Introduction
Malignant tumor growth is a common and serious
disease condition that may affect any number of different organs
and tissues throughout the body, and incur varied and complex
symptoms that are often life threatening. Colorectal cancer (CRC)
is one of the most common gastrointestinal malignancies, and
localizes to the rectum or the junction of the rectum and the
sigmoid colon. In fact, CRC is the fourth most common type of
cancer internationally, following gastric, esophageal, and lung
cancer (1,2).
Malignant tumors are often characterized by both
tissue architecture disruption, and differentiation derangement.
Cell adhesion dysregulation contributes to tumor invasion and
metastasis (3), such that the
abnormal expression of various cell adhesion molecules induces a
loss of cell-cell binding, thereby promoting tumor differentiation
and malignant invasion (4).
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs)
are members of the glycosylphosphatidylinositol (GPI)-linked
immunoglobulin (Ig) superfamily (5).
CEACAM-1 is a CEACAM subtype that is also known as biliary
glycoprotein I or CD66a (6,7). Previous studies have demonstrated that
CEACAM-1 expression is reduced in several tumor types, such as
melanoma, lung, colon and ovarian cancer, compared with the
corresponding normal tissues (8–14). This
suggests that CEACAM-1 may function to inhibit carcinogenesis. In
addition, CEACAM-1 has been reported to promote the apoptosis of
various cells, including pulmonary and mammary epithelial cells,
oral keratinocytes, cancer cells, Jurkat T cells, and
cardiomyocytes (13,15,16).
To date, the expression of CEACAM-1 in CRC has not
been investigated. In the present study, CEACAM-1 expression was
silenced in a CRC cell line, and its effects on cell growth and
apoptosis were examined. The findings provide the first evidence
that decreased CEACAM-1 expression promotes CRC progression.
Materials and methods
Cell culture
HCT-8 cells (Type Culture Collection of the Chinese
Academy of Sciences, Shanghai, China) were maintained (37°C, 5%
CO2) in Dulbecco's modified Eagle's medium supplemented
with 10% fetal calf serum (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 µg/ml penicillin, and 100
µg/ml streptomycin. The cells were passaged every 2–3 days, using
0.02% EDTA and 0.1% trypsin.
Small-interfering RNA (siRNA) design
and cell transfection
Vectors carrying either siRNA specific to CEACAM-1
or a negative control siRNA were generated by GeneChem Co., Ltd.
(Shanghai, China). Three CEACAM-1-specific siRNAs were designed, as
follows: siRNA1 sense, 5′-CAGCCACAGAAAUAAUUUATT-3′ and antisense,
5′-UAAAUUAUUUCUGUGGCUGTT-3′; siRNA2 sense,
5′-CCGUCAAAUUGUAGGAUAUTT-3′ and antisense,
5′-AUAUCCUACAAUUUGACGGTT-3′; and siRNA3 sense,
5′-GAGCUCUUUAUCCCUAACATT-3′ and antisense,
5′-UGUUAGGGAUAAAGAGCUCTT-3′. A siRNA encoding a nonsense sequence
(sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′), was designed and used as a negative
control (GenePharma Co., Ltd., Shanghai, China). HCT-8 cells were
cultured until they reached 50% confluence, washed with PBS, and
transfected with 50 nM CEACAM-1-specific or control siRNA,
according to the manufacturer's instructions. Cells were collected
48 h post-transfection, and maintained in fresh medium for 24 h
prior to further experimentation and/or analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
CEACAM-1 expression was analyzed via RT-qPCR, as
previously described (17). Briefly,
total RNA was extracted from CRC cells, using an RNeasy Mini kit
(Invitrogen; Thermo Fisher Scientific, Inc.). Genomic DNA was
removed from the extracted total RNA via DNase I digestion, and
then the total RNA was reverse transcribed to generate cDNA,
according to the manufacturer's instructions
(PrimeScript™ 1st strand cDNA Synthesis kit; Takara
Biotechnology Co., Ltd., Dalian, China). qPCR was performed in
triplicate in a 10 µl reaction mix consisting of 4 µl template DNA
(0.05 µg/µl), 5 µl SYBR-Green (Takara Biotechnology Co., Ltd.), 0.2
µl each forward and reverse oligonucleotide (10 µM each) and 0.6 µl
deionized water. The thermocycling conditions were as follows: 95°C
for 10 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30
sec, and 72°C for 30 sec. The primers used for qPCR were: CEACAM-1
forward, 5′-CAGGGGCTTCTGCTCACAGC-3′ and reverse,
5′-AGTTGCTTCTTCACAAGAT-3′; β-actin forward,
5′-GGCTGTGGAGACAAAAATGACCTC-3′ and reverse,
5′-AGGCTTGGGCTTGAATGGAGTC-3′. The expression level was estimated
with the 2−ΔΔCq method (18).
