Introduction
Gastric cancer (GC) leads to a major health burden
worldwide (1). Approximately 679,100
novel cases and 498,000 mortalities were estimated in China in 2015
(2). Although the incidence and
mortality rates have declined in the West, the overall 5-year
survival rate remains at ~20% (3).
The need for an effective biomarker for the diagnosis of early
asymptomatic GC remains challenging (4). In current clinical practices, several
serum tumor markers, including CA (carbohydrate antigen) 19-9,
carcinoembryonic antigen and CA125, have been widely used, but
their specificity is ambiguous (5).
Furthermore, the current treatment options for advanced GC lack
specificity, resulting in a large number of side effects and drug
resistance. Thus, there is an urgent requirement to investigate
novel biomarkers and therapeutic targets for GC.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs of length >200 nucleotides that do not encode
proteins (6). Previous studies have
indicated the important function of lncRNAs in cancer development
(7,8).
Sun et al (9) reported that
the lncRNA GClnc1 acted as a scaffold to recruit the WDR5
and KAT2A complex and regulated the transcription of target genes.
Additionally, the lncRNA HOXA11-AS acts as a scaffold to
link chromatin modification factors (PRC2, LSD1 and DNMT1), and
promoted cell proliferation and invasion in GC (10), suggesting that these lncRNAs may serve
as therapeutic targets in GC. Genome-wide analysis has revealed
that lncRNAs exhibit tissue-specific and cancer-specific expression
patterns (11,12). A recent study has identified several
novel circulating lncRNAs for the diagnosis of GC, using
genome-wide lncRNA microarrays (13).
One of those lncRNAs was LINC00857, whose expression was
increased in the tumor tissue and in the serum of patients with GC.
In addition, LINC00857 was upregulated in patients with lung
cancer and was able to promote cell proliferation, colony formation
and invasion (14). However, the
underlying molecular mechanism of LINC00857 in GC
development remains unclear.
In the present study, it was demonstrated that
LINC00857 expression was increased in GC tissues and
associated with poor patient survival rates. Overexpression of
LINC00857 in GC cells promoted cell proliferation and
migration, suggesting that LINC00857 may be used as a novel
biomarker and therapeutic target for GC.
Materials and methods
Patients and clinical samples
A total of 60 patients with GC were enrolled in the
present study, which complied with the Ethics Review Board at the
Yongchuan Hospital of Chongqing Medical University (Chongqing,
China). Written informed consent was obtained from all patients. In
total, 60 pairs of fresh GC tissues and adjacent non-tumor tissues
were collected from patients who had not undergone any treatment at
the Yongchuan Hospital of Chongqing Medical University between
April 2012 and December 2015. The tissue samples were collected in
the operating room and processed within 15 min, and the
non-tumorous samples were taken at a distance of >5 cm from the
tumor. Clinical features of patients with GC included in the
present study are presented in Table
I.
 | Table I.Clinical characterization of patients
with GC. |
Table I.
Clinical characterization of patients
with GC.
