Introduction
Although significant advances have been made toward
the reduction of occupational health hazards associated with lung
cancer, particularly smoking, and in the prevention of various
disorders, lung cancer remains a highly lethal disease. Moreover,
survival rates are not improving owing to late diagnosis and
unsatisfactory monitoring for recurrence (therapy response)
(1,2).
This makes the search for suitable biomarkers one of the highest
priorities in the study of lung cancer (3,4). Abnormal
DNA methylation is an important epigenetic regulator of
tumorigenesis (5). Deciphering common
and specific DNA methylation patterns of cancerous tissue is
essential for understanding tumor development. The potential use of
gene methylation for the detection and diagnosis of lung cancer in
biopsy specimens has been evaluated in several studies (6,7).
Normal mucosa of esophagus-specific 1 (NMES1), also
known as C15orf48, was first identified in a study of human
esophageal squamous cell carcinoma (ESCC) tissues, which showed
that its mRNA and protein levels were reduced in carcinoma samples
(8). Overexpression of NMES1
inhibits cell motility in ESCC cell lines (9), suggesting its suppressive role in
tumorigenesis in the esophagus. Furthermore, NMES1 transcripts have
been found to be inactivated by DNA methylation in invasive
cervical cancer and colon cancer (10,11).
Notably, NMES1 has been identified as a potential candidate for
modifiers of susceptibility to skin tumor promotion by phorbol
ester (12). NMES1 is mainly
expressed in epithelial tissues, including weakly in the lungs
(8), but the precise function of
NMES1 remains unknown. To understand the biological role of NMES1
in lung cancer, pyrosequencing was conducted in the present study
to investigate the methylation status of the NMES1 promoter
in resected primary non-small cell lung cancer (NSCLC), and the
association between these results and clinicopathological
characteristics were assessed.
Materials and methods
Patients and tissue samples
Tumor and corresponding non-malignant lung tissue
specimens (n=178) between January 2002 and July 2010 were provided
by the National Biobank of Korea, Kyungpook National University
Hospital (KNUH; Daegu, South Korea), which is supported by the
Ministry of Health, Welfare and Family Affairs. This study was
conducted with the approval of the Ethics Committee of KNUH
(approval no. 2014-04-210) All materials derived from the National
Biobank of Korea, KNUH, were obtained following approval by the
Institutional Review Board of KNUH and written informed consent was
obtained from all of the participants prior to obtaining the
samples. The clinicopathological characteristics of patients (mean
age 64 years, range 35–83 years) are summarized in Table I. The pathological stage was
determined by applying the seventh edition of the Union for
International Cancer Control and American Joint Committee on Cancer
TNM classification (13).
 | Table I.Association between methylation status
of normal mucosa of esophagus-specific 1 and characteristics of
non-small cell lung cancer patients. |
Table I.
Association between methylation status
of normal mucosa of esophagus-specific 1 and characteristics of
non-small cell lung cancer patients.
| Variables | Methylation, n
(%) | P-valuea |
|---|
| All subjects
(n=178) | 15 (8.4) |
|
| Age, years |
| 0.42 |
| ≤64
(n=80) | 5 (6.3) |
|
| >64
(n=98) | 10 (10.2) |
|
| Sex |
| 0.24 |
| Male
(n=125) | 13 (10.4) |
|
| Female
(n=53) | 2 (3.8) |
|
| Smoking status |
| 0.15 |
| Ever
(n=120) | 13 (10.8) |
|
| Never
(n=58) | 2 (3.4) |
|
| Histological
types |
| 0.05 |
| SCC
(n=85) | 11 (12.9) |
|
| ADC
(n=93) | 4 (4.3) |
|
| Pathological
stage |
| 0.93 |
| Stage I
(n=93) | 8 (8.6) |
|
| Stage
II–IIIA (n=85) | 7 (8.2) |
|
Cell culture and RT-PCR
A normal human lung epithelial cell line (BEAS-2B)
and 12 human lung cancer cell lines (NCI-H522, NCI-H1703,
NCI-H1299, NCI-H2108, NCI-H187, NCI-H2009, NCI-H520, NCI-H23,
NCI-H1373, HCC827, PC9 and A549) were obtained from the American
Type Culture Collection (Manassas, VA, USA). BEAS-2B and the cancer
cell lines were maintained at 37°C with 5% CO2 in
Dulbecco's modified Eagle's medium/F12 and RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
respectively, supplemented with 10% heat-inactivated fetal bovine
serum (Invitrogen; Thermo Fisher Scientific, Inc.). NCI-H187 cells
were treated with the demethylating agent 5-aza-2′-deoxycytidine
(5-AzadC) for 3 days, with culture medium being changed daily.
