Introduction
According to 2015 statistics, lung cancer is
considered to be the most common type of cancer and the leading
cause of cancer-associated mortality in China (1). Similar statistics have been observed
worldwide, with lung cancer being diagnosed in 1.8 million people
in 2012, resulting in 1.6 million cases of mortality, rendering it
the most common cause of cancer-associated mortality in men and the
second most common in women (2).
Adenocarcinoma is the most common form of lung cancer, originating
in peripheral lung tissue and causing almost 40% all lung cancer
cases (3). The most important risk
factor associated with lung adenocarcinoma is tobacco smoking.
Obtaining a reliable diagnosis and identifying prognostic markers
for lung cancer remains a challenge.
The anillin actin binding protein (ANLN) gene
is located on chromosome 7p14.2, and encodes a protein consisting
of 1,124 amino acids that form 4 structural domains, including a
myosin- and actin-binding domain, a RhoA-binding domain and a
C-terminal pleckstrin homology domain (4,5). The ANLN
protein is located in the nucleus, cytoplasm, cytoskeleton,
cleavage furrow and cell cortex, and is expressed in adult
placenta, testis, and the spinal cord, and in numerous fetal organs
(6). ANLN was initially characterized
as a human homologue of anillin, a Drosophila actin-binding protein
that is present in the cortex following breakdown of the nuclear
envelope, and in the cleavage furrow during cytokinesis (7). Anillin serves an important role in
cell-cycle progression and in the assembly of the actin and myosin
contractile ring that separates daughter cells (6). Importantly, anillin has been
demonstrated to be a substrate of the anaphase-promoting
complex/cyclosome (APC/C), a typeof ubiquitin ligase that controls
mitotic progression (8).
As summarized above, anillin is a conserved protein,
which functions in cytoskeletal dynamics during cellularization and
cytokinesis. Previous studies have indicated that knockdown of
anillinresults in cleavage furrow ingression and failure of
cytokinesis in multinucleated monkey BS-C-1 cells (9). The association between cell cycle
regulation and carcinoma is well established. ANLN has been
indicated to be a marker of poor prognosis, and associated with
aggressive cancer phenotypes (10).
The role of anillin as a regulator of the cell cycle serves an