| Variable | Gastric cancer, n
(%) |
|---|
| Sex |
|
| Male | 36 (60) |
|
Female | 24 (40) |
| Age, years |
|
| ≥57 | 31 (52) |
| ≤57 | 29 (48) |
| Histology |
|
|
Adenocarcinoma | 52 (87) |
| Mucinous
adenocarcinoma | 8
(13) |
| TNM stage |
|
| I/II | 18 (30) |
|
III/IV | 42 (70) |
| Lymph node
metastasis |
|
| Yes | 47 (78) |
| No | 13 (22) |
Cell culture
Four GC cell lines (AGS, BGC-823, MKN-45 and
SGC-7901) and a gastric epithelial cell line (GES-1) were purchased
from BeNa Culture Collection (Beijing China). AGS cells were
cultured in Ham's F12 medium (HyClone, GE Healthcare Life Sciences,
Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the
remaining cell lines were cultured in Dulbecco's modified Eagle's
medium/high glucose medium (HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS at 37°C in 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue and cell samples
using TRIzol reagent (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. Total RNA was resuspended in 50 µl
pre-heated (65°C) nuclease-free water, subpackaged into two
nuclease-free tubes and stored at −80°C. A PrimeScript RT reagent
kit (Takara Biotechnology Co., Ltd., Dalian, China) and a Permix Ex
Taq kit (Takara Biotechnology Co., Ltd.) were used to perform
RT-qPCR assays according to the manufacturer's protocol. Briefly,
cDNA was synthesized in a 20 µl reaction volume containing 4 µl 5X
RT buffer, 4 µl nuclease-free water, 1 µl Prime reverse
transcriptase, 1 µl RT primer mixture and 10 µl total RNA following
genomic DNA removal at 37°C for 15 min. qPCR experiments were
performed in a 20 µl reaction volume on a Bio-Rad IQ5 thermocycler
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the
parameters: 95°C for 2 min, 40 cycles of 95°C for 15 sec and 60°C
for 30 sec. The primer sequences for LINC00857 were as
follows: 5′-CCCCTGCTTCATTGTTTCCC-3′ (forward) and
5′-AGCTTGTCCTTCTTGGGTACT-3′ (reverse). The primer sequences for
GAPDH were as follows: 5′-GGTGGTCTCCTCTGACTTCAACA-3′ (forward) and
5′-TCTCTTCCTCTTGTGCTCTTGCT-3′ (reverse). LINC00857
expression was normalized using the 2−ΔΔCq method
(15) relative to the mRNA expression
of GAPDH.
Western blotting
SGC-7901 cells (5×105) were seeded in
6-well plates and transfected with small interfering RNAs (siRNAs).
After 24 h, the cells were harvested and lysed in
radioimmunoprecipitation assay buffer (Roche Diagnostics, Basel,
Switzerland). Total proteins were measured using a BCA kit (Thermo
Fisher Scientific, Inc.), and 20 µg was loaded for 12% SDS-PAGE and
separated, followed by transferred onto polyvinylidene difluoride
(PVDF) membranes and blocked in PBS containing 3% bovine serum
albumin at room temperature for 1 h. Anti-F cyclin D1 rabbit
(dilution, 1:500; cat. no. 2978), anti-cyclin E1 mouse (dilution,
1:500; cat. no. 4129) and anti-GAPDH rabbit (dilution, 1:1,000;
cat. no. 5174) antibodies (Cell Signaling Technology, Inc.,
Danvers, MA, USA) were incubated at 4°C overnight. The
goat-anti-rabbit (cat. no. ZB-2301) or goat-anti-mouse (cat. no.
ZB-2305) HRP-conjugated secondary antibodies (1:10,000; ZSGB-BIO,
Beijing, China) were incubated at room temperature for 1 h.
Finally, the PVDF membranes were visualized using SuperSignal West
Dura Extended Duration Substrate kit (Thermo Fisher Scientific,
Inc.).
Oligonucleotide transfection
SGC-7901 cells (5×105) and MKN 45 cells
(5×105) were seeded in 6-well plates, incubated for 24
h, and then 200 ng/ml specific siRNAs for LINC00857 or
negative control (NC) were transfected into the cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol, and the
cells were harvested after 24 h. The sequences of siRNAs were as
follows: siRNA1, 5′-GGUAAGGGAAGGUGGAGAAUU-3′; siRNA2,
5′-GGCUAUGUGCUGUGAACAAUU-3′.
Cell proliferation
SGC-7901 and MKN-45 cells transfected with
LINC00857 siRNAs or NC were harvested and diluted at a
density of 104 cells/ml, prior to seeding in 96-well
plates. Cell proliferation was analyzed using a Cell Counting Kit-8
kit (CCK-8; Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's protocol. Absorbance at 450 nm was
measured when the reagent reacted for 24, 48, 72 or 96 h at room
temperature.
