Introduction
Esophageal cancer (EC) is the 8th-most prevalent and
the 6th most lethal type of malignant cancer globally; esophageal
squamous cell carcinoma (ESCC) is the predominant pathological type
of EC (1). According to the GLOBOCAN
estimates, in 2012 the incidence of EC varied by >21-fold
internationally (1,2). North-central China is positioned on the
‘esophageal cancer belt’ (3); the
Henan province is representative of this region. Due to the lack of
symptoms at the onset of EC, the majority of patients with EC are
diagnosed and treated at an intermediate or advanced histological
stage, when lymph node or distant metastasis, may have occurred.
Consequently, the 5-year survival rate of EC remains low, at 15–25%
(4,5).
Although treatments for EC, including surgery,
radiation, and chemotherapy, have been greatly improved over the
past decades, the disease continues to be associated with a
relatively poor prognosis. A number of studies have demonstrated
that autologous cellular antigens, including established
serological tumor biomarkers or their corresponding autoantibodies,
could invoke an immune reaction in patients with cancer (6–8). However,
such biomarkers are rapidly degraded or cleared from the
circulatory system (9). Furthermore,
the majority of these biomarkers are insufficient for the early
detection of EC (10,11). Therefore, the discovery of novel tumor
biomarkers is urgently required to detect carcinogenesis at an
early stage as accurately as possible and improve the prognosis of
patients with EC.
MicroRNAs (miRNAs/miRs) are small non-coding
single-stranded RNAs of 20–25 nucleotides. They have been
demonstrated to exist stably in cell-free body fluids, including
the saliva, plasma, serum and urine. The first circulating miRNA
potential biomarker for cancer diagnosis was identified in 2008
(12). MiRNAs continue to attract
research attention for this purpose, and promising diagnostic
markers have been identified for breast cancer (13), liver cancer (14), lung cancer (15), colorectal cancer (16), uterine cancer (17), glioma (18) and gastroesophageal cancer (19); a number of such biomarkers function as
oncogenes or tumor-suppressor genes (20). Numerous studies have associated
various circulating miRNAs with EC (21). However, different research groups have
drawn contrasting conclusions regarding the same plasma miRNAs in
EC. MiR-375 expression was demonstrated to be upregulated in EC by
Li et al (22), and
downregulated by Komatsu et al (23). Other studies demonstrated that the
expression of miR-375 was significantly downregulated in other
types of cancer, particularly in tumors of the digestive tract
(24–27). Verification of these previous
conclusions is required.
In the present study, a meta-analysis of previously
studied circulating miRNAs in plasma or serum was performed to
assess their diagnostic efficiency in EC. The 5 most studied miRNAs
were selected as candidate markers, and their expression levels in
plasma were quantified using reverse transcription
quantitative-polymerase chain reaction (RT-qPCR). The results were
used to verify and evaluate their diagnostic value in a validation
group of ESCC samples.
Subjects and methods
Study population and sample
collection
A total of 125 patients were histopathologically
diagnosed with primary ESCC at the Henan Cancer Hospital (Henan,
China) between April 2013 and December 2014. The patients had
undergone no prior surgery, chemotherapy or radiotherapy. The 7th
edition of the International Union Against Cancer (UICC)
tumor-node-metastasis (TNM) staging system (28) was used for the classification of the
patients with ESCC. Peripheral whole blood was collected from each
patient in EDTA-K2 anti-coagulant tubes prior to any treatment.
Plasma was separated within 2 h of collection by centrifugation at
3,000 × g for 10 min at 4°C. The samples were then stored at −80°C
until required. A total of 125 age- and sex-matched individuals
were selected from the annual health examinations during the same
period, and were enrolled as healthy controls. Patients with
symptoms suggestive of cancer, digestive tract diseases or immune
diseases were excluded. The present study was approved by the
Institute Research Ethics Committee of Henan Cancer Hospital and
Zhengzhou University. Written informed consent was obtained from
all participants.
Meta-analysis of the diagnostic value
of circulating miRNAs to diagnose EC
A systematic literature search in the PubMed and Web
of Science databases was performed, considering published studies
in English or Chinese until April 2016. The major medical subject
heading search terms were as follows: ‘esophageal neoplasms’,
‘microRNAs’, ‘blood’, ‘diagnosis’ and ‘sensitivity and
specificity’, ‘biomarkers’ and ‘tumor’. Manual retrieval was also
applied by tracing references to identify additional records.
All relevant publications were included based on the
following criteria: i) Patients were diagnosed with primary EC; ii)
studies concentrated on circulating miRNAs in serum or plasma; and
iii) studies provided sufficient data for the construction of 2×2
tables. The exclusion criteria were: i) Duplicate studies; ii)
publications not in Chinese or English; iii) reviews,
meta-analyses, case reports, editorials and meeting records; iv)
studies without blood samples, or with samples extracted subsequent
to surgery, chemotherapy or radiotherapy; v) studies that did not
provide sufficient data to calculate the sensitivity, specificity
and AUC of miRNAs and diagnose EC; vi) studies focusing only on the
survival and prognosis of EC; and vii) studies focusing only on the
molecular mechanisms of miRNAs in EC.
Data were extracted from the included studies by two
independent reviewers, and disagreements were resolved through
discussion. The extracted information is provided in Table I. When a training group and a
validation group appeared in one article simultaneously, they were
treated as independent studies in the meta-analysis. From this
meta-analysis, the top 5 most-studied miRNAs were selected as the
candidate markers for further verification.
 | Table I.The main characteristics of eligible
studies in the systematic meta-analysis. |
Table I.
The main characteristics of eligible
studies in the systematic meta-analysis.
|
|
|
| Study population
characteristics |
| Experimental
method |
| Data |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | miR | Trend | Country | Casesa |
Controlsa | Sample type | RNA extraction
reagent/kit | Normalization
control | RT-qPCR type | Cutoff | TP | FP | FN | TN | (Refs.) |
|---|
| Ye et al,
2014 | miR-21 | Up | China | 100 (80/20), 59
(43–88) | 50 | Plasma | mirVANA PARIS | miR-16 | SYBR | YI | 97 | 22 | 3 | 28 | (52) |
| Li et al,
2015 | miR-21 | Up | China | 24b (18/6), 68.5 (47–87) | 19 | Plasma | mirVANA PARIS | miR-1228 | TaqMan | 3.89 | 22 | 6 | 2 | 13 | (53) |
| Li et al,
2015 | miR-21 | Up | China | 38 (30/8) | 19 | Plasma | mirVANA PARIS | miR-1228 | TaqMan | YI | 35 | 8 | 3 | 11 | (22) |
| Wang and Zhang,
2012 | miR-21 | Up | China | 31c (23/8), 61 (46–82) | 39 (30/9), 46
(−) | Plasma | Trizol LS | miR-16 | SYBR | 3.37 | 22 | 12 | 9 | 27 | (40) |
| Komatsu et
al, 2011 | miR-21 | Up | Japan | 50 (44/6) | 20 | Plasma | mirVANA PARIS | Standard curve | TaqMan | 0.22 | 24 | 3 | 26 | 17 | (23) |
| Wu et al,
2014 | miR-223 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 40 | 12 | 23 | 51 | (46) |
| Zhang et al,
2012 | miR-223 | Up | China | 113 (88/25) | 67 | Serum | Phenol/
chloroform | Standard curve | TaqMan | 0.71 | 79 | 13 | 34 | 54 | (36) |
| Zhang et al,
2010 | miR-223 | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 124 | 17 | 25 | 83 | (39) |
| Komatsu et
al, 2011 | miR-375 | Down | Japan | 50 (44/6) | 20 | Plasma | mirVANA PARIS | Standard curve | TaqMan | <0.01 | 39 | 5 | 11 | 15 | (23) |
| Li et al,
2015 | miR-375 | Up | China | 38 (30/8) | 19 | Plasma | mirVANA PARIS | miR-1228 | TaqMan | YI | 31 | 3 | 7 | 16 | (22) |
| Wu et al,
2014 | miR-25 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 47 | 16 | 16 | 47 | (46) |
| Komatsu et
al, 2014 | miR-25 | Up | Japan | 20 | 50 | Plasma | mirVANA PARIS | Standard curve | TaqMan | 0.32 | 17 | 7 | 3 | 43 | (45) |
| Wu et al,
2014 | miR-100 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 48 | 22 | 15 | 41 | (46) |
| Zhang et al,
2010 | miR-100 | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | 0.01 | 95 | 19 | 54 | 81 | (39) |
| Xie et al,