Western blot analysis
CEACAM-1 expression levels were determined via
western blot analysis. CRC cells were washed twice with PBS, and
collected via centrifugation. Proteins were then extracted with
cell lysis buffer (CST Biological Reagents Company, Ltd., Shanghai,
China) containing 1 mM phenylmethylsulfonyl fluoride, and the
protein concentration of each sample was determined using the BCA
protein assay reagent kit (Beyotime Institute of Biotechnology,
Lianyungang, China), and bovine serum albumin was used as a
standard. The samples were denatured, and 50–80 µg were separated
via polyacrylamide gel electrophoresis and transferred onto
nitrocellulose membranes. The membranes were incubated first (4°C,
overnight) with anti-CEACAM-1 (cat. no. AF1857; Novus Biologicals,
Ltd., Cambridge, UK; dilution, 1:200) and anti-β-actin (cat. no.
ab8227; Abcam, Cambridge, UK; dilution, 1:4,000) antibodies. The
membranes were then incubated with horseradish
peroxidase-conjugated rabbit anti-human secondary antibodies (cat.
no. ab6759; Abcam; dilution, 1:3,000) for 12 h at 4°C, and finally
with enhanced chemiluminescence substrate (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The resulting blots were analyzed using
ImageJ software (NIH, Bethesda, MA, USA).
Cell proliferation
Cell proliferation was assessed by a CCK-8 assay
(Roche Diagnostics GmbH, Mannheim, Germany), according to the
manufacturer's instructions. Typically, cells were plated at a
density of 1.5–2×103 cells/well in 96-well plates. After
48 h (to allow cell adherence to occur), cells were incubated with
colorimetric substrates. Colorimetric changes were then measured in
a multi-well spectrophotometer (MR5000 Multiplate Reader; Dynatech,
Denkendorf, Germany), and cell survival following treatment was
expressed as a % of viable cells relative to control cell values.
All experiments were independently conducted three times, and the
results of the three experiments were then averaged.
Cell cycle assay
Cells were fixed, washed with PBS, treated with
RNaseA, and stained (37°C, 30 min) with 25 µg/ml propidium iodide
(PI). The samples were then analyzed via flow cytometry, and the
cell cycle phase distribution was quantified using Modfit Software
(BD Biosciences, Franklin Lakes, NJ, USA). The proliferative index
was calculated to represent the % of cells identified to occur in
the S/G2/M phase.
Apoptosis assay
Cells were collected, stained with Annexin
V-fluorescein isothiocyanate (FITC) and 7-Aminoactinomycin D using
an Annexin V-FITC Apoptosis Detection kit (KeyGen Biotech Co.,
Ltd., Nanjing, China) according to the manufacturer's instructions,
and analyzed via flow cytometry (BD Biosciences).
Statistical analysis
All results are presented as the mean ± standard
error of the mean. Differences between two groups were evaluated by
the unpaired Student's t-test. One-way analysis of variance with
post-hoc analysis by Bonferroni's test was performed to evaluate
the data generated by the cellular viability and apoptosis assays.