Total RNA was extracted from the cultured cells and primary tumor
tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Subsequent to removing residual DNA, first-strand cDNA was
synthesized from total RNA using SuperScript First-Strand Synthesis
System (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. The resulting cDNA was amplified with
sense primer 5′-AGGAACTCATTCCCTTGGTG-3′ and antisense primer
5′-TCCACAGTTTCCCAAGGTTC-3′. The PCR conditions were as follows:
Denaturation at 95°C for 2 min, then 30 cycles of 95°C for 1 min,
58°C for 1 min, 72°C for 1 min, and final extension at 72°C for 5
min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was
amplified with sense primer 5′-CATGACAACTTTGGTATCGTG-3′ and
antisense primer 5′-GTGTCGCTGTTGAAGTCAGA-3′ for the internal
loading control. Amplified products were separated on 2% agarose
gels, visualized with ethidium bromide and photographed by Syngene
DigiGenius Gel Documentation system (Syngene, Frederick, MD, USA).
Band intensities were quantified with ImageJ 1.50i program
(National Institutes of Health, Bethesda, MD, USA) and the relative
amount of NMES1 mRNA, normalized to GAPDH levels, was
expressed as gray values.
Genomic DNA isolation and
pyrosequencing
Genomic DNA was extracted using a QIAamp DNA Mini
kit (Qiagen, Inc., Valencia, CA, USA) and was chemically modified
with the EZ DNA Methylation-Gold kit (Zymo Research Corp., Irvine,
CA, USA) according to the manufacturer's protocol. The methylation
status of NMES1 was quantitatively determined by
pyrosequencing. Briefly, bisulfate-modified DNA was PCR-amplified
using forward primer 5′-TTATAAGTATTTAGGGGGGTTAAGA-3′ and reverse
primer biotin-5′-CCCCCTACAAAACATTCTAC-3′ with the GeneAmp Gold PCR
Reagent kit (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The PCR conditions were as follows: Denaturation at 95°C for 10
min, then 40 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1
min, and final extension at 72°C for 10 min. PCR product quality
and absence of contamination were confirmed by 2% agarose gels with
ethidium bromide staining. Following purification of the PCR
product using Sepharose beads on a PyroMark Vaccum Prep Workstation
(Qiagen, Inc.), pyrosequencing was performed according to the
manufacturer's specifications using a sequencing primer
(5′-TAGGGGGGTTAAGAG-3′) and a PyroMark Q96MD system (Qiagen, Inc.).
The mean methylation index (MI) was calculated from the mean of the
methylation percentage for the eight evaluated CpG sites. To set
the controls for pyrosequencing, CpGenome™ Universal methylated and
unmethylated DNA (Chemicon, Temecula, CA, USA) was used as a
positive and negative control, with stable levels of methylation.
Each pyrosequencing was repeated at least once to confirm the
results.
Statistical analysis
The association between methylation status and
clinicopathological characteristics was analyzed by a χ2
test for categorical variables. A logistic regression test was
conducted to estimate the association between methylation and the
covariates of age, sex, exposure to tobacco smoke and histology.
The overall survival rate (OSR) of NSCLC patients according to
NMES1 methylation status was compared using the Kaplan-Meier
method and the log-rank test. Hazard ratios (HR) and 95% confidence
intervals (CIs) were estimated using the multivariate Cox
proportional hazard model. P<0.05 was considered to indicate a
statistically significant difference.
Results and Discussion
Methylation status and expression of
NMES1 in NSCLC samples
In the present study, pyrosequencing was performed
to analyze the methylation status of the human NMES1 gene in
178 primary NSCLCs and corresponding non-malignant lung tissue
controls. Given the presence of CpG islands (CGIs) in the
NMES1 5′-flanking region, including the first exon, the
pyrosequencing primer was designed so as to encompass 8 CpGs from
−165 to −115 bp upstream of the transcription start site.