2013 | miR-10b | Down | China | 29b (24/5), (61.3±9.5) | 16 (13/3),
(57.5±7.1) | Plasma | mirVANA PARIS | miR-16 | SYBR | 0.56 | 24 | 0 | 5 | 16 | (37) |
| Xu et al,
2015 | miR-10b | Down | China | 50 (27/23) | 50 (30/20) | Serum | mirVANA PARIS | RUN6B | TaqMan | YI | 38 | 8 | 12 | 42 | (49) |
| Zhang et al,
2011 | miR-31 | Up | China | 120 (79/41) 81
(49/32) | 121 (76/45) 81
(43/38) | Serum | miRNeasy Mini | miR-16 | SYBR | 0.01 0.01 | 104 70 | 19 17 | 16 11 | 102 64 | (34) |
| Zhang et al,
2013 | miR-1322 | Up | China | 120 (79/41) 81
(49/32) | 120 (75/45) 81
(43/38) | Serum | miRNeasy Mini | miR-16 | SYBR | 0.01 0.01 | 98 68 | 21 16 | 22 13 | 99 65 | (44) |
| Takeshita et
al, 2013 | miR-1246 | Up | Japan | 101 (89/12) | 46 | Serum | mirVANA PARIS | miR-16 | TaqMan | 1.32 | 72 | 12 | 29 | 34 | (43) |
| Zhang et al,
2010 | miR-10a | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 121 | 20 | 28 | 80 | (39) |
| Zhang et al,
2010 | miR-127 | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 117 | 13 | 32 | 87 | (39) |
| Zhang et al,
2010 | miR-133a | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 97 | 17 | 52 | 83 | (39) |
| Zhang et al,
2010 | miR-148b | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 99 | 13 | 50 | 87 | (39) |
| Zhang et al,
2010 | miR-22 | Up | China | 149 (116/33) | 100 (74/26) | Serum | Trizol | Serum volume | TaqMan | YI | 132 | 14 | 17 | 86 | (39) |
| Li et al,
2015 | miR-16 | Up | China | 38 (30/8) | 19 | Plasma | mirVANA PARIS | miR-1228 | TaqMan | 3.45 | 36 | 8 | 2 | 11 | (22) |
| Li et al,
2015 | miR-185 | Up | China | 38 (30/8) | 19 | Plasma | mirVANA PARIS | miR-1228 | TaqMan | 3.97 | 38 | 8 | 0 | 11 | (22) |
| Liu et al,
2012 | miR-155 | Down | China | 60, (61.9±7.4) | 60, (63.6±8.6) | Plasma | Trizol | RUN6B | SYBR | YI | 38 | 21 | 22 | 39 | (35) |
| Jiang et al,
2015 | miR-218 | Down | China | 106b (69/37) | 60 | Serum | mirVANA PARIS | miR-16 | Molecular
beacon | 0.77 | 76 | 14 | 30 | 46 | (48) |
| He et al,
2015 | miR-20a | Up | China | 70 (46/24), | 40, (61.7±6.9)
(60.5±8.4) | Plasma | Trizol reagent BD
TB-126 | SV40 | SYBR | 4.77 | 45 | 10 | 25 | 30 | (47) |
| He et al,
2015 | let-7a | Down | China | 70 (46/24), | 40, (61.7±6.9)
(60.5±8.4) | Plasma | Trizol reagent BD
TB-126 | SV40 | SYBR | 6.22 | 52 | 6 | 18 | 34 | (47) |
| Hirajim et
al, 2013 | miR-18a | Up | Japan | 106 (87/19) | 54 | Plasma | mirVANA PARIS | Standard curve | TaqMan | 1.99 | 92 | 0 | 14 | 54 | (41) |
| Sharma et
al, 2013 | miR-107 | Down | India | 14 | 17 | Serum | QIAamp Viral RNA
mini | 5s rRNA | SYBR | YI | 9 | 3 | 5 | 14 | (42) |
| Sun et al,
2015 | miR-718 | Down | China | 120 (79/41) | 51 | Plasma | Trizol reagent BD
TB-126 | miR-16 | SYBR | 0.09 | 83 | 17 | 37 | 34 | (51) |
| Guan et al,
2015 | miR-613 | Down | China | 75 (43/32) | 75 | Serum | miRNeasy
serum/plasma | miR-16 | SYBR | 0.88 | 61 | 28 | 14 | 47 | (50) |
| Hui et al,
2015 | miR-129 | Up | China | 69 | 14 | Serum | mirVANA PARIS | miR-1228 | TaqMan | 0.39 | 54 | 4 | 15 | 10 | (38) |
| Hui et al,
2015 | miR-365 | Up | China | 69 | 14 | Serum | mirVANA PARIS | miR-1228 | TaqMan | 5.06 | 56 | 2 | 13 | 12 | (38) |
| Hui et al,
2015 | miR-451 | Up | China | 69 | 14 | Serum | mirVANA PARIS | miR-1228 | TaqMan | 0.57 | 57 | 3 | 12 | 11 | (38) |
| Xu et al,
2015 | miR-29c | Down | China | 50 (27/23) | 50 (30/20) | Serum | mirVANA PARIS | RUN6B | TaqMan | YI | 39 | 7 | 11 | 43 | (49) |
| Xu et al,
2015 | miR-205 | Down | China | 50 (27/23) | 50 (30/20) | Serum | mirVANA PARIS | RUN6B | TaqMan | YI | 38 | 7 | 12 | 43 | (49) |
| Wu et al,
2014 | miR-193 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 47 | 8 | 16 | 55 | (46) |
| Wu et al,
2014 | miR-194 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 54 | 4 | 15 | 10 | (46) |
| Wu et al,
2014 | miR-483 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | YI | 57 | 3 | 12 | 11 | (46) |
| Wu et al,
2014 | miR-337 | Up | China | 63 (55/8),
(62.0±8.3) | 63 (55/8),
(59.7±6.8) | Serum | Trizol | let-7d, 7g and
7i | TaqMan | 0.37 | 54 | 4 | 15 | 10 | (46) |
RNA purification
RNA was purified from 200 µl plasma using a miRNeasy
Serum/Plasma kit (cat. no. 217184; Qiagen GmbH, Hilden, Germany)
and eluted with 14 µl of RNase-free water, according to the
manufacturer's instructions. The yield of RNA was determined by a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Waltham, MA USA). The total extracted RNA was stored at −80°C until
required.
RT-qPCR
Quantification of the 5 selected miRNAs was
performed using RT-qPCR, with 5S rRNA as a reference for
normalization. The reverse transcription of total RNA was performed
using a miScript II Reverse Transcription kit (cat. no. 218161;
Qiagen GmbH) in the GeneAmp® PCR System 9700 (Applied
Biosystems, Foster City, CA, USA). Thermocycling conditions were as
follows: 37°C for 60 min followed by 95°C for 5 min. Each reaction
consisted of 0.5 µg RNA, 1 µl 10× Nucleics mix, 2 µl 5× HiSpec
buffer and 0.5 µl miScript Reverse Transcriptase Mix, with
RNase-free water added for a total volume of 10 µl. The reaction
mix was diluted to 1:10 using RNase-free water and stored at −20°C
until use. The PCR amplification was performed using SYBR Green I
Master on the LightCycler® 480 II Real-time PCR
Instrument (both from Roche Diagnostics GmbH, Basel, Switzerland),
according to the manufacturer's instructions. The thermocycling
conditions of the PCR amplification reactions were as following:
First an initial denaturation for 10 min at 95°C, then 40 cycles of
denaturation at 95°C for 10 sec and annealing at 60°C for 30 sec.
The primer sequences are listed in Table
II. Each reaction was performed in triplicate. The cycle number
at which the fluorescence reached the fixed threshold was referred
to as the cycle threshold (Cq). The expression levels of miRNAs
were calculated using the 2−ΔΔCq method (ΔΔCq =
ΔCqcase - ΔCqcontrol, ΔCq =
CqmiRNA - Cqreference) (29).
 | Table II.Primer sequences used for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction.
| Name | Primer sequence
(5′-3′) |
|---|
| Universal
primer | Unknown (provided
by Qiagen) |
| 5S rRNA |
GGAGACCGCCTGGGAATA |
| hsa-miR-21 |
TAGCTTATCAGACTGATGTTGA |
| hsa-miR-223 |
TGTCAGTTTGTCAAATACCCCA |
| hsa-miR-375 |
TTTGTTCGTTCGGCTCGCGTGA |
| hsa-miR-25 |
CATTGCACTTGTCTCGGTCTGA |
| hsa-miR-100 |
AACCCGTAGATCCGAACTTGTG |
Statistical analysis
Stata 12.0 software (StataCorp LP, College Station,
TX, USA) was used to perform the diagnostic meta-analysis using the
Midas module produced by Dwamena (30). The bivariate mixed effects model
(31) was used to calculate the main
parameters and their corresponding 95% confidence intervals (CIs).
The heterogeneity among studies was assessed by I2
tests, and significant heterogeneity was considered to exist when
I2 >50% (32).
Publication bias was evaluated by Deeks' funnel plots. P<0.1 was
considered to indicate a statistically significantly
difference.
Non-parametric tests (the Mann-Whitney U test and
Kruskal-Wallis test) were used to compare the plasma miRNAs
expression levels. The associations between the concentrations of
plasma miRNAs and clinicopathological features were evaluated by
Fisher's exact test or χ2 test. The analysis of receiver
operating characteristic (ROC) curves was performed to evaluate
diagnostic power, allowing estimation of the area under the ROC
curves (AUCs) with 95% CIs. The Youden index was used to determine
the cutoff value of miRNAs (33).
GraphPad Prism 6 (GraphPad software Inc., La Jolla, CA, USA) was
used to produce figures. A two-sided P<0.05 was considered to
indicate a statistically significant difference.
Results
Subject characteristics
A total of 125 patients with ESCC and 125 age- and
sex-matched healthy controls were included in the study. All
subjects were >40 years old, with ESCC patients ranging from
41–80 and healthy individuals from 40–79; the median age was 63 for
these two groups. The main clinicopathological features of patients
with ESCC are listed in Table III.
No data was available for 4 (3.2%) patients regarding the TNM
stage, 7 (5.6%) regarding the tumor location and 14 (11.2%)
regarding the differentiation grade. There was only data regarding
M-status for a further 3 patients. Therefore, in total, there were
7 patients for which data regarding the T and N stage were not
available.
 | Table III.The association between plasma miRNA
expression levels and clinicopathological factors of patients with
esophageal squamous cell carcinoma. |
Table III.