Additional statistical analyses were performed by Student's
t-tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
Knockdown of CEACAM-1 in HCT-8
cells
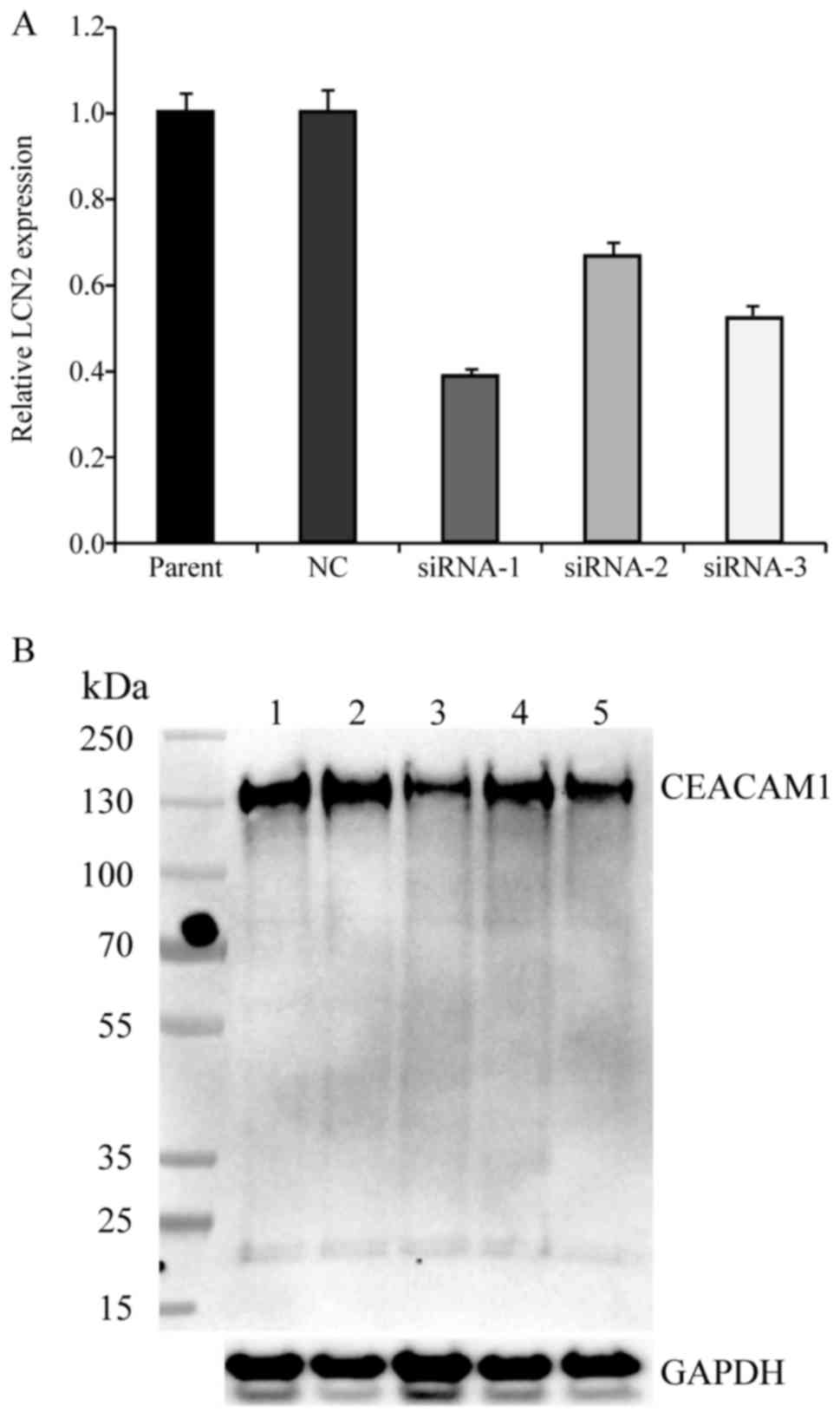
To investigate the function of CEACAM-1 expression
in CRC cells, three separate siRNA sequences, comprising
CEACAM-1-siRNA1, CEACAM-1-siRNA2, and CEACAM-1-siRNA3, were
designed. When testing their efficacy, the three sequences induced
a 61.3, 32.4 and 47.3% decrease in CEACAM-1 mRNA expression,
respectively, in the CEACAM-1-knockdown cells compared with the
negative control HCT-8 cells (Fig.