important role in breast and pancreatic cancer invasion (11,12). ANLN
has been developed as a clinically applicable
immunohistochemistry-based prognostic biomarker for hepatocellular
carcinoma (13). Phylogenetic
analysis aims to understand the conservation characterization of
analyzed proteins. In the present study, the significance of ANLN
protein in human lung adenocarcinoma cancer progression was
investigated using The Cancer Genome Atlas (TCGA) database.
Materials and methods
Data acquisition
The ANLN protein sequences of 11 species (Homo
Sapiens, Chimpanzee, Mouse, Zebrafish, Chicken, Dog, Sheep, Cattle,
Rhesus monkey, Rat and Pig) were downloaded from UniProt
(http://www.uniprot.org/) and the National Center
for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). The mutation statuses
of ANLN were checked using cBioPortal for Cancer Genomics
(http://www.cbioportal.org). Protein
sequence alignment was conducted using Molecular Evolutionary
Genetics Analysis software (version 7; MEGA7) (14). The PyMOL Molecular Graphics System
(version 1.8; Schrödinger, LLC, New York City, NY, USA) was
employed to predict protein functional changes in response to
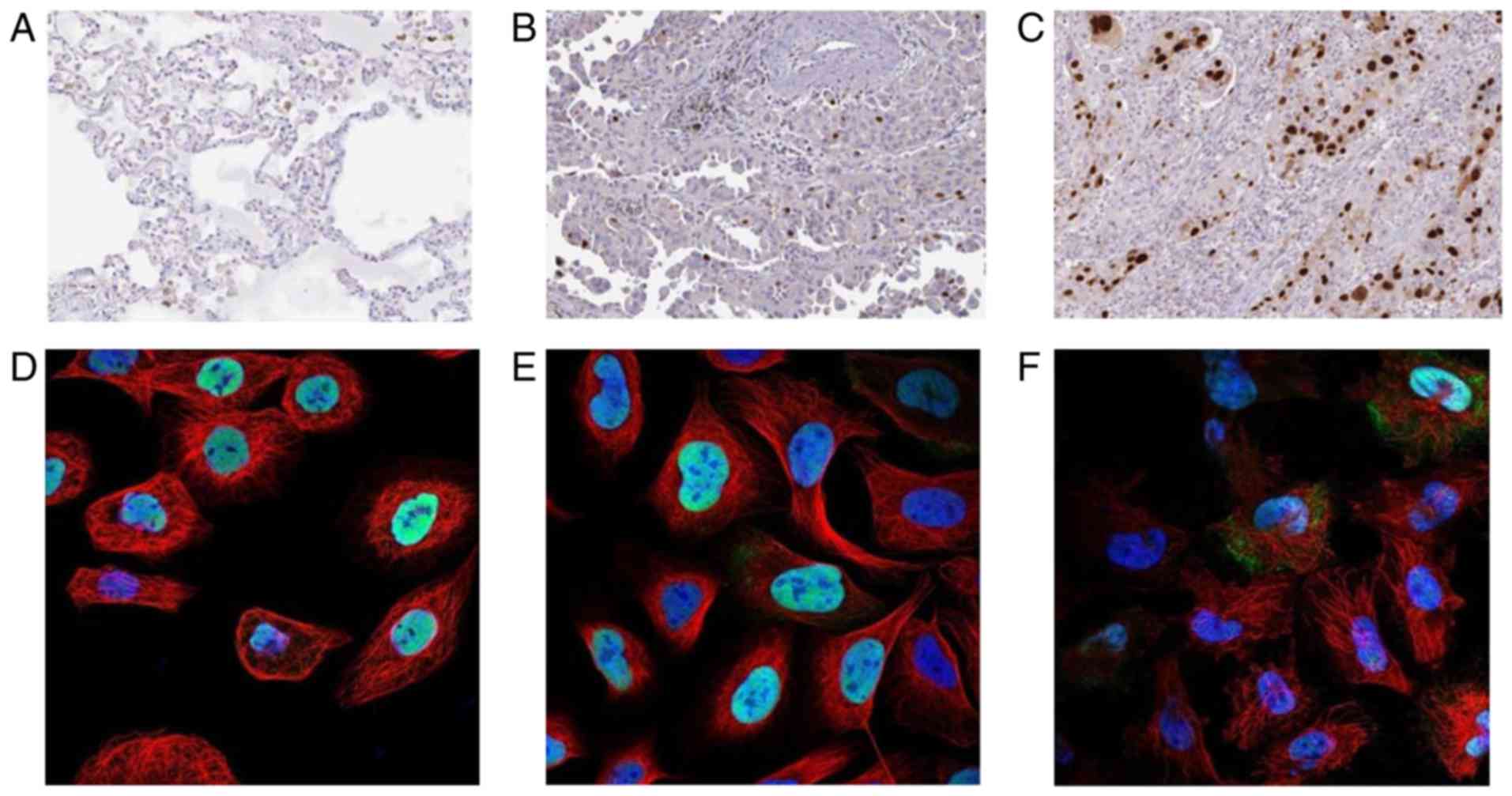
mutations. All immunohistochemical staining images (Fig. 2) were adapted from the Human Protein
Atlas (http://www.proteinatlas.org/humancell). ANLN clinical
and RNA-seq data were downloaded from The Cancer Genome Atlas
(TCGA; https://portal.gdc.cancer.gov/). Gene
set enrichment analysis (GSEA, Desktop version 3.0) (15) was used to determine whether any
prior-defined pathway or set of genes demonstrated statistically
significant differences between two biological states (e.g.,
phenotypes, RNA-seq expression level in current analysis).
Phylogenetic analysis
Phylogenetic relationships in ANLN expression among
the 11 different species were determined using MEGA7 (14). A total of 11 amino acid sequences were
analyzed, and all phylogenetic positions containing missing data
were eliminated. There were a total of 157 positions in the final
dataset. The evolutionary history was inferred using the
Neighbor-Joining method (16). The
phylogenetic tree was drawn to scale, using branch lengths in the
same units as those of the evolutionary distances (number of amino
acid substitutions per site), which were calculated using the
Poisson correction method (17).