The association between plasma miRNA
expression levels and clinicopathological factors of patients with
esophageal squamous cell carcinoma.
|
|
| miR-21 | miR-223 | miR-375 | miR-25 | miR-100 |
|---|
|
|
|
|
|
|
|
|
|---|
| Variable | n | High | Low | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value |
|---|
| Total | 125 | 39 | 86 |
| 42 | 83 |
| 32 | 93 |
| 38 | 87 |
| 36 | 89 |
|
| Sex |
|
|
| 0.213 |
|
| 0.134 |
|
| 0.998 |
|
| 0.802 |
|
| 0.753 |
|
Male | 76 | 21 | 55 |
| 23 | 53 |
| 19 | 57 |
| 21 | 55 |
| 19 | 57 |
|
|
Female | 49 | 18 | 31 |
| 19 | 30 |
| 13 | 36 |
| 17 | 32 |
| 17 | 32 |
|
| Age |
|
|
| 0.959 |
|
| 0.358 |
|
| 0.464 |
|
| 0.716 |
|
| 0.423 |
|
<63 | 59 | 20 | 39 |
| 20 | 39 |
| 16 | 43 |
| 19 | 40 |
| 16 | 43 |
|
|
≥63 | 66 | 19 | 47 |
| 22 | 44 |
| 16 | 50 |
| 19 | 47 |
| 20 | 46 |
|
| Tumor location |
|
|
| 0.097 |
|
| 0.123 |
|
| 0.668 |
|
| 0.003 |
|
| 0.928 |
|
Upper | 22 | 7 | 15 |
| 8 | 14 |
| 5 | 17 |
| 6 | 16 |
| 4 | 18 |
|
|
Middle | 65 | 17 | 48 |
| 16 | 49 |
| 20 | 45 |
| 16 | 49 |
| 24 | 41 |
|
|
Lower | 31 | 15 | 16 |
| 18 | 13 |
| 4 | 27 |
| 15 | 16 |
| 6 | 25 |
|
| T stage |
|
|
| 0.006 |
|
| 0.002 |
|
| <0.001 |
|
| 0.001 |
|
| <0.001 |
|
Tis-T1 | 50 | 25 | 25 |
| 27 | 32 |
| 3 | 47 |
| 25 | 25 |
| 5 | 45 |
|
| T2 | 20 | 6 | 14 |
| 4 | 16 |
| 10 | 10 |
| 3 | 17 |
| 8 | 12 |
|
|
T3-4 | 48 | 8 | 40 |
| 11 | 37 |
| 14 | 34 |
| 9 | 39 |
| 18 | 30 |
|
| N stage |
|
|
| 0.122 |
|
| 0.001 |
|
| <0.001 |
|
| <0.001 |
|
| <0.001 |
|
Negative | 86 | 33 | 53 |
| 38 | 48 |
| 16 | 70 |
| 34 | 52 |
| 20 | 66 |
|
|
Positive | 32 | 6 | 26 |
| 4 | 28 |
| 11 | 21 |
| 3 | 29 |
| 11 | 21 |
|
| M stage |
|
|
| 0.906 |
|
| 0.158 |
|
| 0.075 |
|
| 0.513 |
|
| 0.126 |
|
Negative | 116 | 38 | 78 |
| 41 | 75 |
| 27 | 89 |
| 36 | 80 |
| 31 | 85 |
|
|
Positive | 5 | 1 | 4 |
| 1 | 4 |
| 2 | 3 |
| 1 | 4 |
| 1 | 4 |
|
| TNM stage |
|
|
| 0.001 |
|
| <0.001 |
|
| <0.001 |
|
| <0.001 |
|
| <0.001 |
|
0/I | 54 | 29 | 25 |
| 32 | 22 |
| 3 | 51 |
| 28 | 26 |
| 6 | 48 |
|
| II | 38 | 4 | 34 |
| 6 | 32 |
| 16 | 22 |
| 7 | 31 |
| 16 | 22 |
|
|
III/IV | 29 | 6 | 23 |
| 4 | 25 |
| 10 | 19 |
| 2 | 27 |
| 10 | 19 |
|
| Differentiation
grade |
|
|
| 0.453 |
|
| 0.654 |
|
| 0.016 |
|
| 0.772 |
|
| 0.012 |
|
Well | 15 | 7 | 8 |
| 6 | 9 |
| 1 | 14 |
| 3 | 12 |
| 1 | 14 |
|
|
Moderate | 46 | 14 | 32 |
| 14 | 32 |
| 9 | 37 |
| 17 | 29 |
| 11 | 35 |
|
|
Poor | 50 | 16 | 34 |
| 20 | 30 |
| 16 | 34 |
| 13 | 37 |
| 18 | 32 |
|
Characteristics of the studies
included in the meta-analysis
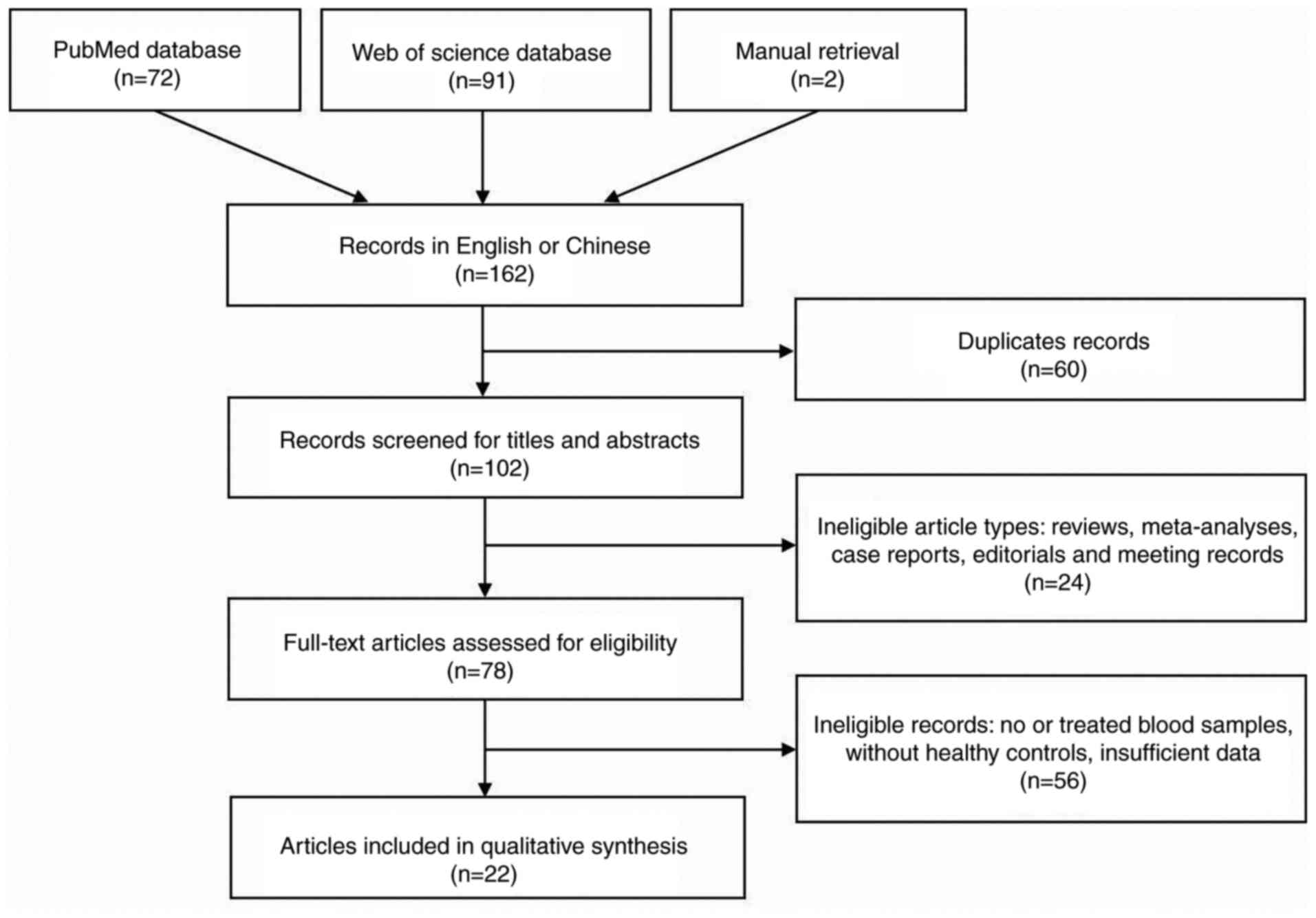
With non-English/Chinese studies excluded, a total
of 162 articles were retrieved from the 2 databases and manual
retrieval (Fig. 1). Among them, 140
were subsequently excluded according to the exclusion criteria. As
a consequence, 22 articles (22,23,34–53)
were eventually included in the meta-analysis. Among them, 7
articles (22,38,39,22)
studied more than one miRNA. A total of 2 articles (34,44)
focused on different miRNAs in the same subjects and contained a
validation group. Thus, this meta-analysis contained 45 studies and
33 miRNAs, 1,589 patients with EC and 1,112 healthy
individuals.
The main characteristics of each study are listed in
Table I. The vast majority of studies
considered only ESCC, with just 21 patients from 3 studies
(37,48,53)
presenting with EAC (esophageal adenocarcinoma). In addition, one
study (40) did not provide patient
histological types. All studies were published between 2010 and
2016 in Asia, of which 39 were published in China, 5 in Japan and 1
in India. MiR-21, miR-375, miR-223, miR-25, miR-100 and miR-10b
were reported twice or more in different articles.
The majority of the miRNAs were reported as
overexpressed in ESCC. However, different studies arrived at
opposing conclusions regarding the expression of miR-375. All
eligible studies in this meta-analysis used RT-qPCR to determine
the expression levels of miRNAs in plasma (n=16) or serum (n=29),
of which 13 studies performed RT-qPCR with SYBR-Green while 32
studies used probes. The expression levels of miRNAs were
relatively or absolutely quantified using the 2−ΔCq or
2−ΔΔCq methods. The methods for normalization varied
from total serum volume to reference controls, including SV40,
RUN6B, 5S rRNA, the combination of let-7d, 7g and 7i, miR-16,
miR-1228 or other synthetic miRNAs for the construction of standard
curves. Cutoff values were set according to the Youden index and
varied from <0.01 to 6.22. The overall quality of the included
studies in the meta-analysis was assessed using the Quality
Assessment of Diagnostic Accuracy Studies 2 (54), and the result was relatively
moderate.