Consistently, Arai et al (9)
previously demonstrated a high degree of DNA methylation within the
same region in ESCC. Accurate and reproducible estimates of
methylated cytosine content were obtained in all tested samples,
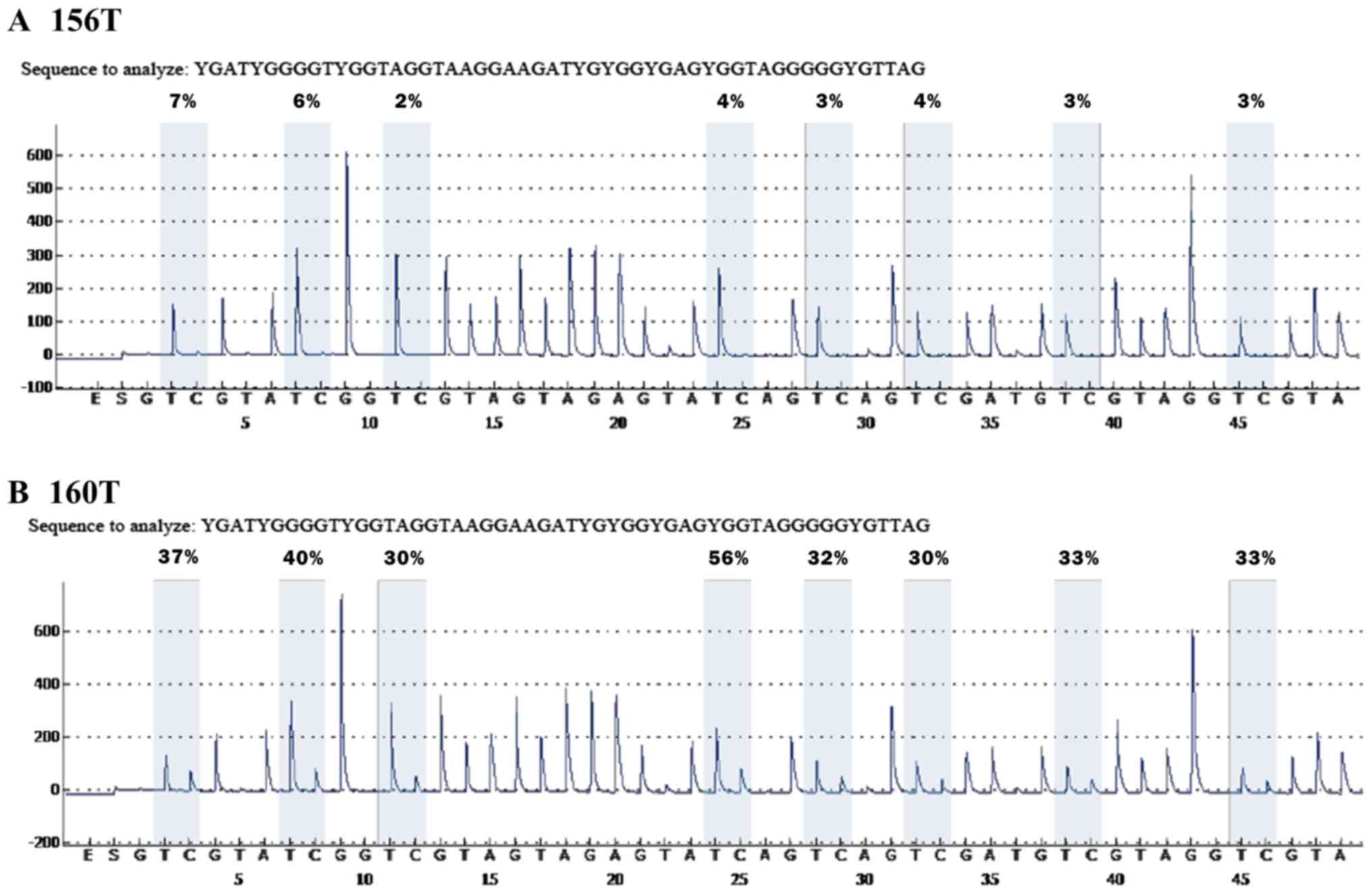
and representative pyrograms are shown in Fig. 1. Pyrosequencing of the representative
PCR products showed that all cytosines at non-CpG sites were
converted to thymines, ruling out the possibility of incomplete
bisulfite conversion. Considering a mean MI of 7.10% for all
non-malignant lung tissues, 7.10 was set as a cut-off point for a
methylation-positive classification (data not shown). NMES1
methylation was detected exclusively in malignant tissues, at a
frequency of 8.4% (15/178), suggesting that NMES1 promoter
methylation may be a tumor-associated event during NSCLC
tumorigenesis. These results represent the first demonstration of
aberrant methylation of NMES1 in primary tumors of NSCLC
patients. Furthermore, using an average MI of 28.95% for 15
methylated samples as a cut-off value (data not shown),
methylation-positive tumors were divided into two groups, weak
methylation (7.10≤MI<28.95) and strong methylation (MI≥28.95).