Statistical analysis
All statistical analyses were conducted using R
software (Version 3.3.1, http://www.R-project.org/). Monte Carlo simulation
(18) was employed to compare RNA-seq
expression levels between patients with lung adenocarcinoma and
normal control tissues. χ2 was used to analyze
differences in survival. The false discovery rate (FDR) method was
employed to conceptualize the rate of type I errors in null
hypothesis testing when conducting multiple comparisons in GSEA
analysis (19).
Results
ANLN mutations and evolutionary
conservation in lung adenocarcinoma
The identified mutations of ANLN in cancer
include amplification, deletion and single nucleotide polymorphism
(SNP) mutations (missense, nonsense and splice errors) (TCGA
database, https://cancergenome.nih.gov/). The mutation rate has
been demonstrated to differ according to the type of cancer,
ranging from 19.6% in prostate cancer (20) to 0.2% in clear cell renal cell
carcinoma (21). A total of 27
mutations were identified in 446 patients with lung adenocarcinoma
(Fig. 1A and B) using TCGA, including
12 amplifications, 2 deletions and 13 SNP mutations. The 13 SNP
mutations included 4 G>T, 3 G>A, 2 G>C, 1 C>T and 1
C>A substitutions, among which 5 mutations influenced conserved
amino acids sequences (Fig. 1A). A
phylogenetic tree was created from the evolutionary analysis
performed using MEGA7 software (Fig.
1C). The optimal phylogenetic tree is presented, with a sum
branch length of 0.897. The ANLN protein sequences from Homo
sapiens and Chimpanzee are closely associated, indicating a
recent common ancestor. Protein function prediction conducted with
PyMOL suggested that these mutations (S594N, G659V, I1014V and
R1026L) may influence ionic bonds and β-folding (Fig. 1D) in the ANLN protein. However,
whether these mutations cause gain or loss function remains to be
elucidated.