Evaluation of the meta-analysis
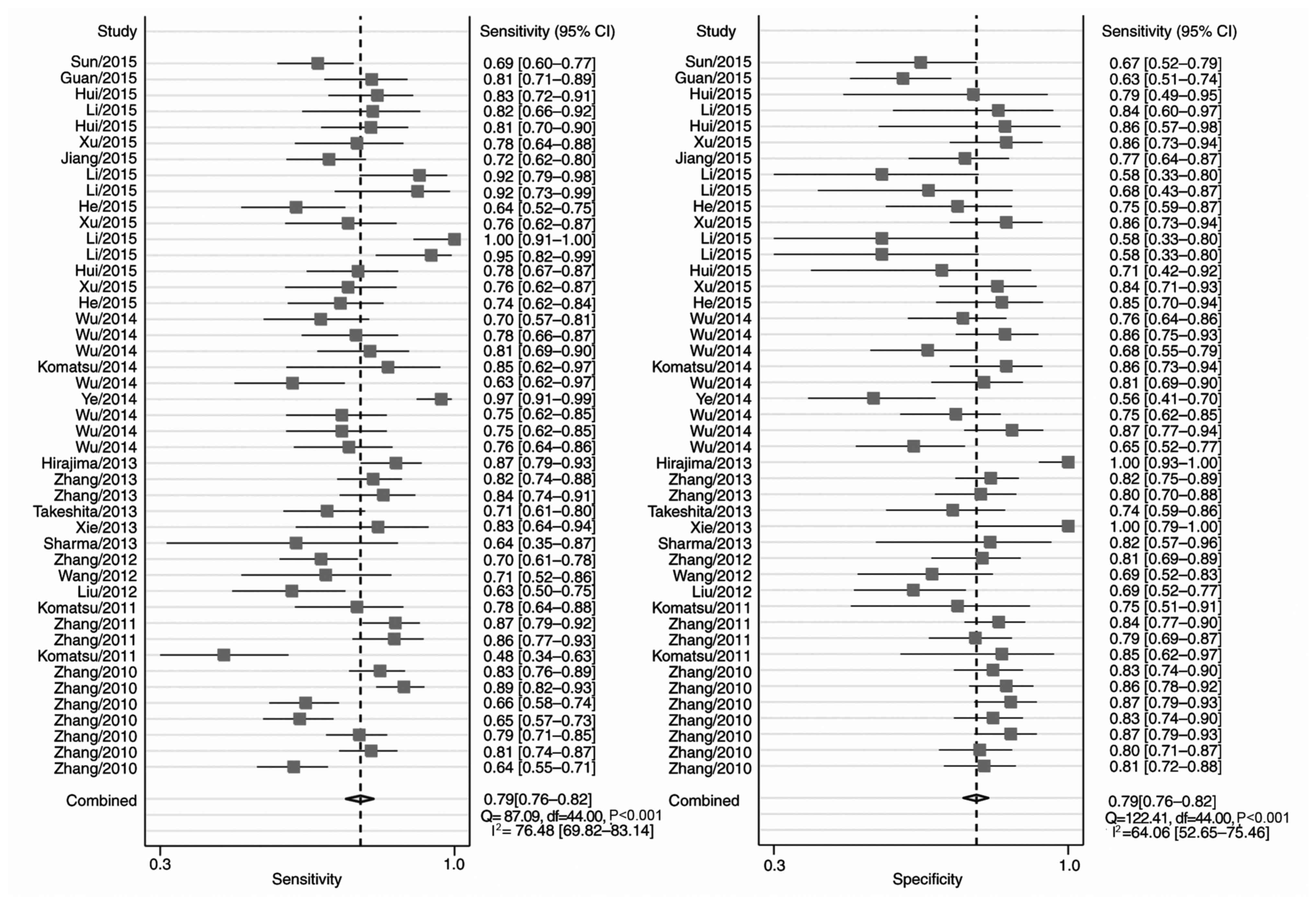
Fig. 2 includes the
forest plots for 45 studies of the sensitivity and specificity of
miRNA detection for EC diagnosis. Heterogeneity between studies in
sensitivity and specificity were recorded; the I2 values
were 76 and 64%, respectively. Overall pooled sensitivity and
specificity were 0.79 (95% CI, 0.76–0.82 for both), and the AUC was
0.86 (95% CI, 0.83–0.89), indicating a relatively high accuracy. As
the majority of patients presented with ESCC, the pooled ESCC
estimates were close to the overall estimates (Table IV).
 | Table IV.Summary of the estimates of
diagnostic value, with 95% CIs. |
Table IV.
Summary of the estimates of
diagnostic value, with 95% CIs.
| Category | Sensitivity | Specificity | LRP | LRN | DOR | AUC |
|---|
| Country |
|
China | 0.79
(0.76–0.82) | 0.79
(0.76–0.81) | 3.7 (3.3–4.2) | 0.26
(0.23–0.30) | 14 (11–18) | 0.86
(0.82–0.89) |
|
Non-China | 0.72
(0.58–0.82) | 0.88
(0.68–0.96) | 5.8 (1.9–17.7) | 0.32
(0.19–0.54) | 18 (4–87) | 0.85
(0.81–0.88) |
| Sample type |
|
Serum | 0.76
(0.72–0.79) | 0.81
(0.75–0.85) | 3.9 (3.0–5.1) | 0.30
(0.25–0.35) | 13 (9–19) | 0.84
(0.81–0.87) |
|
Plasma | 0.82
(0.76–0.86) | 0.78
(0.75–0.82) | 3.8 (3.2–4.4) | 0.24
(0.19–0.30) | 16 (12–22) | 0.86
(0.83–0.89) |
| ESCC only | 0.79
(0.75–0.82) | 0.79
(0.76–0.82) | 3.8 (3.3–4.4) | 0.27
(0.23–0.31) | 14 (11–18) | 0.86
(0.83–0.89) |
| Overall | 0.79
(0.76–0.82) | 0.79
(0.76–0.82) | 3.8 (3.3–4.3) | 0.27
(0.23–0.31) | 14 (11–18) | 0.86
(0.83–0.89) |
Since only one study was performed in India, the
studies were divided into 2 subgroups: China and non-China.
Subgroup analyses based on countries and sample types were
performed (Table IV). The China
subgroup had a higher pooled sensitivity compared with the
non-China subgroup (0.79 vs. 0.72), but a lower pooled specificity
(0.79 vs. 0.88). Regarding sample types, the pooled sensitivity was
0.82 vs. 0.76, and the specificity 0.78 vs. 0.81, when comparing
plasma to serum. Heterogeneity was observed within each subgroup,
suggesting that country and sample types were not the major sources
of heterogeneity in the meta-analysis. In the meta-regression
analyses, differences in the pooled sensitivity and specificity
were statistically significant for the following covariates: The
number of cases and controls, the method used to perform RT-qPCR
and sample type. Hence, the sources of inter-study heterogeneity
remain unclear.
The Deeks' funnel plot was used to explore the
potential publication bias. The P-value of the slope coefficient
was 0.881, indicating no distinct asymmetry and a relatively low
probability of publication bias in the meta-analysis.
Comparison of the 5 candidate miRNAs
in patients with ESCC compared with healthy controls
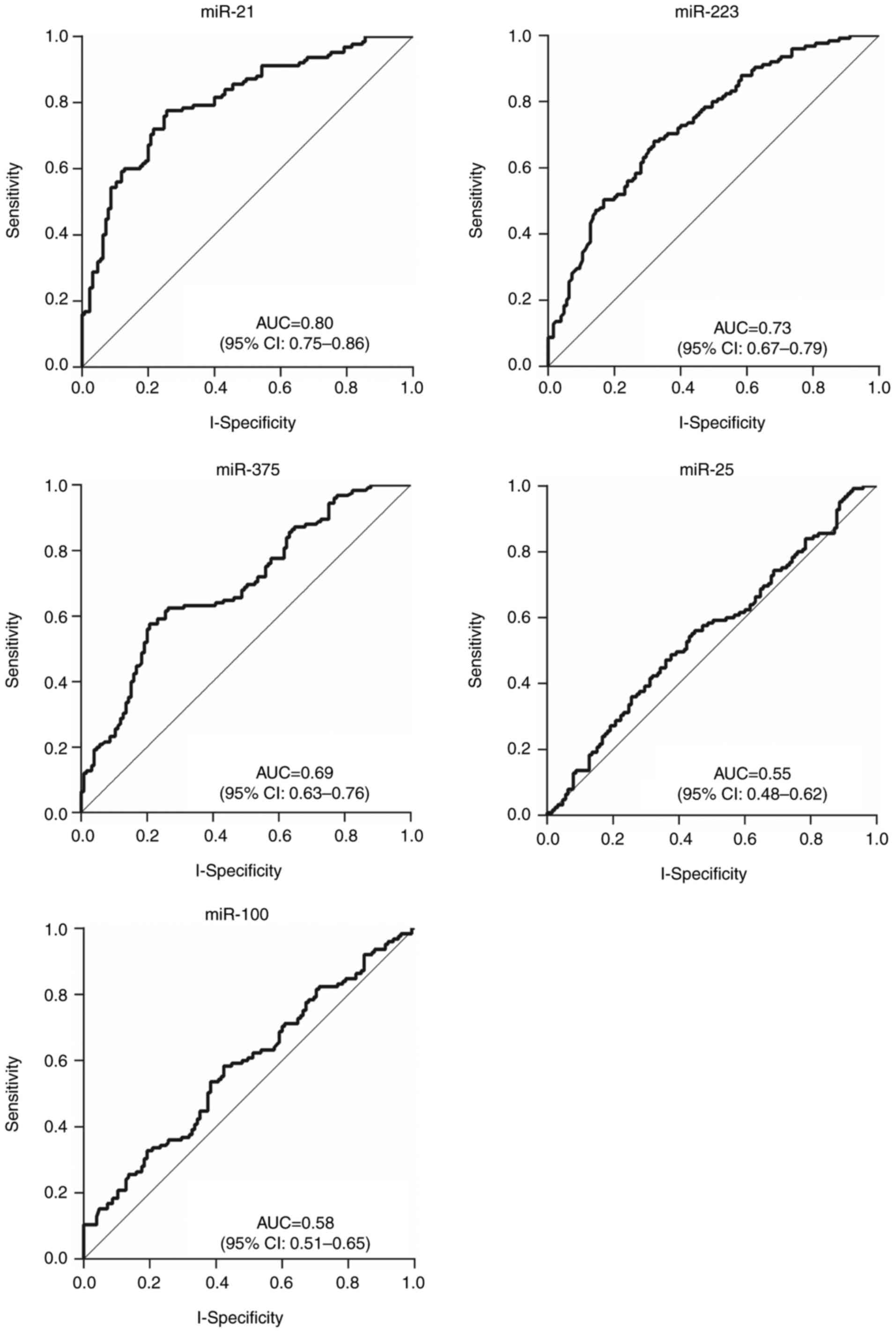
The plasma expression levels of the 5 candidate
miRNAs, miR-21, miR-223, miR-100, miR-25, and miR-375, were
determined using RT-qPCR. Analyses of ROC curves were conducted to
evaluate their diagnostic values. Although the results revealed
statistically significant differences in the expression of the 4
miRNAs (P=0.025 for miR-100, and P<0.001 for miR-21, miR-223,
and miR-375), only those ≥2-fold upregulated or ≤0.5-fold
downregulated between the blood of patients with ESCC and controls
were considered to be meaningful. Consequently, miR-21 and miR-223
expression was determined to be upregulated whereas miR-375 was
downregulated in patients with ESCC compared with healthy
individuals (Table V). The
corresponding AUCs were 0.80 (95% CI, 0.75–0.86) for miR-21, 0.73
(95% CI, 0.67–0.79) for miR-223 and 0.69 (95% CI, 0.63–0.76) for
miR-375, respectively (Table V;
Fig. 3).