Accordingly, 10 cases were assigned to the weak methylation group
and 5 to the strong methylation group. Although there is no
reasonable rationale to use median MI for assigning certain
specimens to the weak or the strong methylation groups, this
approach was followed based on the observation of a previous study
(13). Originally, Shaw et al
(14) divided methylated samples into
weak- and strong-methylation groups based on a median split, which
is necessary to silence mRNA expression of the corresponding genes.
In our previous study, using the log-rank test for patient OSR
through a series of methylation levels measured by pyrosequencing
revealed that the lowest P-value is observed around the median
level (15).
To determine whether promoter methylation could be
involved in the regulation of NMES1 expression, NMES1
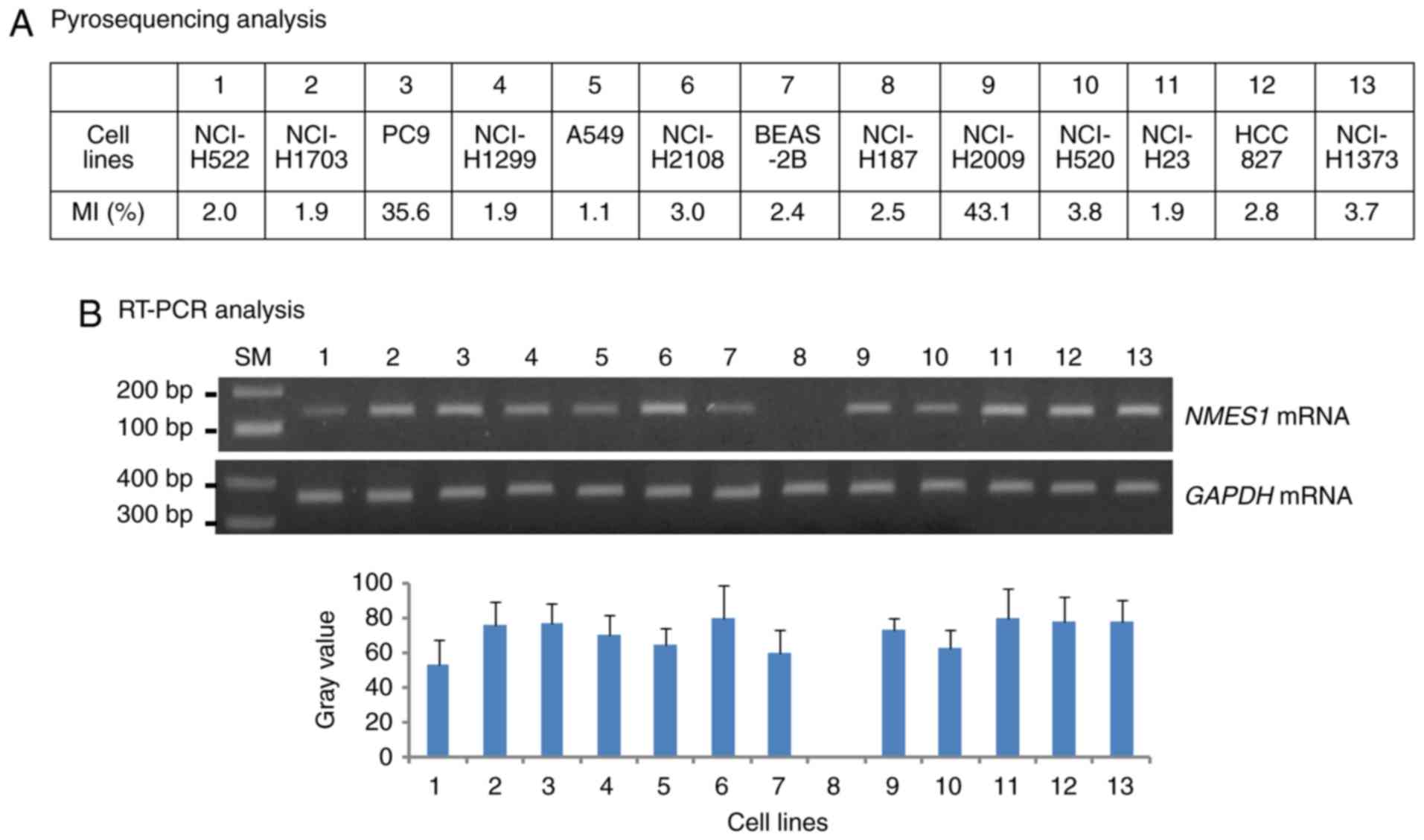
mRNA levels were analyzed in 13 human lung cancer cell lines.