1A). The results from western blot analysis revealed a
concordant reduction in CEACAM-1 protein expression levels compared
with the control group (Fig. 1B). The
CEACAM-1-siRNA1 sequence was selected for use in subsequent
experiments, since it induced the greatest reduction in
CEACAM-1 expression.
CEACAM-1 downregulation inhibits cell
proliferation
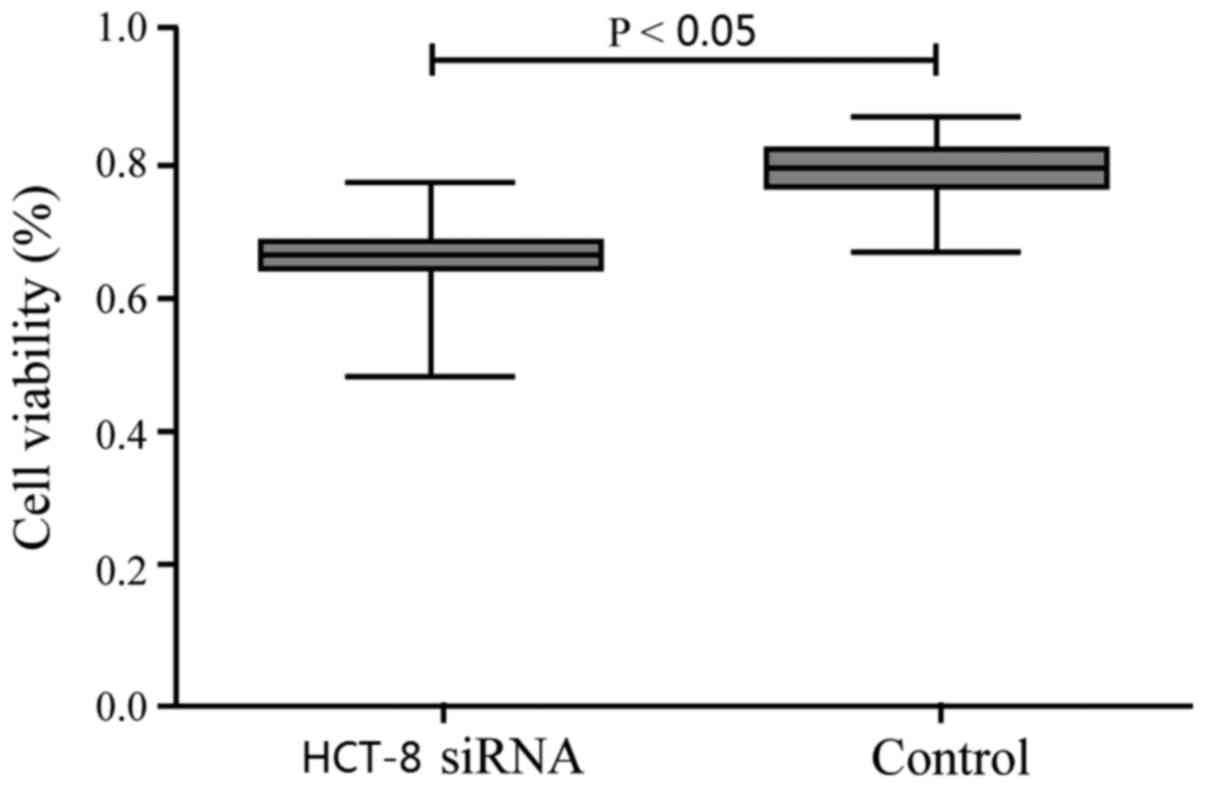
CEACAM-1 knockdown significantly decreased the
numbers of viable HCT-8 cells compared with the control group
(Fig. 2), suggesting that CEACAM-1
silencing inhibited CRC cell proliferation.