 | Table V.Diagnostic values of the 5 candidate
miRNAs in differentiating patients with esophageal squamous cell
carcinoma from controls. |
Table V.
Diagnostic values of the 5 candidate
miRNAs in differentiating patients with esophageal squamous cell
carcinoma from controls.
| Biomarker | Fold change | P-value | Sensitivity | Specificity | AUC (95% CI) |
|---|
| miR-21 | 3.13 | 0.000 | 0.74 | 0.78 | 0.80
(0.75–0.86) |
| miR-223 | 3.88 | 0.000 | 0.68 | 0.68 | 0.73
(0.67–0.79) |
| miR-100 | 0.72 | 0.164 | 0.58 | 0.58 | 0.58
(0.51–0.65) |
| miR-25 | 1.44 | 0.025 | 0.54 | 0.57 | 0.55
(0.48–0.62) |
| miR-375 | 0.37 | 0.000 | 0.78 | 0.59 | 0.69
(0.63–0.76) |
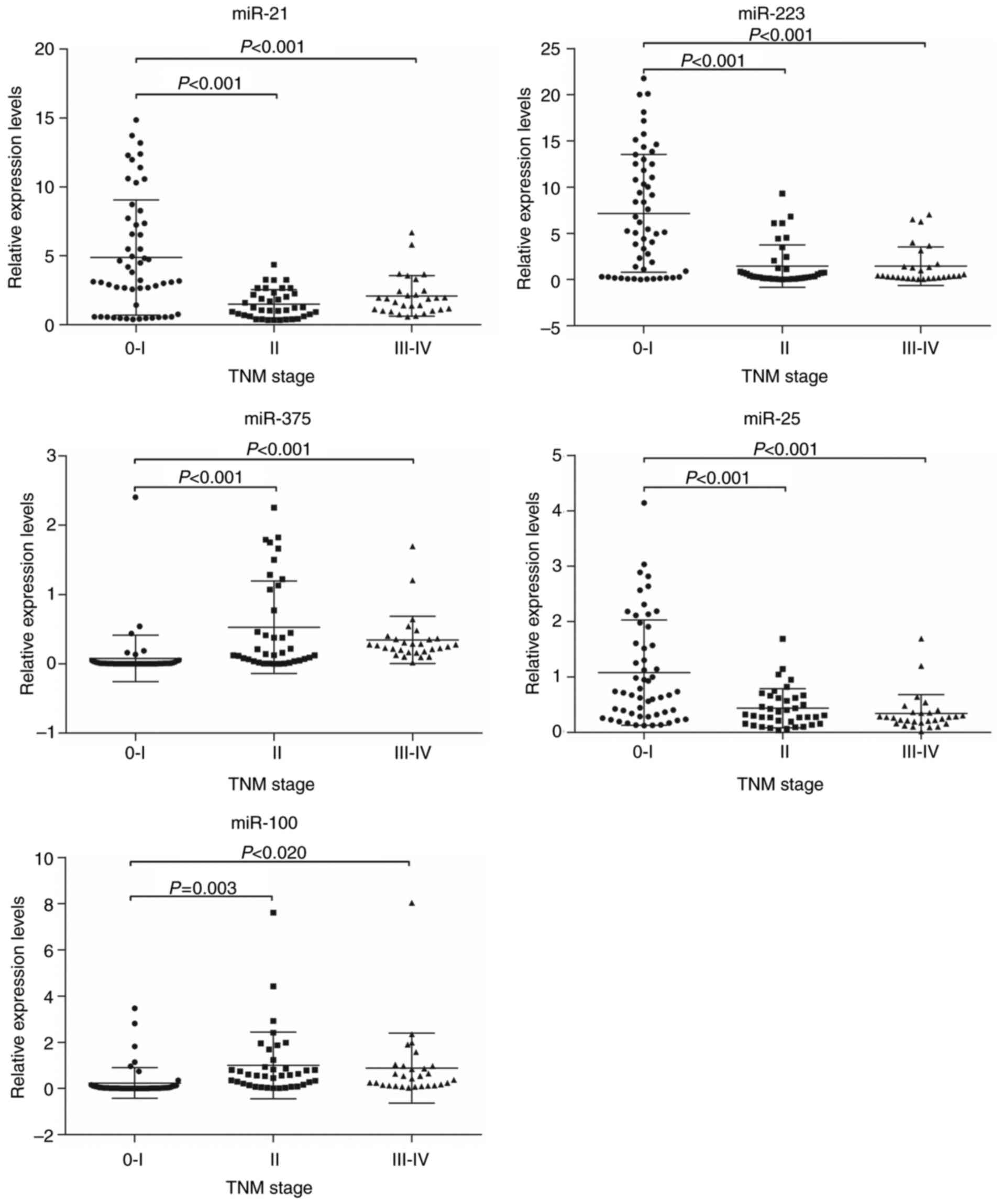
Associations of the expression levels
of plasma miRNAs with ESCC clinicopathological factors
Based on the mean expression level, the patients
were divided into a high-expression group and a low-expression
group for each miRNA. Age and sex were not associated with the
expression of any miRNA (P>0.05). The expression level of miR-25
was significantly associated with tumor location (P<0.001), and
was higher for tumors of the lower-esophagus than those in the
upper- and middle-esophagus. MiR-375 and miR-100 were associated
with differentiation grade (P=0.019 and P=0.010, respectively), but
there was no significant association between the expression of the
other miRNAs and the differentiation grade. The expression levels
of all 5 plasma miRNAs were also significantly associated with the
TNM stage, particularly the T stage. However, the M stage was not
associated with the expression of any miRNA (Table IV). The statistical association of
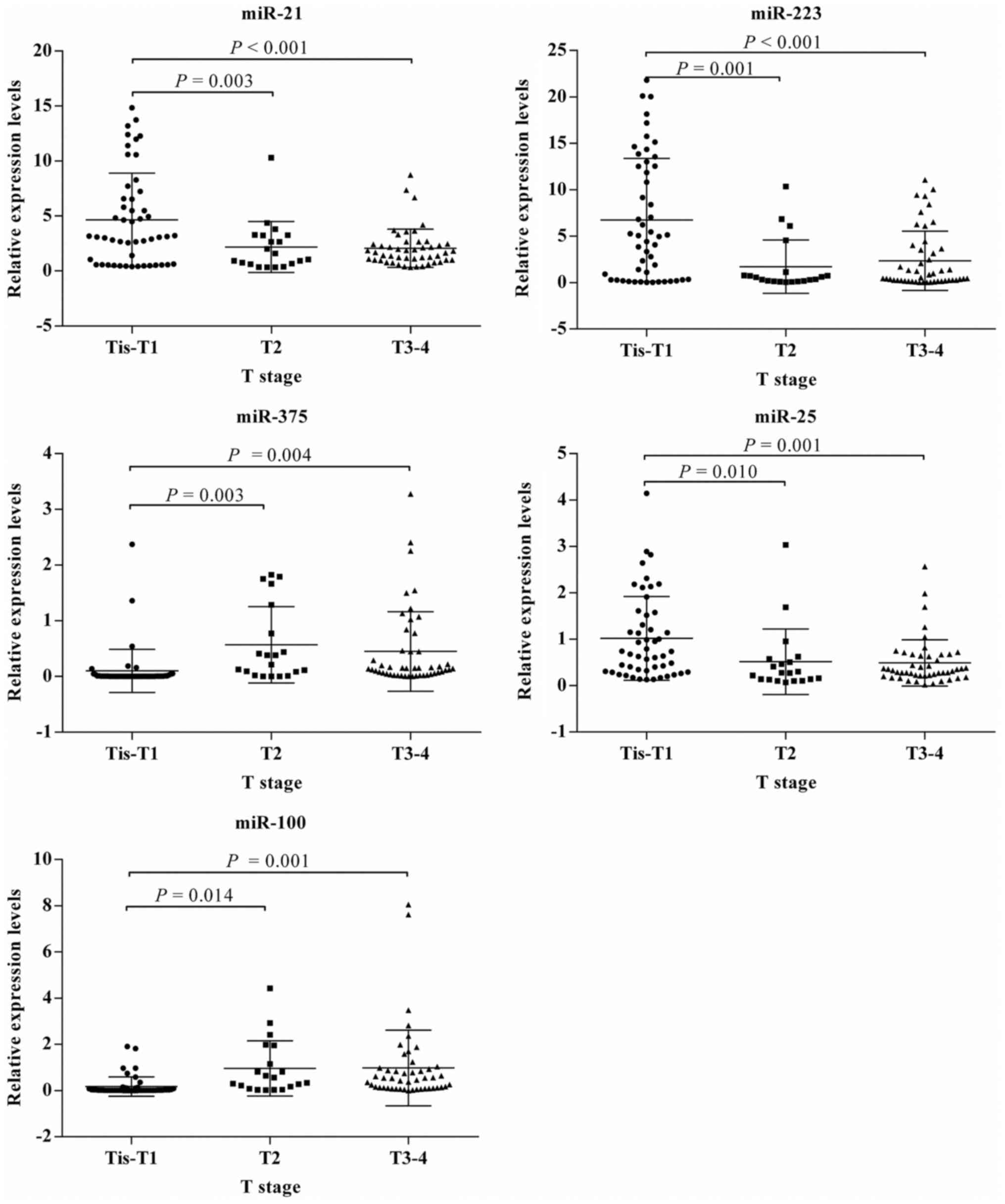
miRNA expression level with TNM stage (Fig. 4) and T stage (Fig. 5) were consistent for all the miRNAs
considered. The expression levels of miR-21, miR-223, and miR-25
were higher, whereas miR-100 and miR-375 were lower in patients
with stage 0–I or Tis-T1 tumors than those in other stages.