Methylation status combined with RT-PCR findings showed that
NMES1 mRNA was present in PC9 and NCI-H2009 cells with
methylated alleles (MI=35.6 and MI=43.1, respectively), but was
absent in NCI-H187 cells without methylated alleles (MI=2.5)
(Fig. 2A and B). Moreover, NCI-H187
cells failed to restore their native expression level following
5-AzadC treatment (data not shown). The correlation between CGI
hypermethylation and NMES mRNA expression in weak and strong
methylation groups was assessed. However, the NMES1
expression could not be evaluated according to methylation degree
(such as negative, weak and strong) in cancer tissues owing to no
preparation of total RNAs to ideally match all malignant tissues
with weak or strong methylation (data not shown). These results
suggest that CGI hypermethylation may not be associated with
NMES1 silencing; instead, other mechanisms may control its
expression. This observation does not fit into the classical
paradigm showing an inverse correlation between DNA methylation and
gene expression (16). Unexpectedly,
integration of DNA methylation profiles and mRNA expression data in
lung adenocarcinoma has indicated that approximately one-third of
genes, which are differentially methylated between tumors and
normal tissue, show concurrent changes in gene expression (17). Accordingly, regulation through DNA
methylation is likely more complex than previously anticipated. The
identification of long-range DNA methylation, spreading of DNA
methylation and DNA methylation just outside CGIs add increasing
intricacy to the association between DNA methylation and gene
expression (18,19). Alternatively, different CGIs or CpG
sites within a CGI could have differential effects on gene
expression (20,21). Moreover, single CGI methylation is not
sufficient to maintain silencing (22). It could be speculated that other sites
instead of the regions analyzed in the present study may be
directly associated with NMES1 expression. Subsequently, the
5 CpGs that were located at the −397 to −357 region by
pyrosequencing were investigated, showing no significant
hypermethylation in malignant tissues compared with that in
non-malignant tissues (data not shown). An attempt was made to
compare the methylation of 6 CpGs at the transcription start site,
but no pyrosequencing primer with a high score was available (data
not shown). Therefore, additional research is required to clarify
the mechanism regulating NMES1 expression.
Association of NMES1 promoter
methylation with clinicopathological parameters and clinical