Statistical analysis
Data were analyzed using SPSS software (version
19.0; IBM Corp., Armonk, NY, USA) and GraphPad Prism (version 6.0;
GraphPad Software, Inc., La Jolla, CA, USA). All data are presented
as the mean ± standard deviation. The differences between two
groups were analyzed using the unpaired Student's t-test. One-way
analysis of variance with post hoc comparison using Sidak's
multiple comparisons test was used for multiple groups. Patient
survival was calculated using the Kaplan-Meier estimator method.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Increased expression of LINC00857 is
associated with poor survival rate in patients with GC
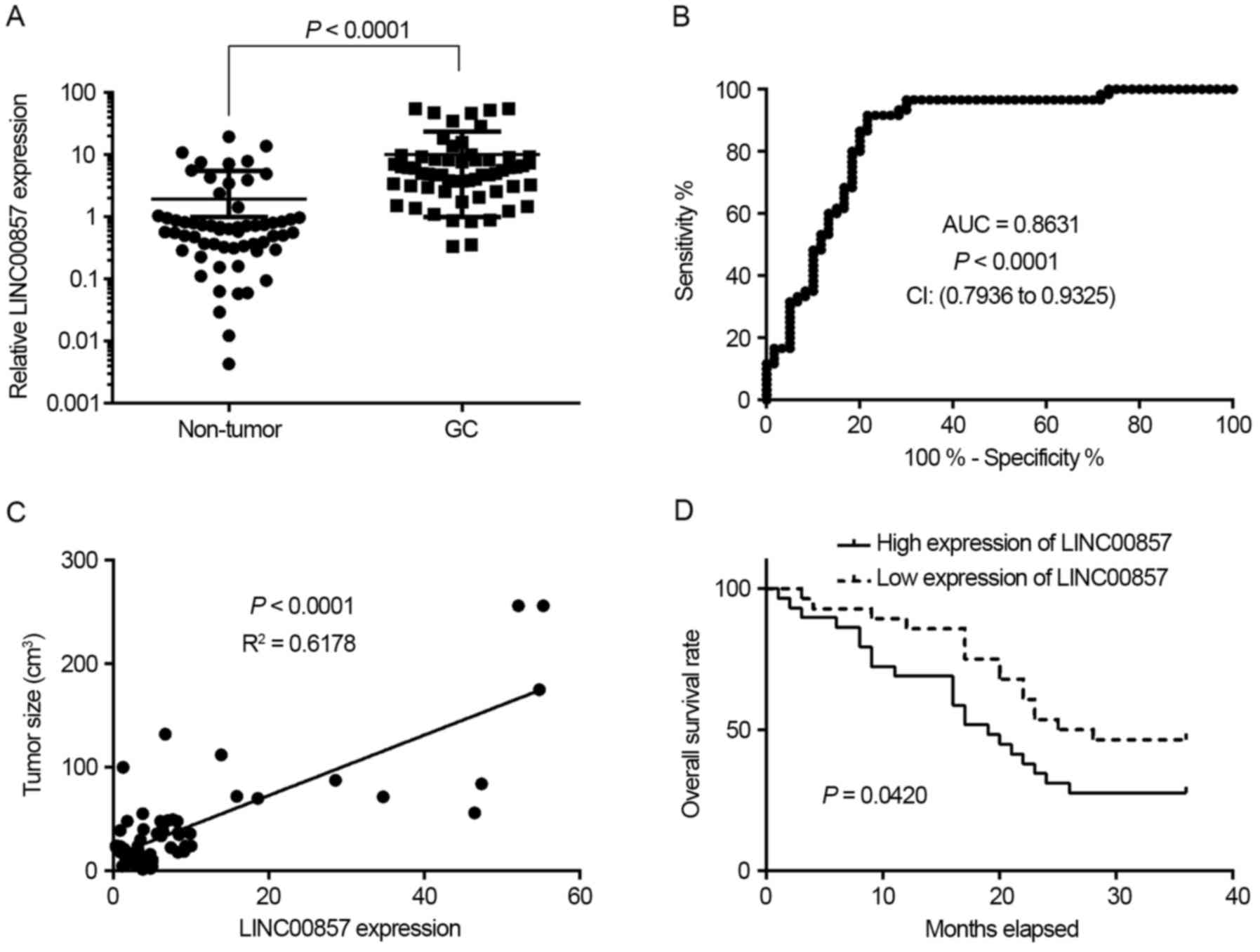
To investigate LINC00857 expression in GC, 60
pairs of GC tissues and adjacent non-tumor tissues were collected.