Diagnostic value of the 5 candidate
miRNAs for early ESCC
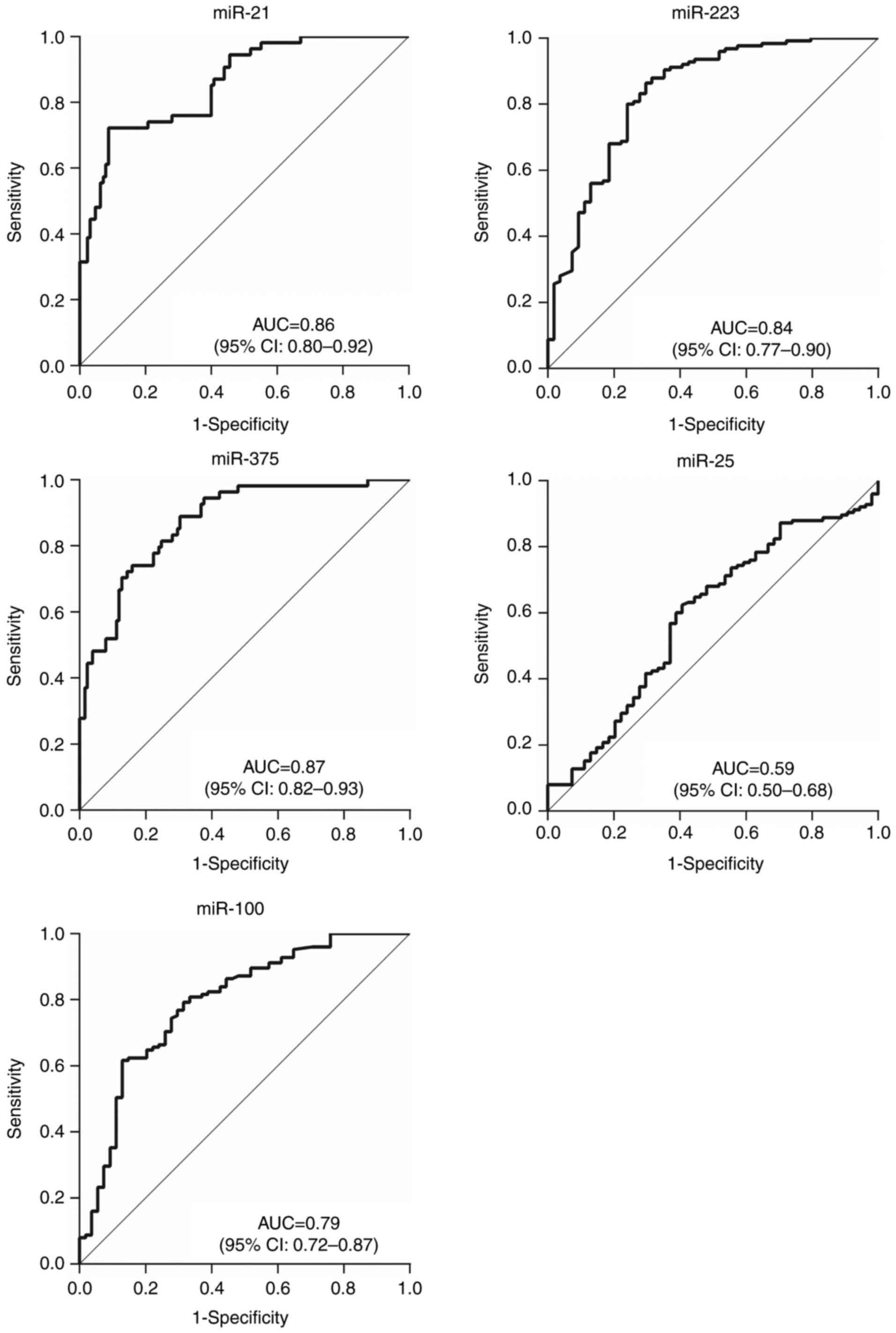
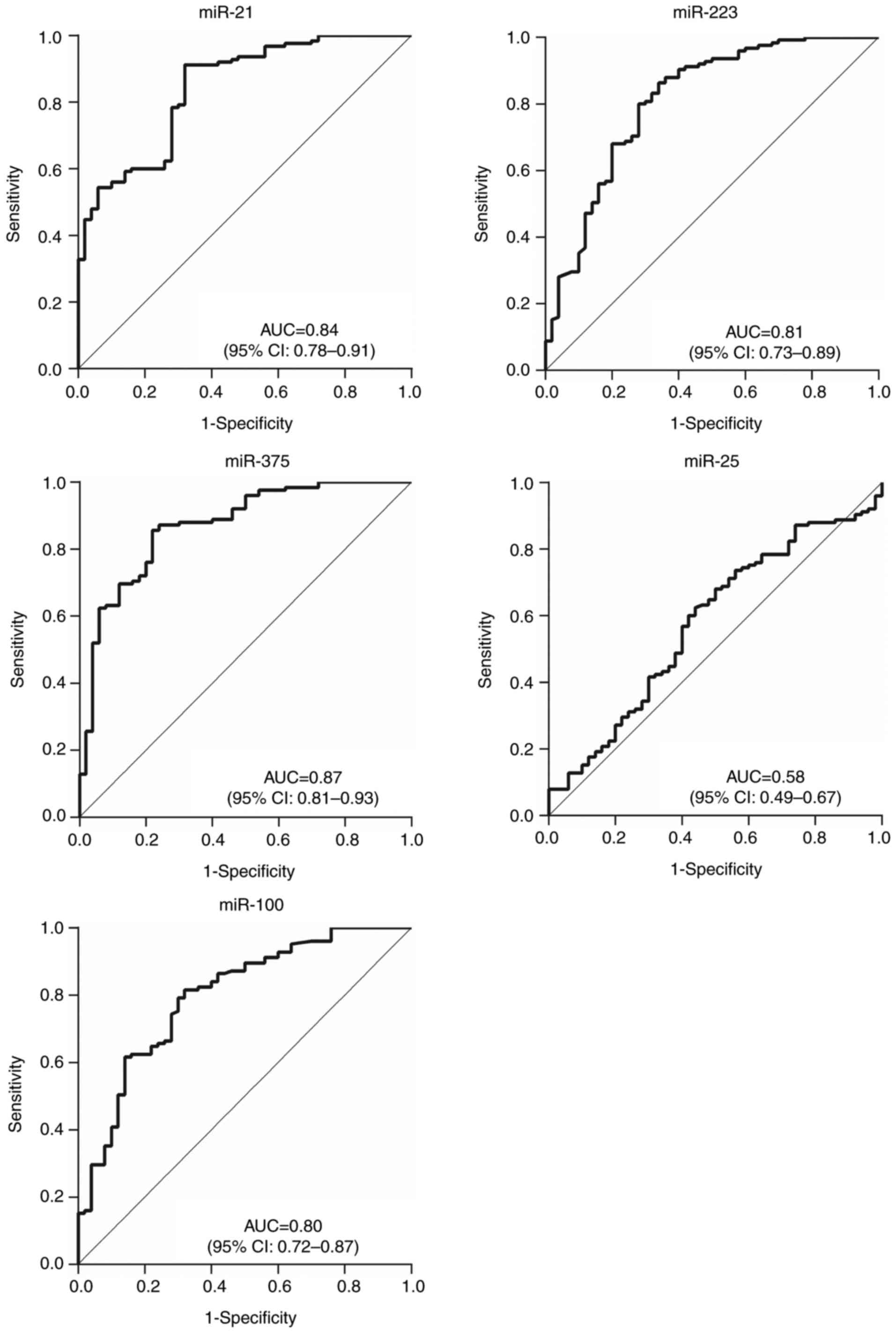
ROC curves were constructed and analyzed to evaluate
the diagnostic values of the 5 candidate miRNAs for differentiating
patients with ESCC in stages 0–I (Fig.
6) and Tis-T1 (Fig. 7) from
healthy individuals. The results indicated that the 3 abnormally
expressed miRNAs had relatively high AUCs of 0.86 (95% CI,
0.80–0.92) for miR-21, 0.83 (95% CI, 0.77–0.90) for miR-223 and
0.87 (95% CI, 0.82–0.93) for miR-375 when comparing patients in
stage 0–I to healthy patients; the AUCs when comparing patients
with Tis-T1 tumors to all healthy patients were 0.84 (95% CI,
0.78–0.91), 0.81 (95% CI, 0.73–0.89) and 0.87 (95% CI, 0.81–0.93)
respectively. This indicates that these miRNAs could be effective
in providing early diagnosis. The diagnostic value of plasma
miR-100 was also improved, from AUC 0.58 (95% CI, 0.51–0.65) for
all patients with ESCC, to 0.79 (95% CI, 0.72–0.87) for stage 0–I
and 0.80 (95% CI, 0.72–0.87) for stage Tis-T1.