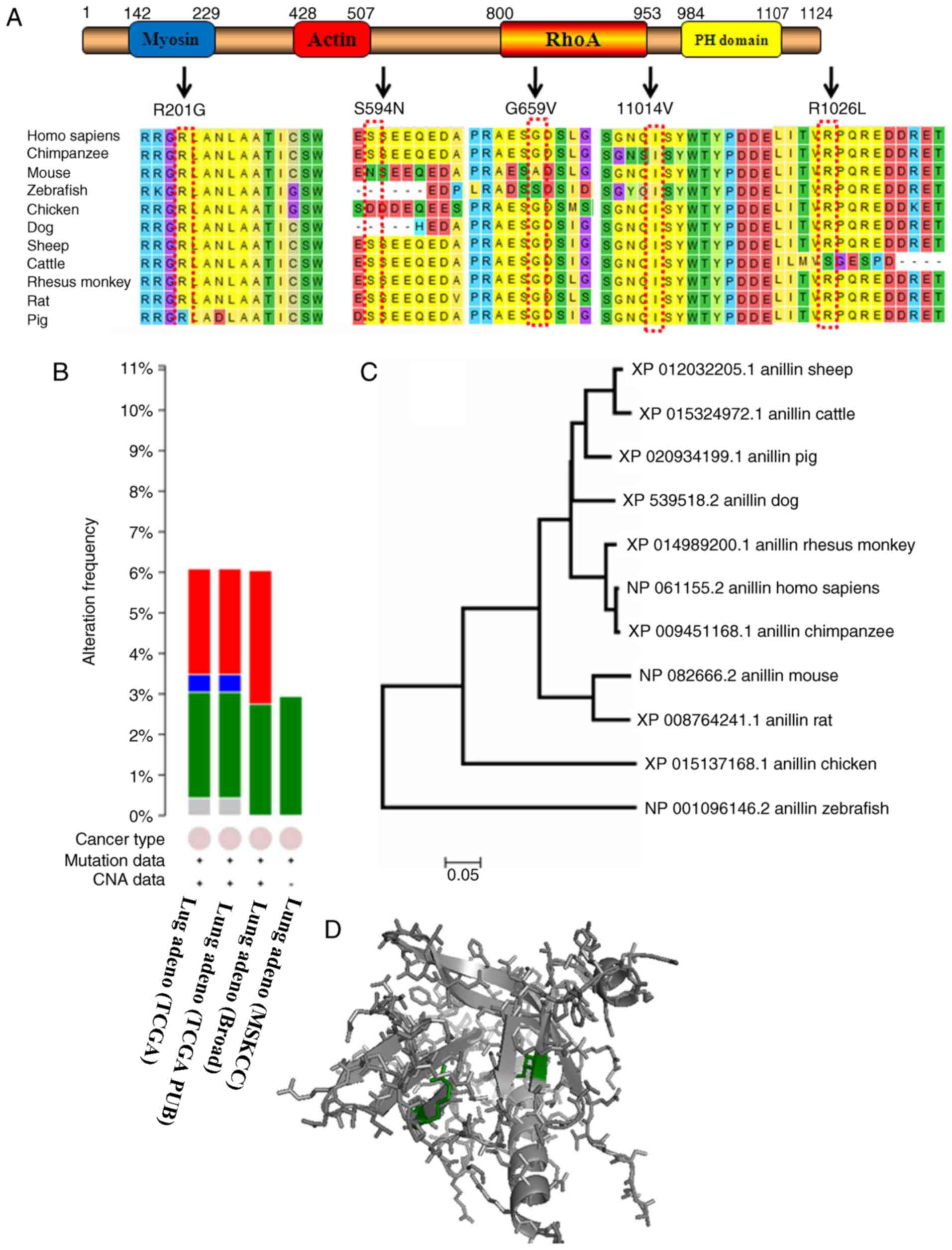 | Figure 1.(A) The primary structure of the ANLN
protein and how 5 mutations alter the sequence in 11 species using
MEGA7.0 software. (B) ANLN mutations in patients with lung
adenocarcinoma using cBioPortal (Four lung adenocarcinoma
researches from TCGA database, including Lung adenocarcinoma
(TCGA), Lung adenocarcinoma (TCGA PUB), Lung adenocarcinoma (Broad)
and Lung adenocarcinoma (MSKCC) citied in the TCGA database. Red
section is amplification, green is mutation, blue is deep deletion,
and grey is multiple alterations). (C) The phylogenetic
relationships regarding the ANLN protein among 11 species using
MEGA7.0 software. (D) Prediction of changes in the ANLN
protein as a result of mutations S594N, G659V, I1014V and R1026L
using PyMOL software. Green indicates alternative positions of
mutations S594N, G659V, I1014V and R1026L. ANLN, anillin actin
binding protein. |
ANLN expression in lung
adenocarcinoma
Immunohistochemical staining of the ANLN protein
indicated that it is located in the nucleus, and that patients with
lung adenocarcinoma exhibit increased ANLN expression compared with
healthy controls (Fig. 2).
ANLNRNA-seq levels were increased in patients with lung
adenocarcinoma compared with normal control tissues (Fig. 3A; Monto Carlo Simulation, P<0.001)
and were significantly associated with high Tumor-Node-Metastasis
pathological stage (Fig. 3B;
P<0.05), suggesting that ANLN can predict the disease
progression and outcome of patients with lung adenocarcinoma.
Kaplan-Meier survival curve analysis demonstrated that the curves
for high and low expression of ANLN were significantly different,
and that high expression of ANLN was predictive of a poor outcome
(Fig. 3C). However, it ANLN RNA-seq
levels were not demonstrated to be different between patients with
and without ANLN mutations (Fig.
3D).