outcomes
NMES1 promoter methylation was significantly
more frequent in squamous cell carcinoma than in adenocarcinoma
(P=0.05; Table I). However, no
significant correlation was observed between methylation and any
other factors, including age, sex, smoking status and pathological
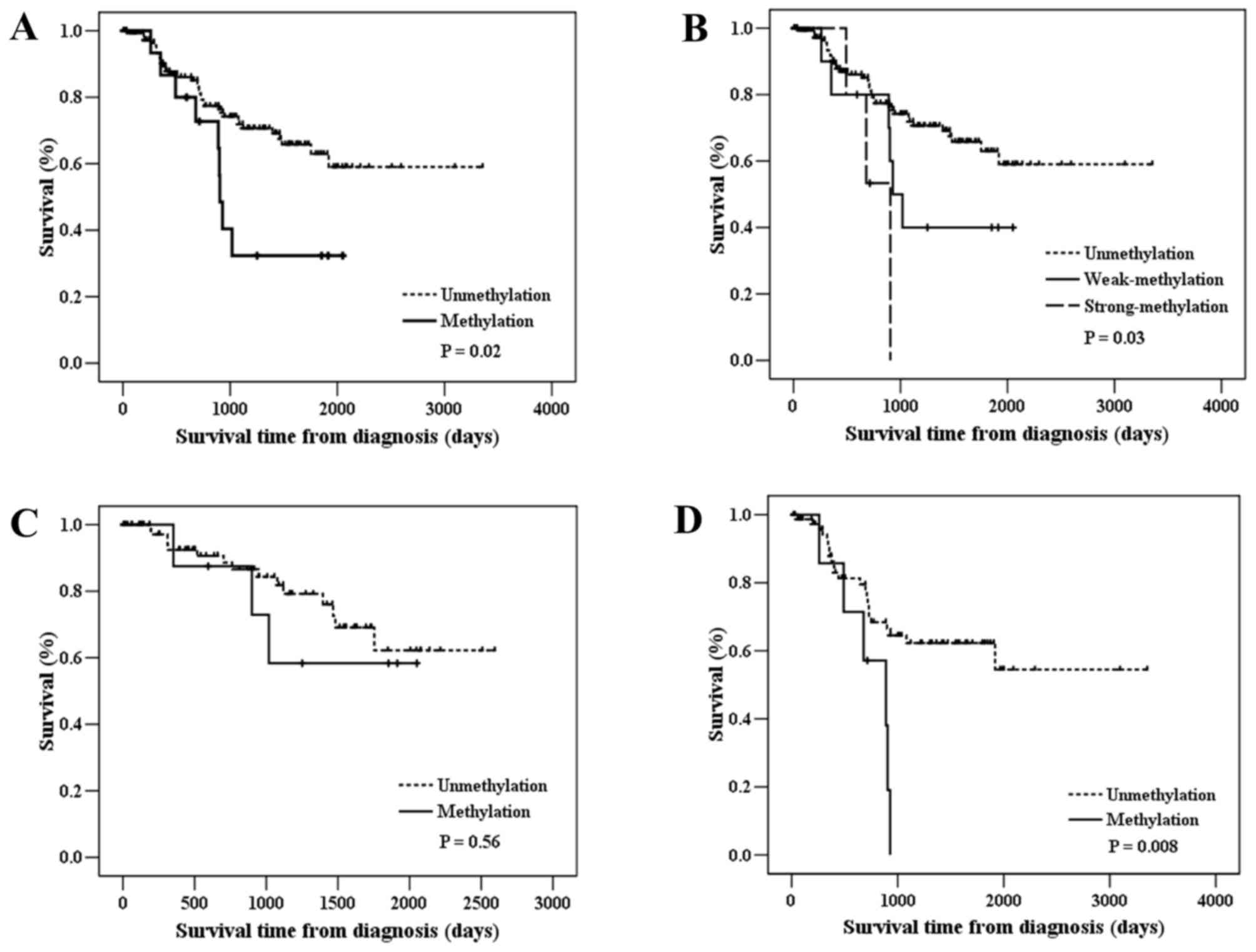
stage (Table I). Next, Kaplan-Meier
survival analysis was performed to determine the prognostic
potential of NMES1 methylation. Notably,
methylation-positive patients had worse OSR compared with
methylation-negative patients [log-rank P (PLR)=0.02]
(Table II and Fig. 3A). Notably, patients with strong
methylation exhibited significantly reduced OSR, compared with
those with weak methylation (HRadj, 3.05 vs. 2.45;
Ptrend=0.02) (Table II
and Fig. 3B), indicating that CGI
methylation levels could influence the OSR. When stratified
according to clinicopathological patient characteristics,
NMES1 methylation was significantly associated with
unfavorable survival in patients with stage II–IIIA disease
(PLR=0.008), but not in patients with stage I disease
(Fig. 3C and D). To evaluate
NMES1 methylation as an independent prognostic factor in
NSCLC, the data was further analyzed using the Cox proportional
hazards regression adjusting for possible confounders of survival.
NMES1 methylation was significantly associated with worse
OSR in all patients [adjusted HR (HRadj), 2.62; 95% CI, 1.20–5.69;
P=0.02]. It is now known that NMES1 regulates cancer cell motility
and is a component of mitochondrial respiratory chain complex IV
(9,23). Importantly, a growing body of evidence
has demonstrated that tumor cells have defective mitochondrial
respiration due to their dominant glycolytic metabolism (24). However, the precise function of NMES1
remains elusive. It is also difficult to imagine how methylated
NMES1 would have any consequence in the tumorigenesis of
lung cancer if mRNA expression and methylated DNA levels do not
match. In addition to gene transcription repression, DNA
methylation may also be important for alternative splicing, gene
mutation and chromatin remodeling (25–27). Taken
together, these results suggest that NMES1 may serve an
important role in lung cancer pathogenesis and its methylation
could be considered a prognostic marker for NSCLC patients. The
present observations may offer novel insights for the clinical
management of pyrosequencing-derived methylation levels.
 | Table II.Overall survival according to normal
mucosa of esophagus-specific 1 methylation in non-small cell lung
cancer patients. |
Table II.