Discussion
It has been demonstrated that miRNAs are remarkably
stable, reproducible and consistent in clinical studies (55). Furthermore, they can be easily
detected by microarray or RT-qPCR for EC diagnosis, which is
advantageous over conventional detection methods for EC, including
endoscopic and pathologic examination. Numerous studies have
explored the associations between miRNAs and various cancers.
However, a large proportion of this research was performed in
tissues or cell lines, and studies in the context of bodily fluids
are rare. Compared with other bodily fluids, blood is generally
more applicable as it is easily attained in a relatively
noninvasive manner and can be stored for long periods. Circulating
miRNAs are highly resistant to RNase A-digestion and other adverse
environments, including repeated freeze-thaw cycles, prolonged
storage at room temperature and extreme pH conditions (56). Thus, circulating miRNAs have provided
a promising scope for the discovery of diagnostic biomarkers since
2008 (12).
Considering that a number of circulating miRNAs have
been identified for the detection of EC, a meta-analysis was
conducted in the present study to evaluate the diagnostic values of
circulating miRNAs in plasma or serum in EC. The results revealed a
relatively high accuracy, with an AUC of 0.86. In the subgroup and
meta-regression analyses, it was demonstrated that the covariates
of country, sample type, size of cases and controls, and the
detection method used to perform RT-qPCR varied in the pooled
sensitivity and specificity. However, heterogeneity remained
evident in all subgroups based on these covariates, indicating that
the major sources of heterogeneity were not identified through
these analyses.
Although the expression levels of the circulating
miRNAs were all quantified using RT-qPCR, the heterogeneity
identified in the meta-analysis may be due to the varying RT-qPCR
methods and quantification methods of the studies analyzed. The
reagent used for RNA extraction from serum or plasma also varied,
as did the standardized control (57). Furthermore, only miRNAs significantly
overexpressed or reduced in expression were selected from the
eligible studies in the present meta-analysis. For example, Li
et al (22) selected 9 miRNAs
to evaluate their diagnostic values, but only 4 differentially
expressed miRNAs were identified in the study. Furthermore, all
studies were designed as case-controls, which may also contribute
to the heterogeneity. Ethnicity would be more likely to be the
source of heterogeneity instead of country, as all study subjects
were confined to Asia.
The present meta-analysis concentrated only on
patients who had not undergone surgery, chemotherapy or
radiotherapy in order to achieve a better reflection of the
diagnostic efficiency of circulating miRNAs. Wang et al
(58) concluded that the sample type
was a source for heterogeneity, potentially due to the negligence
of the value of I2 in subgroup and
meta-regression analyses.
Since the majority of miRNAs considered in the
studies included in the meta-analysis were reported only once, the
top 5 most-studied miRNAs (miR-21, miR-25, miR-100, miR-223 and
miR-375) were selected as candidate markers. According to the
meta-analysis, all 5 miRNAs have been identified as overexpressed
in EC, other than miR-375, for which there are opposing results
regarding its expression (22,23). The
present study used specifically selected patient samples and
RT-qPCR to validate the previous results. Only the expression
levels of plasma miR-21, miR-223 and miR-375 were significantly
different in patients with ESCC compared with controls. Among them,
miR-223 was the most significantly upregulated (fold change, 3.88)
and miR-21 had the highest diagnostic value (AUC, 0.80). None of
the miRNAs were associated with sex or age, which was in accordance
with previous reports (21,58,59). All 5
miRNAs were associated with the T and N TNM stages, with the
exception of miR-21, which was associated only with T stage. M
stage was not associated with any of the 5 miRNAs. Only miR-25 was
found to be associated with tumor location, with tumors from the
lower esophagus exhibiting a higher expression of miR-25.
Considering that a patient's prognosis is improved
by an early diagnosis, ROC curves were drawn to evaluate the
diagnostic values of plasma miRNAs in differentiating patients with
early ESCC (in stages 0–I and Tis-T1) from healthy controls.
Potentially due to the grouping method, except for miR-25, the
expression levels of all the miRNAs in patients with 0–I or Tis-T1
tumors were significantly and consistently altered compared to
patients with medium (stage II) and advanced (stage III–IV), or T2
and T3-4 tumors. The AUC for miR-100 increased from 0.58 in all
patients with ESCC to 0.79 in stage 0–I and 0.80 in stage Tis-T1,
and the AUC for miR-375 increased from 0.69 in all patients with
ESCC to 0.87 in stage 0–I and 0.87 in stage Tis-T1. Thus, it is
concluded that plasma miR-21, miR-223, miR-100 and miR-375 have the
potential to be used as biomarkers for the diagnosis of patients
with ESCC, particularly in the early stages of disease.
MiR-21 has been studied in various types of cancer;
it is hypothesized that it functions as an oncogene and is
upregulated in various types of solid tumor. It is widely involved
in cell growth, proliferation, invasion, intravasation and
metastasis by targeting tumor suppressor genes, including phosphate
and tensin homolog (60,61), programmed cell death 4 (62,63),
tropomyosin 1 (64), Sprouty2
(65), acidic nuclear phosphoprotein
32 and SWI/SNF-related, matrix associated, actin dependent
regulator of chromatin A4 (66).
Previous studies have demonstrated that miR-21 was associated with
chemoresistance (61,67) as well as prognosis (23,68) in
ESCC.
MiR-375 is predominantly expressed in the pancreatic
islets (24). Its expression has been
reported as downregulated in multiple types of malignancy,
including pancreatic adenocarcinomas (69), gastric cancer (24), head and neck squamous cell carcinoma
(70) and liver cancer (71). Janus kinase 2, MAX interactor 1, and
aryl hydrocarbon receptor have been identified as targets of
miR-375 that are involved in the regulation of carcinogenesis
(24,72). There is considerable evidence that
miR-375 is a tumor suppressor gene (26). The present study corroborates this
hypothesis, and not the findings of Li et al (22). One possible reason for these differing
conclusions may be error in the original experiment or the
subsequent analysis. The expression level of miR-375 may have been
interfered with by other factors, for example, blood sample
pollution. Alternatively, miR-375 may serve distinct roles
(oncogene or tumor suppressor gene) in different types of cancer,
as is the case for miR-223 and miR-25. Further research is required
to explore the mechanism of miR-375 in ESCC.
Previous studies have indicated that miR-223 and
miR-25 are overexpressed in many types of cancer, and may promote
cell proliferation, migration and invasion. However, miR-223
expression has been reported as downregulated in primary small cell
lung cancer (73), and miR-25 may
inhibit the proliferation of colon cancer and anaplastic thyroid
carcinoma cells, thus acting as a tumor suppressor gene (74,75).
The present study focused only on the diagnostic
value of circulating miRNAs in EC. A range of miRNAs have been
identified to exhibit altered expression in EC. These potential
biomarkers may be superior in sensitivity to the established
serologic biomarkers, including squamous cell carcinoma antigen
(SCCA) and carcinoembryonic antigen (CEA) (10,11).
However, issues remain regarding the applicability of circulating
miRNAs as diagnostic biomarkers. No clear consensus exists for the
normalization of miRNA detection in serum or plasma, nor does a
consistent method for analyzing RT-qPCR data, which leads to the
poor repeatability of results. This is a limitation when
determining the value of miRNAs for diagnosis. Therefore, it
remains unclear which single miRNA, or panel of miRNAs, could be
used effectively for the early detection of ESCC. Thus, further
efforts are required to set the relevant standards to avoid futile
and repetitive studies in the future. The survival rate for ESCC
remains poor even following surgery, radiotherapy and chemotherapy.
Although it has not yet been satisfactorily achieved, alterations
to miRNA expression have the potential to be developed into novel
therapies in the future. Considerable efforts are required before
miRNA-based methods can be clinically applied.
In conclusion, the present study verified that
miR-21 and miR-223 were over-expressed, whereas miR-375 expression
was reduced, in patients with ESCC compared with healthy
individuals. The identified miRNAs may serve as non-invasive
biomarkers for the diagnosis of ESCC, particularly early ESCC.
MiR-100 is also a promising biomarker for the early diagnosis of
ESCC. Further large-scale prospective studies are required to
confirm the clinical diagnostic value of circulating miRNAs in
ESCC.
Acknowledgements
The present study was supported by grants from the
General Program of National Natural Science Foundation of China
(grant no. 81372371), the Science and Technology Programs for
Tackling Key Problems in Henan province (grant nos. 162102310037
and 201503200), the Zhongyuan Scholar Program (grant no.
162101510006) and the Major Project of Science and Technology in
Henan province (grant no. 161100311400).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hiyama T, Yoshihara M, Tanaka S and
Chayama K: Genetic polymorphisms and esophageal cancer risk. Int J
Cancer. 121:1643–1658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Law S and Wong J: The current management
of esophageal cancer. Adv Surg. 41:93–119. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang JY, Looi KS and Tan EM:
Identification of tumor-associated antigens as diagnostic and
predictive biomarkers in cancer. Methods Mol Biol. 520:1–10. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu BC, Wang LD and Pei H:
Tumor-associated autoantibody measurement for screening esophageal
cancer. J Clin Exp Med. 2007.
|
|
8
|
Dai L, Ren P, Liu M, Imai H, Tan EM and
Zhang J: Using immunomic approach to enhance tumor-associated
autoantibody detection in diagnosis of hepatocellular carcinoma.
Clin Immunol. 152:127–139. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anderson KS and LaBaer J: The sentinel
within: Exploiting the immune system for cancer biomarkers. J
Proteome Res. 4:1123–1133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mroczko B, Kozłowski M, Groblewska M,
Łukaszewicz M, Nikliński J, Jelski W, Laudański J, Chyczewski L and
Szmitkowski M: The diagnostic value of the measurement of matrix
metalloproteinase 9 (MMP-9), squamous cell cancer antigen (SCC) and
carcinoembryonic antigen (CEA) in the sera of esophageal cancer
patients. Clin Chim Acta. 389:61–66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Łukaszewicz-Zając M, Mroczko B, Kozłowski
M, Nikliński J, Laudański J and Szmitkowski M: Higher importance of
interleukin 6 than classic tumor markers (carcinoembryonic antigen
and squamous cell cancer antigen) in the diagnosis of esophageal
cancer patients. Dis Esophagus. 25:242–249. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asaga S, Kuo C, Nguyen T, Terpenning M,
Giuliano AE and Hoon DS: Direct serum assay for microRNA-21
concentrations in early and advanced breast cancer. Clin Chem.