RT-qPCR experiments demonstrated that LINC00857 expression
was significantly increased in GC tissues compared with non-tumor
tissues (P<0.0001; Fig. 1A).
Additionally, receiver operating characteristic (ROC) curve
analysis indicated that LINC00857 expression signature
exhibited an increased area under the curve (AUC) value of 0.8631
with a sensitivity of 91.67% and a specificity of 78.33% in GC
tissues (Fig. 1B). Further analysis
revealed that LINC00857 expression was positively associated
with tumor size (Fig. 1C).
Furthermore, survival rate analysis indicated that patients with GC
with increased expression of LINC00857 exhibited a poorer
survival rate compared with those with low expression of
LINC00857 (Fig. 1D). These
results suggest that LINC00857 expression may serve as a
novel biomarker for the prognosis of GC.
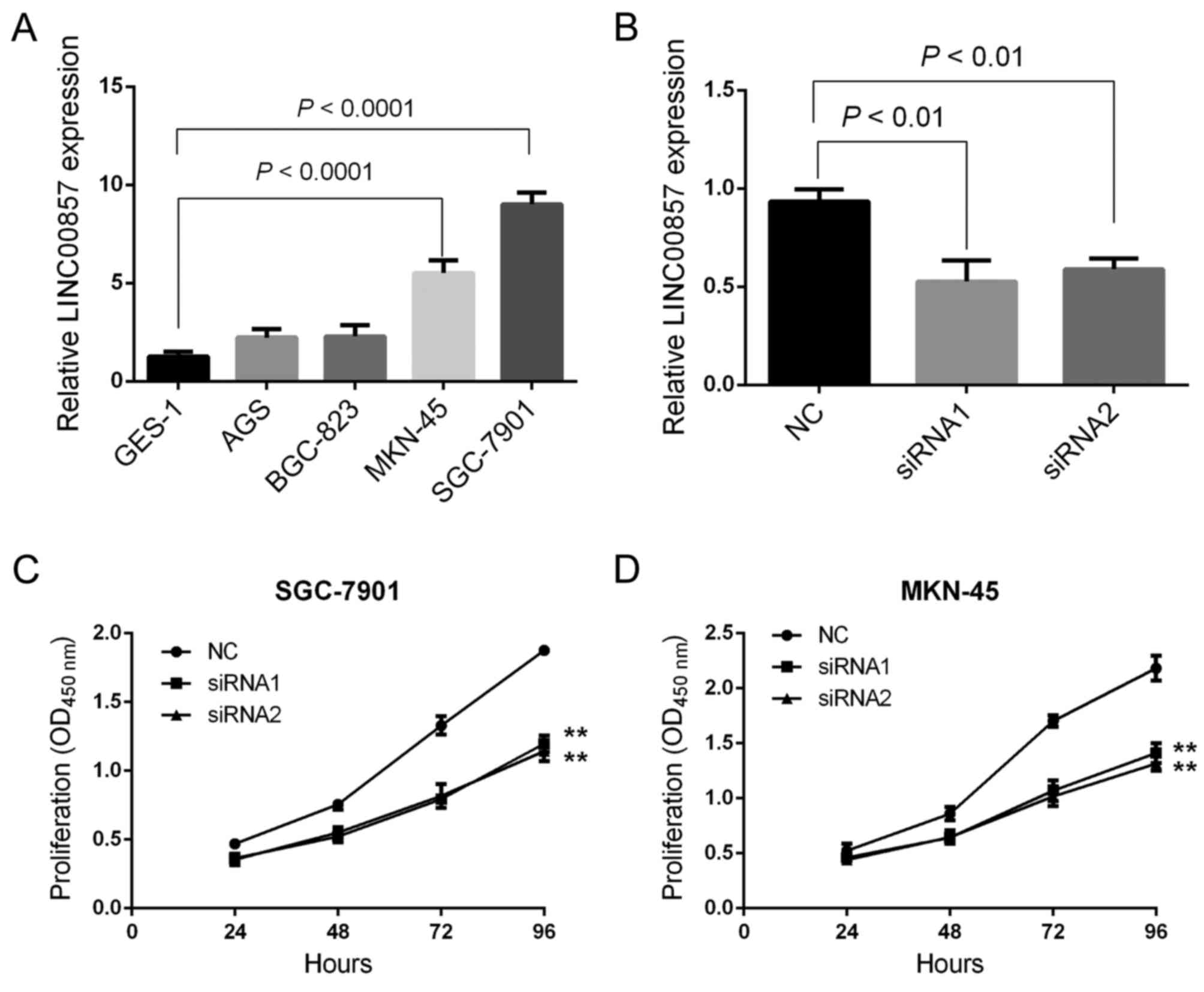
Downregulation of LINC00857 suppresses GC cell
proliferation. To determine the function of LINC00857 in the
development of GC, certain GC cell lines, including GES-1, AGS,
BGC-823, MKN-45 and SGC-7901, were used. RT-qPCR experiments
revealed that LINC00857 expression was significantly
increased in MKN-45 and SGC-7901 cells compared with that in the
remaining cell lines. In particular, the lowest expression of