Overall survival according to normal
mucosa of esophagus-specific 1 methylation in non-small cell lung
cancer patients.
|
|
|
|
|
| Crude | Adjusted |
|---|
|
|
|
|
|
|
|
|
|---|
| Variables | Cases, n | Mortality, n
(%)a | 5-year survival
rateb |
PLR | HR (95% CI) | P-value | HR (95%
CI)c | P-value |
|---|
| All subjects | 178 |
|
|
|
|
|
|
|
|
Non-methylation | 163 | 38 (23.3) | 63 |
| 1.00 |
| 1.00 |
|
| Methylation | 15 | 9 (60.0) | 32 | 0.02 | 2.30
(1.11–4.76) | 0.03 | 2.62
(1.20–5.69) | 0.02 |
|
Non-methylation | 163 | 38 (23.3) | 63 | 0.04 | 1.00 |
| 1.00 |
|
| Weak
methylation | 10 | 6 (60.0) | 40 |
| 1.98
(0.84–4.68) | 0.12 | 2.45
(0.99–6.11) | 0.05 |
| Strong
methylation | 5 | 3 (60.0) | 0 |
| 3.44
(1.04–11.39) | 0.04 | 3.05
(0.84–11.06) | 0.09 |
|
Ptrend |
|
|
|
| 0.01 |
| 0.02 |
|
The present study has several limitations. First,
the retrospective design and small number of sample cases could
confer potential selection bias in results interpretation. Second,
there is a shortage of information on qPCR and NMES1
expression according to methylation degree in the tumor tissues.
Finally, there is no reasonable rationale to use median MI for
assigning certain specimens to the weak or the strong methylation
groups.
The present study has shown that the NMES1
promoter was methylated exclusively in tumor tissues of NSCLCs and
that its methylation was associated significantly with unfavorable
OSR in those patients. Although the current study did not offer a
complete overview due the small sample size and lack of information
on protein expression, it is the first report to demonstrate
aberrant methylation of NMES1 in NSCLC and it provides
clinical evidence to support the tumor-suppressing role of
NMES1 in NSCLC. Future studies with larger sample sizes are
required to confirm this conclusion.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Korea Health Technology R&D Project through the Korea Health
Industry Development Institute, funded by the Ministry of Health
and Welfare (grant no. HI4C0402).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
DSK contributed to the experimental design and
implementation, performed the experiments and data analysis, and
drafted the manuscript. WKL performed statistical analyses. JYP
contributed to experiment implementation, interpreted the patient
data and modified the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Kyungpook National University Hospital (2014-04-210).
Written informed consent was obtained from all participants or
their families prior to obtaining the samples.
Consent for publication
All participants provided written informed consent
for publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McIntyre A and Ganti AK: Lung cancer-A
global perspective. J Surg Oncol. 115:550–554. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
I H and Cho JY: Lung cancer biomarkers.
Adv Clin Chem. 72:107–170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vargas AJ and Harris CC: Biomarker
development in the precision medicine era: Lung cancer as a case
study. Nat Rev Cancer. 16:525–537. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liloglou T, Bediaga NG, Brown BR, Field JK
and Davies MP: Epigenetic biomarkers in lung cancer. Cancer Lett.
342:200–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walter K, Holcomb T, Januario T, Yauch RL,
Du P, Bourgon R, Seshagiri S, Amler LC, Hampton GM and S Shames D:
Discovery and development of DNA methylation-based biomarkers for
lung cancer. Epigenomics. 6:59–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou J, Wang H, Lu A, Hu G, Luo A, Ding F,
Zhang J, Wang X, Wu M and Liu Z: A novel gene, NMES1, downregulated
in human esophageal squamous cell carcinoma. Int J Cancer.