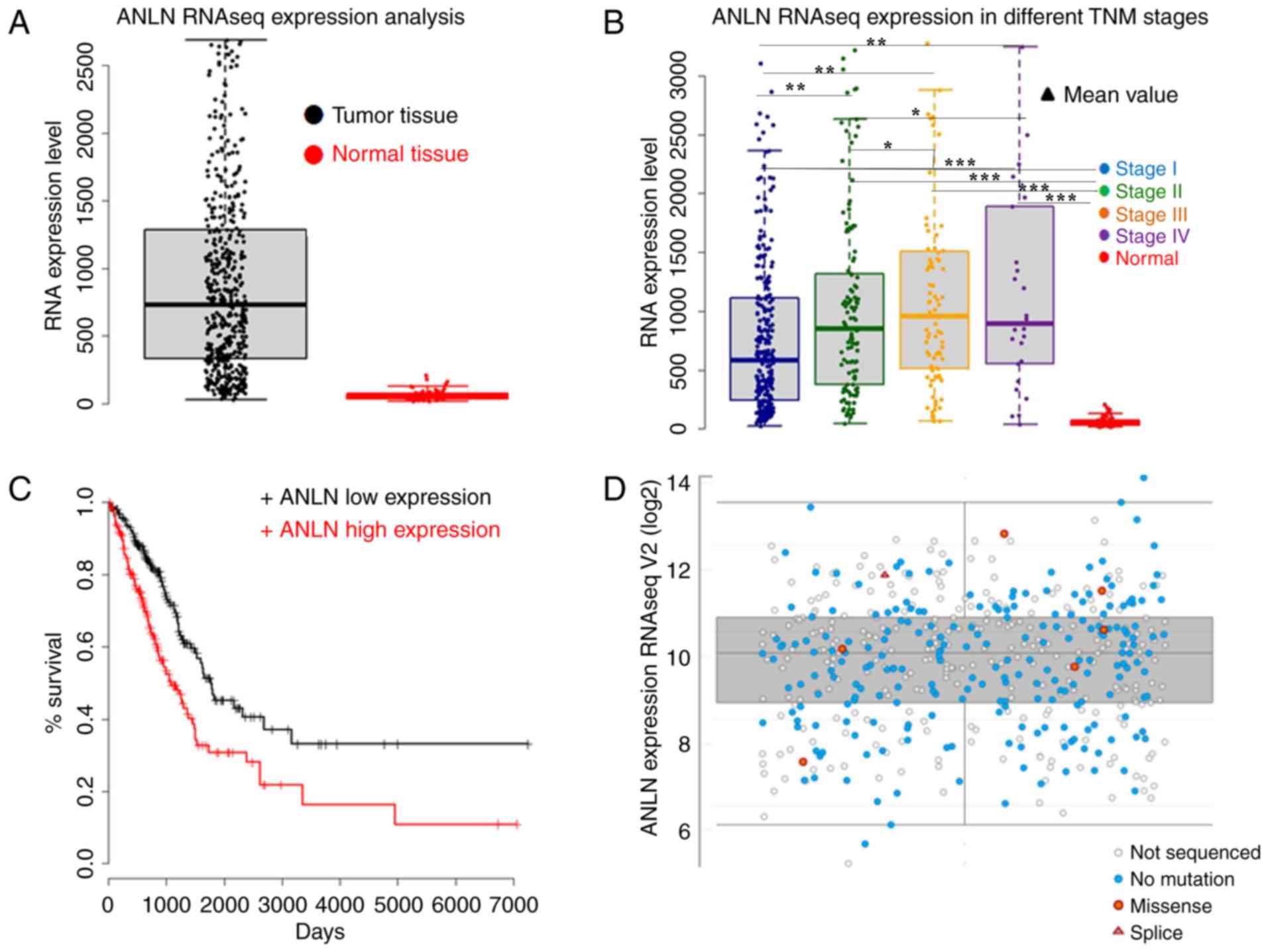 | Figure 3.(A) Comparison of ANLN RNA-seq
expression between tumor tissues and normal tissues (Monte Carlo
Simulation, P<0.001). (B) ANLN RNA-seq expression
increases with TNM stage (Monte Carlo Simulation, Stage I vs. Stage
II, P<0.01; Stage I vs. Stage III, P<0.01; Stage I vs. Stage
IV, P<0.01; Stage II vs. Stage III, P<0.05; Stage II vs.