CEACAM-1 downregulation increases cell
apoptosis
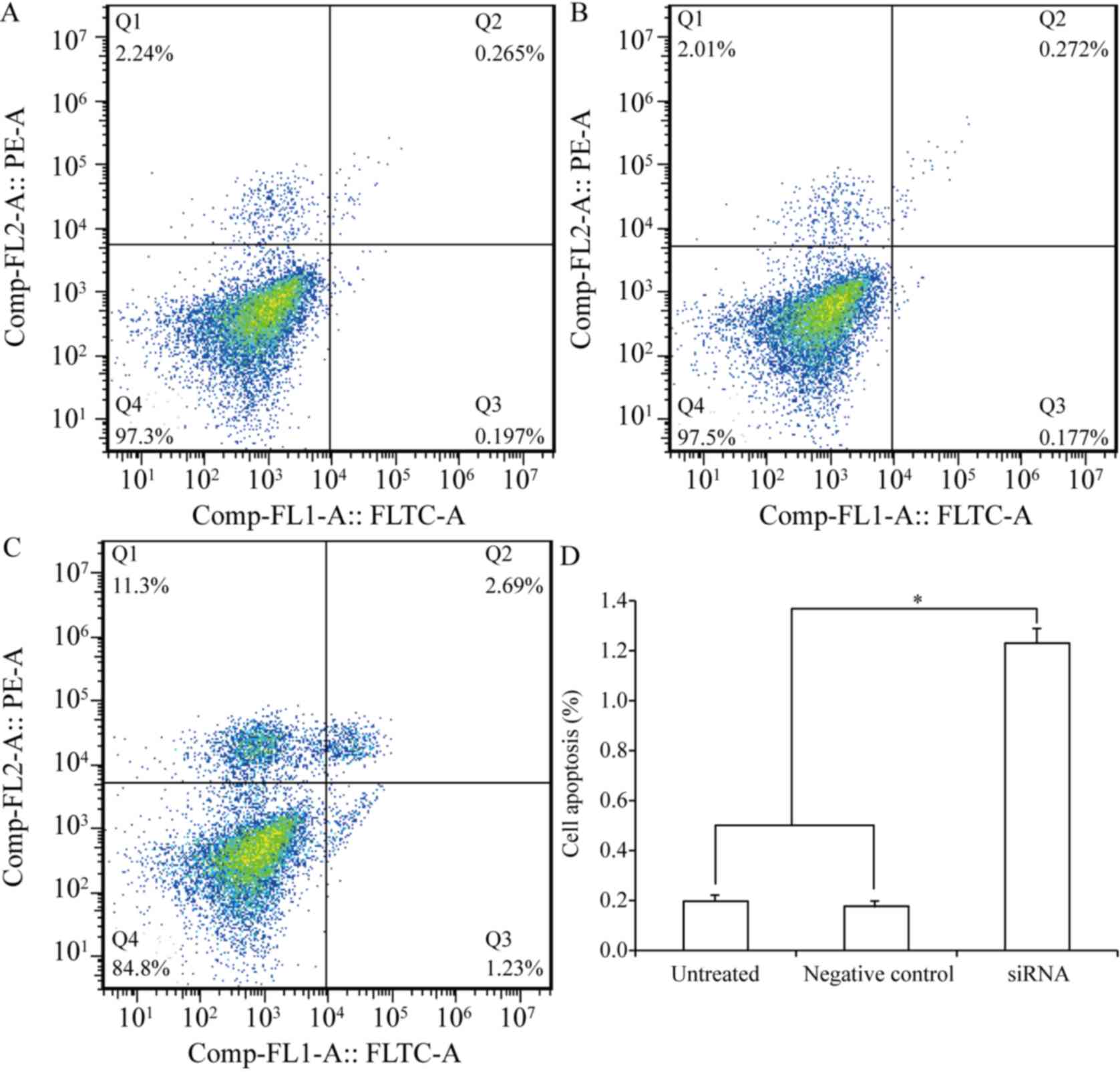
Next, the effect of CEACAM-1 silencing on apoptosis
was determined in CRC cells. The results from Annexin V cytometric
analysis revealed that CEACAM-1 downregulation resulted in a
significant increase in the apoptotic rate of HCT-8 cells (1.23% in
the CEACAM-1-knockdown group compared with 0.197% in the control
group; P<0.01; Fig. 3). The
experiment was performed in triplicate.