101:311–316. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arai M, Imazeki F, Sakai Y, Mikata R, Tada
M, Seki N, Shimada H, Ochiai T and Yokosuka O: Analysis of the
methylation status of genes up-regulated by the demethylating
agent, 5-aza-2′-deoxycytidine, in esophageal squamous cell
carcinoma. Oncol Rep. 20:405–412. 2008.PubMed/NCBI
|
|
10
|
Sova P, Feng Q, Geiss G, Wood T, Strauss
R, Rudolf V, Lieber A and Kiviat N: Discovery of novel methylation
biomarkers in cervical carcinoma by global demethylation and
microarray analysis. Cancer Epidemiol Biomarkers Prev. 15:114–123.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spisák S, Kalmár A, Galamb O, Wichmann B,
Sipos F, Péterfia B, Csabai I, Kovalszky I, Semsey S, Tulassay Z
and Molnár B: Genome-wide screening of genes regulated by DNA
methylation in colon cancer development. PLoS One. 7:e462152012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Riggs PK, Angel JM, Abel EL and Digiovanni
J: Differential gene expression in epidermis of mice sensitive and
resistant to phorbol ester skin tumor promotion. Mol Carcinog.
44:122–136. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Detterbeck FC, Boffa DJ and Tanoue LT: The
new lung cancer staging system. Chest. 136:260–271. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shaw RJ, Hall GL, Lowe D, Bowers NL,
Liloglou T, Field JK, Woolgar JA and Risk JM: CpG island
methylation phenotype (CIMP) in oral cancer: Associated with a
marked inflammatory response and less aggressive tumour biology.
Oral Oncol. 43:878–886. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee SM, Lee WK, Kim DS and Park JY:
Quantitative promoter hypermethylation analysis of RASSF1A in lung
cancer: Comparison with methylation-specific PCR technique and
clinical significance. Mol Med Rep. 5:239–244. 2012.PubMed/NCBI
|
|
16
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bert SA, Robinson MD, Strbenac D, Statham
AL, Song JZ, Hulf T, Sutherland RL, Coolen MW, Stirzaker C and
Clark SJ: Regional activation of the cancer genome by long-range
epigenetic remodeling. Cancer Cell. 23:9–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim DS, Lee SM, Yoon GS, Choi JE and Park
JY: Infrequent hypermethylation of the PTEN gene in Korean
non-small cell lung cancers. Cancer Sci. 101:568–572. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lessi F, Beggs A, de Palo M, Anti M,
Macarone Palmieri R, Francesconi S, Gomes V, Bevilacqua G,
Tomlinson I and Segditsas S: Down-regulation of
serum/glucocorticoid regulated kinase 1 in colorectal tumours is
largely independent of promoter hypermethylation. PLoS One.
5:e138402010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Braga LC, Silva LM, Ramos AP, Piedade JB,
Vidigal PV, Traiman P and da Silva Filho AL: Single CpG island
methylation is not sufficient to maintain the silenced expression
of CASPASE-8 apoptosis-related gene among women with epithelial
ovarian cancer. Biomed Pharmacother. 68:87–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Floyd BJ, Wilkerson EM, Veling MT, Minogue
CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, et al:
Mitochondrial protein interaction mapping identifies regulators of
respiratory chain function. Mol Cell. 63:621–632. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garcia-Heredia JM and Carnero A: Decoding
Warburg's hypothesis: Tumor-related mutations in the mitochondrial
respiratory chain. Oncotarget. 6:41582–41599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maunakea AK, Chepelev I, Cui K and Zhao K:
Intragenic DNA methylation modulates alternative splicing by
recruiting MeCP2 to promote exon recognition. Cell Res.
23:1256–1269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hitchins MP, Rapkins RW, Kwok CT,
Srivastava S, Wong JJ, Khachigian LM, Polly P, Goldblatt J and Ward
RL: Dominantly inherited constitutional epigenetic silencing of
MLH1 in a cancer-affected family is linked to a single nucleotide
variant within the 5′UTR. Cancer Cell. 20:200–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Forn M, Muñoz M, Tauriello DV,
Merlos-Suárez A, Rodilla V, Bigas A, Batlle E, Jordà M and Peinado
MA: Long range epigenetic silencing is a trans-species mechanism
that results in cancer specific deregulation by overriding the
chromatin domains of normal cells. Mol Oncol. 7:1129–1141. 2013.
View Article : Google Scholar : PubMed/NCBI
|