Stage IV, P<0.05; Stage III vs. Stage IV, P>0.05; All Stage
vs. Normal, P<0.001). (C) Survival curve analysis of ANLN
RNA-seq expression. The two curves are significantly difference
(P=0000138). (D) ANLN RNA-seq expression in patients with
different mutations or without mutation. ANLN, anillin actin
binding protein; RNA-seq, RNA sequencing; TNM,
Tumor-Node-Metastasis. ***P<0.001; **P<0.01; *P<0.05. |
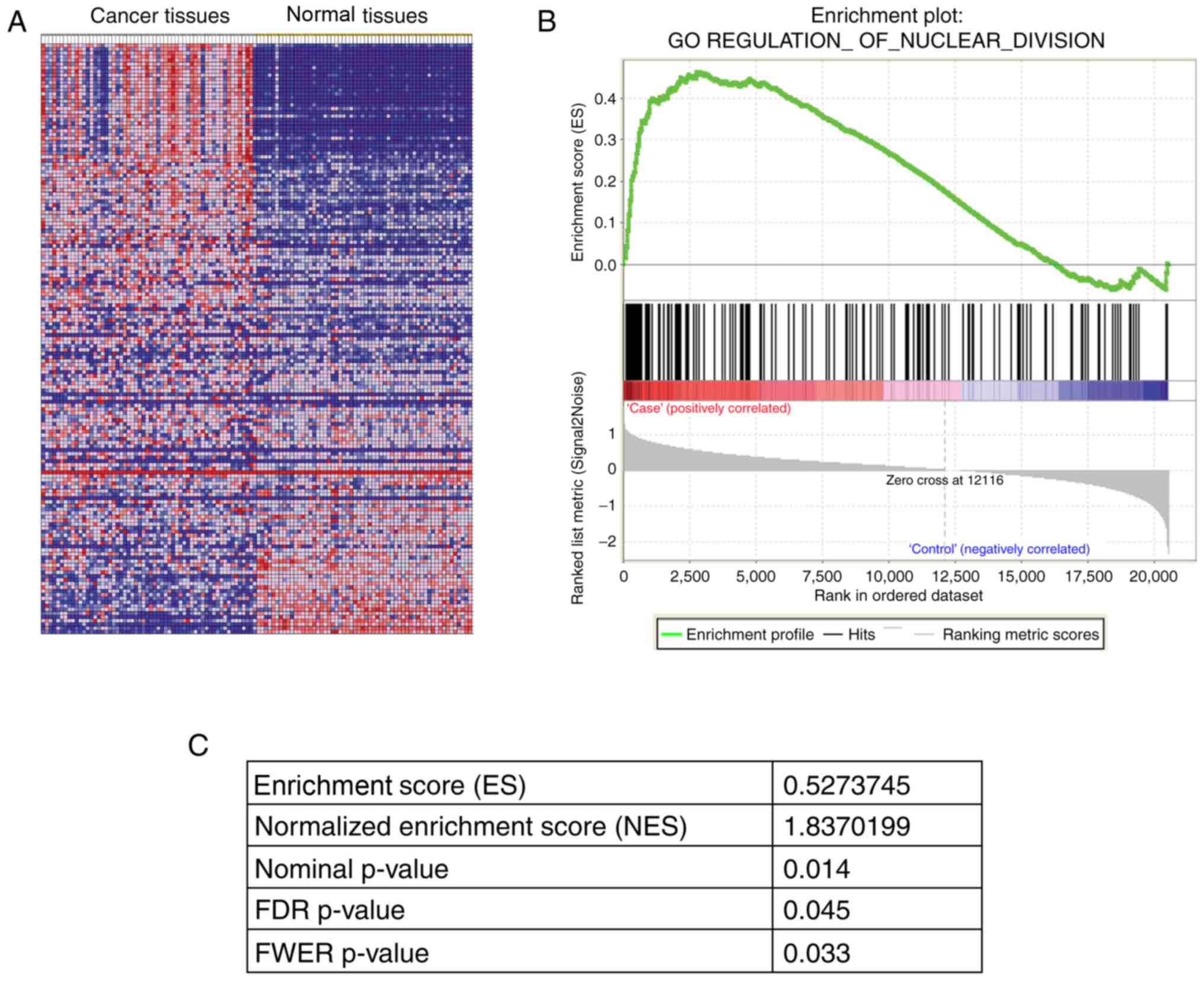
ANLN specified pathway in lung
adenocarcinoma
A total of 59 pairs of lung adenocarcinoma RNA-seq
data were extracted for further pathway analysis. A total of 24
Gene Ontology sets and 178 Kyoto Encyclopedia of Genes and Genomes
pathways were demonstrated to be associated with ANLN expression
(All analysis was done by using GSEA software). The results of GSEA
indicated that the pathway of ‘regulation of nuclear division’
(Fig. 4A and B) was enriched in
patients with lung adenocarcinoma (FDR, q=0.023). The enrichment
score was 0.464 and the normalized enrichment score was 1.834
(Fig. 4C).
