: Extracellular
vesicle long noncoding RNA as potential biomarkers of liver cancer.
Brief Funct Genomics. 15:249–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolha L, Ravnik-Glavač M and Glavač D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Misawa A, Takayama K, Urano T and Inoue S:
Androgen-induced long noncoding RNA (lncRNA) Socs2-As1 promotes
cell growth and inhibits apoptosis in prostate cancer cells. J Biol
Chem. 291:17861–1780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue S, Li QW, Che JP, Guo Y, Yang FQ and
Zheng JH: Decreased expression of long non-coding RNA NBAT-1 is
associated with poor prognosis in patients with clear cell renal
cell carcinoma. Int J Clin Exp Pathol. 8:3765–3774. 2015.PubMed/NCBI
|
|
17
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bruix J and Sherman M; Practice Guidelines
Committee: American association for the study of liver diseases,
management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chun YH, Kim SU, Park JY, Kim DY, Han KH,
Chon CY, Kim BK, Choi GH, Kim KS, Choi JS and Ahn SH: Prognostic
value of the 7th edition of the AJCC staging system as a clinical
staging system in patients with hepatocellular carcinoma. Eur J
Cancer. 47:2568–2575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Forner A, Reig ME, de Lope CR and Bruix J:
Current strategy for staging and treatment: The BCLC update and
future prospects. Semin Liver Dis. 30:61–74. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: Gsea-p: A desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Singal AG, Pillai A and Tiro J: Early
detection, curative treatment, and survival rates for
hepatocellular carcinoma surveillance in patients with cirrhosis: A
meta-analysis. PLoS Med. 11:e10016242014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Poon D, Anderson BO, Chen LT, Tanaka K,
Lau WY, van Cutsem E, Singh H, Chow WC, Ooi LL, Chow P, et al:
Management of hepatocellular carcinoma in Asia: Consensus statement
from the asian oncology summit 2009. Lancet Oncol. 10:1111–1118.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo Y, Wang J, Zhang L, Shen S, Guo R,
Yang Y, Chen W, Wang Y, Chen G and Shuai X: Theranostical
nanosystem-mediated identification of an oncogene and highly
effective therapy in hepatocellular carcinoma. Hepatology.
63:1240–1255. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cui SX, Zhang YS, Chu JH, Song ZY and Qu
XJ: Des-gamma-carboxy prothrombin (DCP) antagonizes the effects of
gefitinib on human hepatocellular carcinoma cells. Cell Physiol
Biochem. 35:201–212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hiraoka A, Ishimaru Y, Kawasaki H, Aibiki
T, Okudaira T, Toshimori A, Kawamura T, Yamago H, Nakahara H, Suga
Y, et al: Tumor Markers AFP, AFP-L3, and DCP in hepatocellular
carcinoma refractory to transcatheter arterial chemoembolization.
Oncology. 89:167–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun M and Kraus WL: From discovery to
function: The expanding roles of long noncoding RNAs in physiology
and disease. Endocr Rev. 36:25–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Knoll M, Lodish HF and Sun L: Long
non-coding RNAs as regulators of the endocrine system. Nat Rev
Endocrinol. 11:151–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tantai J1, Hu D1, Yang Y1 and Geng J1:
Combined identification of long non-coding RNA XIST and HIF1A-AS1
in serum as an effective screening for non-small cell lung cancer.
Int J Clin Exp Pathol. 8:7887–7895. 2015.PubMed/NCBI
|
|
33
|
Jin C, Shi W, Wang F, Shen X, Qi J, Cong
H, Yuan J, Shi L, Zhu B, Luo X, et al: Long non-coding RNA HULC as
a novel serum biomarker for diagnosis and prognosis prediction of
gastric cancer. Oncotarget. 7:51763–51772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Amorim M, Salta S, Henrique R and Jerónimo
C: Decoding the usefulness of non-coding RNAs as breast cancer
markers. J Transl Med. 14:2652016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chauhan R and Lahiri N: Tissue- and
serum-associated biomarkers of hepatocellular carcinoma. Biomark
Cancer. 8:37–55. 2016.PubMed/NCBI
|