LINC00857 was observed in GES-1 cells (Fig. 2A). Since the highest expression of
LINC00857 was observed in MKN-45 and SGC-7901 cells, those
cell lines were selected for the subsequent gene transfection
experiments as described previously (14). RT-qPCR experiments indicated that the
two sets of siRNAs significantly knocked down the expression of
LINC00857 in MKN-45 and SGC-7901 cells (Fig. 2B). Cell proliferation was analyzed
using a CCK-8 assay and the results indicated that LINC00857
downregulation significantly inhibited the proliferative capacity
of MKN-45 and SGC-7901 cells (Fig. 2C and
D), suggesting that LINC00857 may promote GC
development.
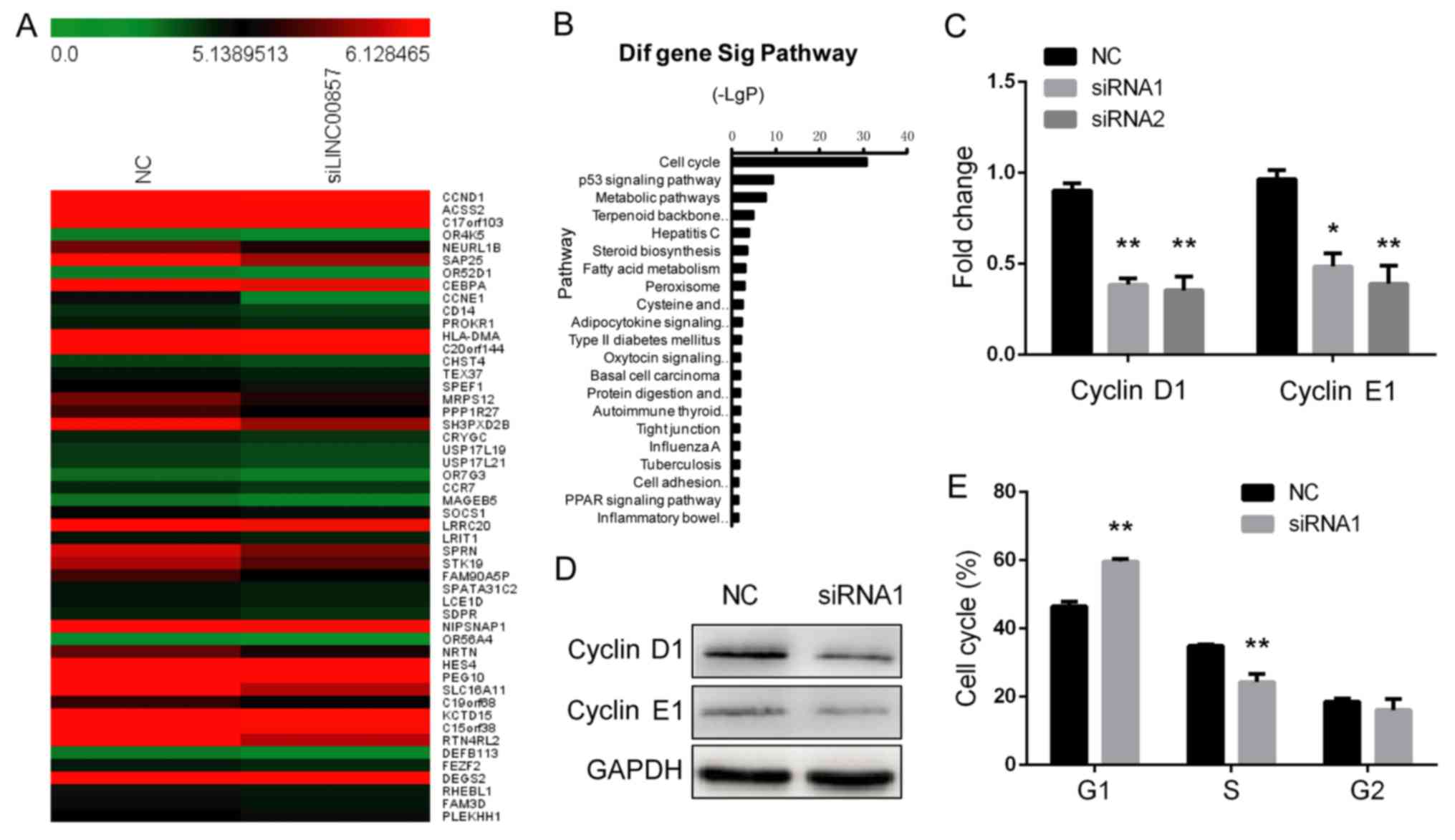
LINC00857 activates multiple signaling
pathways involved in the cell cycle. To investigate the molecular
mechanisms by which LINC00857 knockdown suppresses cell
proliferation, a whole transcriptome analysis using Affymetrix
ST2.1 exon arrays was performed in LINC00857-knockdown
SGC-7901 cells. SGC-7901 cells transfected with a negative control
were used additionally. A hierarchical clustering analysis
demonstrated the aberrant expression of multiple transcripts
between the two groups (Fig. 3A).
Pathway analysis revealed that LINC00857 led to a
significant inactivation of the cell cycle signaling pathway