Discussion
The present study indicates that ANLN may be a
potential diagnostic and prognostic biomarker for lung
adenocarcinoma based on the following findings. A 6% incidence of
genetic change in ANLN was identified in patients with lung
adenocarcinoma. Approximately 50% amplifications (12/27) lead to
high expression of ANLN (Fig. 1B),
however, the association between other mutations and ANLN protein
function require further investigation. Immunohistochemical
staining of ANLN in patients with lung adenocarcinoma revealed that
it was more highly expressed in adenocarcinoma tissues than in
normal tissues, which was also supported by the RNA-seq comparison.
The ANLN RNA-seq expression level was associated with TNM
stage in lung adenocarcinoma, as well as overall survival rate,
suggesting that high expression of ANLN in lung
adenocarcinoma is predictive of poor prognosis. GSEA identified
that the pathway of ‘regulation of nuclear division’, involving
ANLN, was significantly upregulatedin subjects with lung
adenocarcinoma compared with normal controls.
Suzuki et al (22) have investigated the significance of
ANLN gene in lung cancer using cDNA microarray. In their
latest functional experiment, the growth of non-small-cell lung
carcinoma (NSCLC) was suppressed using ANLN small
interfering RNAs (22). Furthermore,
induction of exogenous overexpression of ANLN increased the
migratory ability of mammalian cells via interaction with RhoA.
Univariate analysis of gene expression in 66 patients with squamous
cell carcinoma who had undergone surgical resection identified that
ANLN had significant prognostic value (22). This was positively validated in an
independent study with a separate cohort of 26 patients (23). All of the above results were supported
by the present study.
The regulation of nuclear division pathway,
involving ANLN denotes a cell cycle process that comprises the
steps by which the nucleus of a eukaryotic cell divides, including
the condensation of chromosomal DNA into a highly compact form
(24). Inheritance of a defective
genome and its mitotic proliferation can lead to pathological
conditions, including numerous types of cancer (25). The role of anillin as a cell cycle
regulator has been demonstrated to serve an important role in
carcinoma invasion (26). However,
the detailed mechanisms behind these effects require further
investigation.
Limitations of the present study include its
retrospective nature, and the use of external databases. The
majority of patients on TCGA database were of European descent, and
further research using a boarder patient spectrum is required.
In conclusion, the present study established that
ANLN protein is a potential prognostic marker for lung
adenocarcinoma. However, further investigation is required to
elucidate the mechanism of the involvement of ANLN in cell
cycle-regulation and the development of adenocarcinoma.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
In the current investigation, the ANLN protein
sequences were downloaded from UniProt (uniprot.org/) and also the National Center for
Biotechnology Information (NCBI; ncbi.nlm.nih.gov/). ANLN clinical and RNA-seq data
were downloaded from The Cancer Genome Atlas (TCGA; portal.gdc.cancer.gov/).