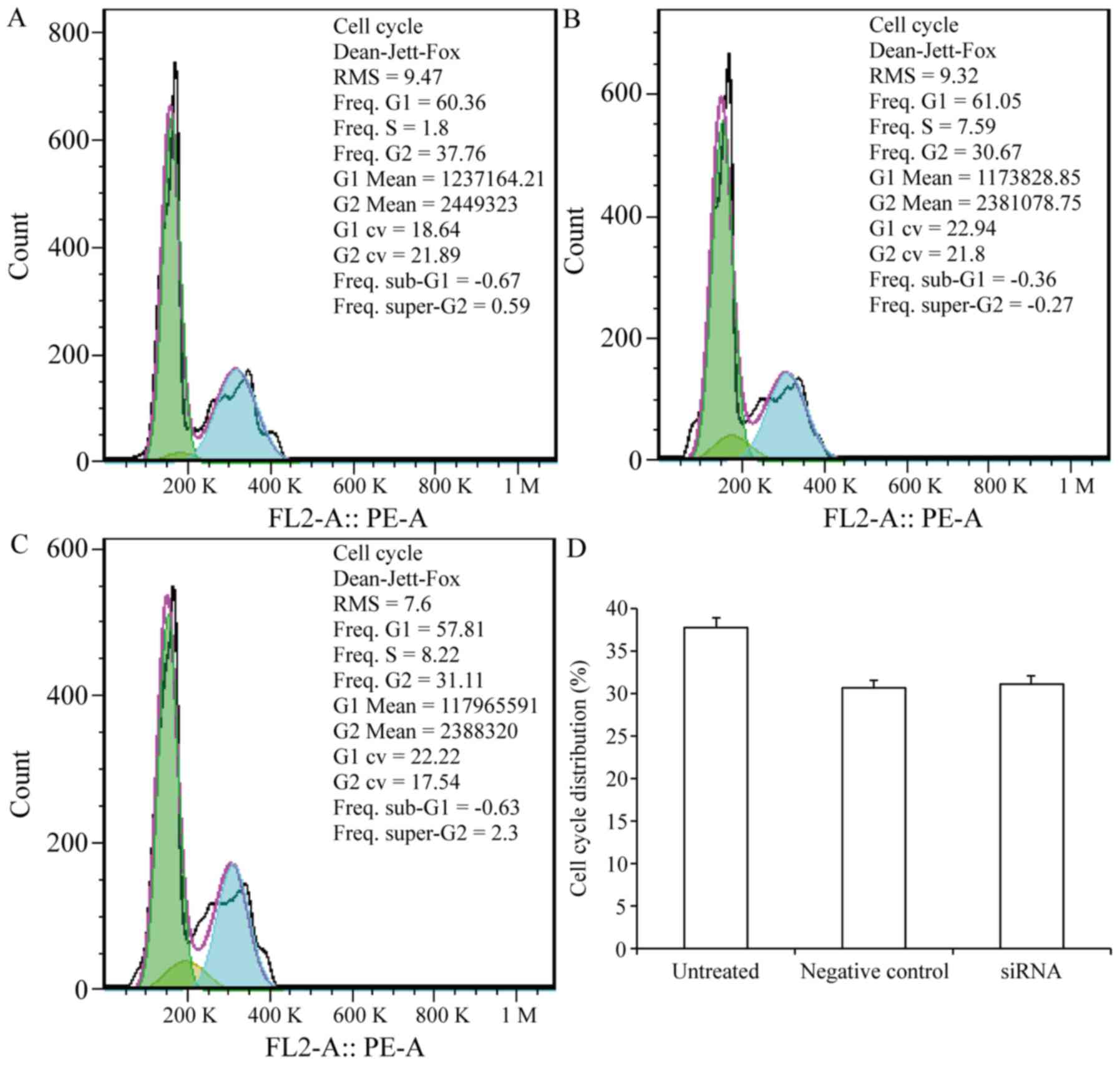
CEACAM-1 might function to inhibit carcinogenesis,
which is often associated with both cell cycle arrest and
activation of the cell death pathway. Therefore, the cell cycle
phase distribution was examined in the CEACAM-1-knockdown cells to
determine whether CEACAM-1 knockdown has an effect in cell cycle
arrest. The results revealed no significant difference in the % of
G2/M-phase cells that occurred in the CEACAM-1-knockdown
compared with the control group (P>0.05; Fig. 4). The experiment was performed in
triplicate.