(Fig. 3B). RT-qPCR and western blot
analysis confirmed that the mRNA and protein expression of cyclin
D1 and cyclin E1 were significantly decreased in
LINC00857-knockdown SGC-7901 cells compared with in control
cells (Fig. 3C and D). Furthermore,
flow cytometric analysis indicated that LINC00857-knockdown
SGC-7901 cells were arrested in G1 phase (Fig. 3E). These data suggest that
LINC00857 is able to regulate G1/S transition in
GC cells.
Discussion
In the present study, it was demonstrated that
increased expression of LINC00857 was associated with the
poor survival rate and tumor size of patients with GC. ROC analysis
revealed that LINC00857 expression may serve as an
independent biomarker for the diagnosis of GC. A cell proliferation
assay revealed that knockdown of LINC00857 significantly
inhibited the proliferative capacity of GC cells in vitro.
Furthermore, genome-wide analysis demonstrated that
LINC00857 knockdown resulted in the inactivation of the cell
cycle signaling pathway. RT-qPCR and western blot analysis also
confirmed that the mRNA and protein expression of cyclin D1 and
cyclin E1 were decreased in LINC00857-knockdown SGC-7901
cells. Furthermore, flow cytometric analysis indicated that
knockdown of LINC00857 inhibited G1/S transition
in SGC-7901 cells.