Authors' contributions
XL, WZ and YW performed the sequencing and
statistical analyses and manuscript preparing. SL was responsible
for study conception and design.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization (WHO): Global
status report on alcohol and health 2014. WHO; Geneva; 2014
|
|
3
|
Wu-Williams AH, Dai XD, Blot W, Xu ZY, Sun
XW, Xiao HP, Stone BJ, Yu SF, Feng YP and Ershow AG: Lung cancer
among women in north-east China. Br J Cancer. 62:982–987. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strausberg RL, Feingold EA, Grouse LH,
Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler
GD, Altschul SF, et al: Generation and initial analysis of more
than 15,000 full-length human and mouse cDNA sequences. Proc Natl
Acad Sci USA. 99:16899–16903. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Straight AF, Field CM and Mitchison TJ:
Anillin binds nonmuscle myosin II and regulates the contractile
ring. Mol Biol Cell. 16:193–201. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oegema K, Savoian MS, Mitchison TJ and
Field CM: Functional analysis of a human homologue of the
Drosophila actin binding protein anillin suggests a role in
cytokinesis. J Cell Biol. 150:539–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Monzo P, Gauthier NC, Keslair F, Loubat A,
Field CM, Le Marchand-Brustel Y and Cormont M: Clues to
CD2-associated protein involvement in cytokinesis. Mol Biol Cell.
16:2891–2902. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oegema K, Savoian MS, Mitchison TJ and
Field CM: Functional analysis of a human homologue of the
Drosophila actin binding protein anillin suggests a role in
cytokinesis. J Cell Biol. 150:539–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ronkainen H, Hirvikoski P, Kauppila S and
Vaarala MH: Anillin expression is a marker of favourable prognosis
in patients with renal cell carcinoma. Oncol Rep. 25:129–133.
2011.PubMed/NCBI
|
|
11
|
Wang Z, Chen J, Zhong MZ, Huang J, Hu YP,
Feng DY, Zhou ZJ, Luo X, Liu ZQ, Jiang WZ and Zhou WB:
Overexpression of ANLN contributed to poor prognosis of
anthracycline-based chemotherapy in breast cancer patients. Cancer
Chemother Pharmacol. 79:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Olakowski M, Tyszkiewicz T, Jarzab M, Krol
R, Oczko-Wojciechowska M, Kowalska M, Kowal M, Gala GM, Kajor M,
Lange D, et al: NBL1 and anillin (ANLN) genes over-expression in
pancreatic carcinoma. Folia Histochem Cytobiol. 47:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim H, Kim K, Yu SJ, Jang ES, Yu J, Cho G,
Yoon JH and Kim Y: Development of biomarkers for screening
hepatocellular carcinoma using global data mining and multiple
reaction monitoring. PLoS One. 8:e634682013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kumar S, Stecher G and Tamura K: MEGA7:
Molecular evolutionary genetics analysis version 7.0 for bigger
datasets. Mol Biol Evol. 33:1870–1874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Subramanian A, Kuehn H, Gould J, Tamayo P
and Mesirov JP: GSEA-P: A desktop application for gene set
enrichment analysis. Bioinformatics. 23:3251–3253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saitou N and Nei M: The neighbor-joining
method: A new method for reconstructing phylogenetic trees. Mol
Biol Evol. 4:406–425. 1987.PubMed/NCBI
|
|
17
|
Zuckerkandl E and Pauling L: Evolutionary
divergence and convergence in proteins. Evol Genes Proteins.
97:97–166. 1965. View Article : Google Scholar
|
|
18
|
Mahadevan S: Monte Carlo Simulation. In:
Mechanical Engineering. Marcel Dekker. New York, NY and Basel; pp.
123–146. 1997
|
|
19
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statistical Soc Ser B (Methodological).
57:289–300. 1995.
|
|
20
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of clear cell renal cell
carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Skrzypski M, Jassem E, Taron M, Sanchez
JJ, Mendez P, Rzyman W, Gulida G, Raz D, Jablons D, Provencio M, et
al: Three-gene expression signature predicts survival in
early-stage squamous cell carcinoma of the lung. Clin Cancer Res.
14:4794–4799. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nigg EA: Mitotic kinases as regulators of
cell division and its checkpoints. Nat Rev Mol Cell Biol. 2:21–32.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brinkley BR: Managing the centrosome
numbers game: From chaos to stability in cancer cell division.
Trends Cell Biol. 11:18–21. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park MT and Lee SJ: Cell cycle and cancer.
J Biochem Mol Biol. 36:60–65. 2003.PubMed/NCBI
|