Discussion
To date, the function of CEACAM-1 expression in
malignant tumors remains unclear. Previous studies have
investigated CEACAM expression via either immunohistochemical or
serum expression analyses (19,20). To
elucidate its functional role in CRC, the present study assessed
the effect of silencing CEACAM-1 expression on the viability and
proliferation of a CRC cell line CEACAM-1 belongs to a diverse
family of GPI-linked Igs that combine the structural features of
the Ig superfamily with the functional properties of cadherins
(21).
In the present study, CEACAM-1 expression was
inhibited in the HCT-8 cell line via three custom-designed siRNA
sequences, achieving a maximal reduction in CEACAM-1 mRNA
expression by 61.3% compared with the control cells, and a similar
reduction in CEACAM-1 protein production. Subsequently, it was
determined that CEACAM-1 downregulation significantly inhibited
cell proliferation and promoted cell apoptosis, suggesting that
CEACAM-1 may have a potential clinical use in the treatment of CRC.
Consistent with these results, a previous study has demonstrated
that CEACAM-1 knockdown results in the decreased proliferation and
migration of human pancreatic adenocarcinoma Pac 5061 cells
(17). Thus, it can be concluded that
CEACAM-1 is likely an important modulator of CRC.
Collectively, the results of the present study
demonstrated that CEACAM-1 downregulation in CRC cells
significantly inhibited cell proliferation and promoted apoptosis.
Thus, CEACAM-1 may be a critical mediator of CRC cell growth and
progression, and as a result, a promising potential target for
novel CRC treatment strategies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Key Project
of Universities and Colleges in Henan, China (grant no.
15A310033).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
Z-MH designed, analyzed the experiments and wrote
the manuscript. H-MH and Y-WS performed the experiments and
co-wrote the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Zeng H and Zhang S: The
incidence and mortality of major cancers in China, 2012. Chin J
Cancer. 35:732016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosso T, Malvezzi M, Bosetti C, Bertuccio
P, Negri E and La Vecchia C: Cancer mortality in Europe, 1970–2009:
An age, period, and cohort analysis. Eur J Cancer Prev. 27:88–102.
2016. View Article : Google Scholar
|
|
3
|
Cavallaro U and Christofori G: Cell
adhesion in tumor invasion and metastasis: Loss of the glue is not
enough. Biochim Biophys Acta. 1552:39–45. 2001.PubMed/NCBI
|
|
4
|
Pignatelli M and Vessey CJ: Adhesion
molecules: Novel molecular tools in tumor pathology. Hum Pathol.