Recent evidence suggests that a number of lncRNAs
were able to serve as potential biomarkers for GC (16–19). In a
previous study by Zhang et al (13), lncRNA microarray profiling was applied
to identify several novel lncRNAs, including LINC00857, for
the diagnosis of GC. These data are consistent with the results of
the present study in that LINC00857 exhibited an increased
AUC value in GC tissues compared with that of non-tumor tissues.
The association between LINC00857 expression and the
survival rate of patients with GC was further assessed, and it was
demonstrated that patients with GC with increased expression of
LINC00857 exhibited a poor survival rate. These results
suggest that LINC00857 expression may serve as a biomarker
for the prognosis of GC.
Although a number of lncRNAs have been annotated,
functional interpretation remains limited. Currently, the
biological effect of LINC00857 on GC development remains
largely unclear. The results of the present study demonstrated that
LINC00857 knockdown significantly inhibited GC cell
proliferation through regulating cell cycle signaling pathways,
confirming the results of previous studies (14). Furthermore, LINC00857
expression might be regulated by cyclin D1 and cyclin E1, which
induces G1 arrest in GC cells. Previous studies have
also demonstrated that the downregulation of cyclin D1 and cyclin
E1 significantly inhibited GC cell proliferation (20,21).
Nevertheless, the molecular mechanisms by which LINC00857
regulates GC cell proliferation require investigation in future
studies. Future in vivo experiments are also required to
confirm the results of the present study.
In conclusion, the results of the present study
demonstrated that LINC00857 expression is associated with
the poor survival rate of patients with GC and that downregulating
LINC00857 expression suppresses GC cell proliferation in
vitro through cell cycle arrest.
Acknowledgements
The authors thank Professor Zhao Yun for providing
support for the reverse transcription-quantitative polymerase chain
reaction technology.
Funding
The present study was supported by the Yongchuan
Science & Technology Commission (grant nos. 2014rc9004 and
2013nc8017).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FH designed the present study and wrote the
manuscript. FH and KP performed the experiments and analyzed the
data. MJR, FWZ and TWY collected the samples of the patients and
analyzed the data.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics Review
Board at the Yongchuan Hospital of Chongqing Medical University
(Chongqing, China).
Consent for publication
The authors declare that the patients have provided
written informed consent for the publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang AP, Liu J, Lei HY, Zhang QW, Zhao L
and Yang GH: CA72-4 combined with CEA, CA125 and CAl9-9 improves
the sensitivity for the early diagnosis of gastric cancer. Clin
Chim Acta. 437:183–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng S, Xiao YF, Tang B, Hu CJ, Xie R,
Yang SM and Li BS: Long noncoding RNA in digestive tract cancers:
Function, mechanism, and potential biomarker. Oncologist.
20:898–906. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu
TC, Tang JY, Bao YJ, Hu Y, Lin Y, et al: LncRNA GClnc1 promotes
gastric carcinogenesis and may act as a modular scaffold of WDR5
and KAT2A complexes to specify the histone modification pattern.
Cancer Discov. 6:784–801. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: LncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1 and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y,
Liang W, Hu C, Liu Y, Li J, et al: Genome-wide lncRNA microarray
profiling identifies novel circulating lncRNAs for detection of
gastric cancer. Theranostics. 7:213–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, He Y, Liu W, Bai S, Xiao L, Zhang
J, Dhanasekaran SM, Wang Z, Kalyana-Sundaram S, Balbin OA, et al:
Non-coding RNA LINC00857 is predictive of poor patient survival and
promotes tumor progression via cell cycle regulation in lung
cancer. Oncotarget. 7:11487–11499. 2016.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCTmethod. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang
Q, Ren M, Chen L, Yuan D, Zhang Y, et al: The lncRNA MALAT1 is a
novel biomarker for gastric cancer metastasis. Oncotarget.
7:56209–56218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leung SY, Ho C, Tu IP, Li R, So S, Chu KM,
Yuen ST and Chen X: Comprehensive analysis of 19q12 amplicon in
human gastric cancers. Mod Pathol. 19:854–863. 2006. View Article : Google Scholar : PubMed/NCBI
|