25:849–856. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oettle H, Post S, Neuhaus P, Gellert K,
Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C,
et al: Adjuvant chemotherapy with gemcitabine vs observation in
patients undergoing curative-intent resection of pancreatic cancer:
A randomized controlled trial. JAMA. 297:267–277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rojas M, Fuks A and Stanners CP: Biliary
glycoprotein, a member of the immunoglobulin supergene family,
functions in vitro as a Ca2(+)-dependent intercellular adhesion
molecule. Cell Growth Differ. 1:527–533. 1990.PubMed/NCBI
|
|
7
|
Beauchemin N, Draber P, Dveksler G, Gold
P, Gray-Owen S, Grunert F, Hammarström S, Holmes KV, Karlsson A,
Kuroki M, et al: Redefined nomenclature for members of the
carcinoembryonic antigen family. Exp Cell Res. 252:243–249. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arabzadeh A, Chan C, Nouvion AL, Breton V,
Benlolo S, DeMarte L, Turbide C, Brodt P, Ferri L and Beauchemin N:
Host-related carcinoembryonic antigen cell adhesion molecule 1
promotes metastasis of colorectal cancer. Oncogene. 32:849–860.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thöm I, Schult-Kronefeld O, Burkholder I,
Schuch G, Andritzky B, Kastendieck H, Edler L, Wagener C, Bokemeyer
C, Schumacher U and Laack E: Expression of CEACAM-1 in pulmonary
adenocarcinomas and their metastases. Anticancer Res. 29:249–254.
2009.PubMed/NCBI
|
|
10
|
Thies A, Moll I, Berger J, Wagener C,
Brümmer J, Schulze HJ, Brunner G and Schumacher U: CEACAM1
expression in cutaneous malignant melanoma predicts the development
of metastatic disease. J Clin Oncol. 20:2530–2536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dango S, Sienel W, Schreiber M, Stremmel
C, Kirschbaum A, Pantel K and Passlick B: Elevated expression of
carcinoembryonic antigen-related cell adhesion molecule 1
(CEACAM-1) is associated with increased angiogenic potential in
non-small-cell lung cancer. Lung Cancer. 60:426–433. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoshikawa M, Morine Y, Ikemoto T, Imura S,
Higashijima J, Iwahashi S, Saito YU, Takasu C, Yamada S, Ishikawa
D, et al: Elevated preoperative serum CEA level is associated with
poor prognosis in patients with hepatocellular carcinoma through
the epithelial-mesenchymal transition. Anticancer Res.
37:1169–1175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li N, Yang JY, Wang XY, Wang HT, Guan BX
and Zhou CJ: Carcinoembryonic antigen-related cell adhesion
molecule 1 is expressed and as a function histotype in ovarian
tumors. Ann Diagn Pathol. 20:7–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gebauer F, Wicklein D, Horst J, Sundermann
P, Maar H, Streichert T, Tachezy M, Izbicki JR, Bockhorn M and
Schumacher U: Carcinoembryonic antigen-related cell adhesion
molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer.
PLoS One. 9:e1130232014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y and Shively JE: CEACAM1 regulates
Fas-mediated apoptosis in Jurkat T-cells via its interaction with
β-catenin. Exp Cell Res. 319:1061–1072. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu GX, Xie Q, Zhou CJ, Zhang XY, Ma BL,
Wang CQ, Wei FC, Qu X and Sun SZ: The possible roles of
OPN-regulated CEACAM1 expression in promoting the survival of
activated T cells and the apoptosis of oral keratinocytes in oral
lichen planus patients. J Clin Immunol. 31:827–839. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Beauchemin N and Arabzadeh A:
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs)
in cancer progression and metastasis. Cancer Metastasis Rev.
32:643–671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Simeone DM, Ji B, Banerjee M, Arumugam T,
Li D, Anderson MA, Bamberger AM, Greenson J, Brand RE, Ramachandran
V and Logsdon CD: CEACAM1, a novel serum biomarker for pancreatic
cancer. Pancreas. 34:436–443. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blumenthal RD, Leon E, Hansen HJ and
Goldenberg DM: Expression patterns of CEACAM5 and CEACAM6 in
primary and metastatic cancers. BMC Cancer. 7:22007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hauck W, Nédellec P, Turbide C, Stanners
CP, Barnett TR and Beauchemin N: Transcriptional control of the
human biliary glycoprotein gene, a CEA gene family member
down-regulated in colorectal carcinomas. Eur J Biochem.
223:529–541. 1994. View Article : Google Scholar : PubMed/NCBI
|