57:84–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang L, Cheng Q, Zhang BH and Zhang MZ:
Circulating microRNAs as biomarkers in hepatocellular carcinoma
screening: A validation set from China. Medicine (Baltimore).
94:e6032015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Z, Zhao C and Wang Z: MicroRNAs as
ideal biomarkers for the diagnosis of lung cancer. Tumour Biol.
35:10395–10407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan L, Zhao W, Yu H, Wang Y, Liu Y and Xie
C: A comprehensive meta-analysis of MicroRNAs for predicting
colorectal cancer. Medicine (Baltimore). 95:e27382016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hou C, Tan G and Feng S: Clinical
significance of microRNA expressions in diagnosing uterine cancer
and predicting lymph node metastasis. Tumour Biol. 35:10789–10798.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu S, Guan J and Liu Y: Identification of
microRNAs as novel biomarkers for glioma detection: A meta-analysis
based on 11 articles. J Neurol Sci. 348:181–187. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song JH and Meltzer SJ: MicroRNAs in
pathogenesis, diagnosis, and treatment of gastroesophageal cancers.
Gastroenterology. 143:35–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu F, Tian T, Xia LL, Ding Y, Cormier RT
and He Y: Circulating miRNAs as novel potential biomarkers for
esophageal squamous cell carcinoma diagnosis: A meta-analysis
update. Dis Esophagus. 30:1–9. 2017. View Article : Google Scholar
|
|
22
|
Li B, Yu Q, Shi Z, Li P and Fu S:
Circulating microRNAs in esophageal squamous cell carcinoma:
Association with locoregional staging and survival. Int J Clin Exp
Med. 8:7241–7250. 2015.PubMed/NCBI
|
|
23
|
Komatsu S, Ichikawa D, Takeshita H,
Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H,
Shiozaki A, et al: Circulating microRNAs in plasma of patients with
oesophageal squamous cell carcinoma. Br J Cancer. 105:104–111.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y,
Yao H, Liu X, Ke Y, Si J and Zhou T: MiR-375 frequently
downregulated in gastric cancer inhibits cell proliferation by
targeting JAK2. Cell Res. 20:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F,
Shen Y, Lu W, Wan X and Xie X: miR-375 is down-regulated in
squamous cervical cancer and inhibits cell migration and invasion
via targeting transcription factor SP1. Am J Pathol. 179:2580–2588.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M and
Moriyama M: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumors. 7th edition.
Wiley-Blackwell; Oxford, UK: 2009
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dwamena B: MIDAS: Stata module for
meta-analytical integration of diagnostic test accuracy studies.
Stat Softw Components. 14:2007.
|
|
31
|
Reitsma JB, Glas AS, Rutjes AW, Scholten
RJ, Bossuyt PM and Zwinderman AH: Bivariate analysis of sensitivity
and specificity produces informative summary measures in diagnostic
reviews. J Clin Epidemiol. 58:982–990. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Akobeng AK: Understanding diagnostic tests
3: Receiver operating characteristic curves. Acta Paediatr.
96:644–647. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo
L and Lu SH: The oncogenetic role of microRNA-31 as a potential
biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond).
121:437–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu R, Liao J, Yang M, Shi Y, Peng Y, Wang
Y, Pan E, Guo W, Pu Y and Yin L: Circulating miR-155 expression in
plasma: A potential biomarker for early diagnosis of esophageal
cancer in humans. J Toxicol Environ Health A. 75:1154–1162. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Li Y, Wang C, Guan X, Chen X,
Zeng K and Zhang C and Zhang C: Sequencing and identification of
mircORNAs from a genome-wide expression profile in serum of
esophageal squamous cell carcinoma. J Clin Lab Sci. 2012.
|
|
37
|
Xie Z, Chen G, Huang J and Li Z: The
diagnostic significance of plasma miR-10 for esophageal cancer.
Guangdong Med J. 34:2465–2468. 2013.
|
|
38
|
Hui B, Chen X, Hui L, Xi R and Zhang X:
Serum miRNA expression in patients with esophageal squamous cell
carcinoma. Oncol Lett. 10:3008–3012. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang C, Wang C, Chen X, Yang C, Li K,
Wang J, Dai J, Hu Z, Zhou X, Chen L, et al: Expression profile of
microRNAs in serum: A fingerprint for esophageal squamous cell
carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang B and Zhang Q: The expression and
clinical significance of circulating microRNA-21 in serum of five
solid tumors. J Cancer Res Clin Oncol. 138:1659–1666. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hirajima S, Komatsu S, Ichikawa D,
Takeshita H, Konishi H, Shiozaki A, Morimura R, Tsujiura M, Nagata
H, Kawaguchi T, et al: Clinical impact of circulating miR-18a in
plasma of patients with oesophageal squamous cell carcinoma. Br J
Cancer. 108:1822–1829. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sharma P, Saraya A, Gupta P and Sharma R:
Decreased levels of circulating and tissue miR-107 in human
esophageal cancer. Biomarkers. 18:322–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Takeshita N, Hoshino I, Mori M, Akutsu Y,
Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et
al: Serum microRNA expression profile: miR-1246 as a novel
diagnostic and prognostic biomarker for oesophageal squamous cell
carcinoma. Br J Cancer. 108:644–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang T, Zhao D, Wang Q, Yu X, Cui Y, Guo
L and Lu SH: MicroRNA-1322 regulates ECRG2 allele specifically and
acts as a potential biomarker in patients with esophageal squamous
cell carcinoma. Mol Carcinog. 52:581–590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Komatsu S, Ichikawa D, Hirajima S,
Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H,
Shiozaki A, et al: Plasma microRNA profiles: Identification of
miR-25 as a novel diagnostic and monitoring biomarker in
oesophageal squamous cell carcinoma. Br J Cancer. 111:1614–1624.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X,
Zhang Y, Chen X, Wang J, Zen K, et al: Diagnostic and prognostic
implications of a serum miRNA panel in oesophageal squamous cell
carcinoma. PLoS One. 9:e922922014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He FC, Meng WW, Qu YH, Zhou MX, He J, Lv P
and Ming L: Expression of circulating microRNA-20a and let-7a in
esophageal squamous cell carcinoma. World J Gastroenterol.
21:4660–4665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jiang Z, Song Q, Yang S, Zeng R, Li X,
Jiang C, Ding W, Zhang J and Zheng Y: Serum microRNA-218 is a
potential biomarker for esophageal cancer. Cancer Biomark.
15:381–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu H, Yao Y, Meng F, Qian X, Jiang X, Li
X, Gao Z and Gao L: Predictive value of serum miR-10b, miR-29c, and
miR-205 as promising biomarkers in esophageal squamous cell
carcinoma screening. Medicine (Baltimore). 94:e15582015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guan S, Wang C, Chen X, Liu B, Tan B, Liu
F, Wang D, Han L, Wang L, Huang X, et al: miR-613: A novel
diagnostic and prognostic biomarker for patients with esophageal
squamous cell carcinoma. Tumour Biol. 37:4383–4391. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun L, Dong S, Dong C, Sun K, Meng W, Lv
P, Yin H, Ming L and He F: Predictive value of plasma miRNA-718 for
esophageal squamous cell carcinoma. Cancer Biomark. 16:265–273.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ye M, Ye P, Zhang W, Rao J and Xie Z:
Diagnostic values of salivary versus and plasma microRNA-21 for
early esophageal cancer. Nan Fang Yi Ke Da Xue Xue Bao. 34:885–889.
2014.(In Chinese). PubMed/NCBI
|
|
53
|
Li BX, Shi ZL, Yu Q and Fu S: Dynamic
monitoring of miR-21 in peripheral blood before and after
radiotherapy in patients with esophageal carcinoma and its clinical
implication. Tumor. 35:550–555. 2015.
|
|
54
|
Whiting PF, Rutjes AW, Westwood ME,
Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA and Bossuyt
PM: QUADAS-2 Group: QUADAS-2: A revised tool for the quality
assessment of diagnostic accuracy studies. Ann Intern Med.
155:529–536. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of microRNAs in living cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Redshaw N, Wilkes T, Whale A, Cowen S,
Huggett J and Foy CA: A comparison of miRNA isolation and RT-qPCR
technologies and their effects on quantification accuracy and
repeatability. Biotechniques. 54:155–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Y, Wang Q, Zhang N, Ma H, Gu Y, Tang
H, Xu Z and Gao Y: Identification of microRNAs as novel biomarkers
for detecting esophageal squamous cell carcinoma in Asians: A
meta-analysis. Tumor Biol. 35:11595–11604. 2014. View Article : Google Scholar
|
|
59
|
Wan J, Wu W, Che Y, Kang N and Zhang R:
Insights into the potential use of microRNAs as a novel class of
biomarkers in esophageal cancer. Dis Esophagus. 29:412–420. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H,
Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, et al: Role of
microRNA-21 and effect on PTEN in Kazakh's esophageal squamous cell
carcinoma. Mol Biol Rep. 38:3253–3260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sayed D, Rane S, Lypowy J, He M, Chen IY,
Vashistha H, Yan L, Malhotra A, Vatner D and Abdellatif M:
MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Schramedei K, Mörbt N, Pfeifer G, Läuter
J, Rosolowski M, Tomm JM, von Bergen M, Horn F and Brocke-Heidrich
K: MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4.
Oncogene. 30:2975–2985. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kurashige J, Kamohara H, Watanabe M,
Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y and
Baba H: Serum microRNA-21 is a novel biomarker in patients with
esophageal squamous cell carcinoma. J Surg Oncol. 106:188–192.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Komatsu S, Ichikawa D, Takeshita H,
Konishi H, Nagata H, Hirajima S, Kawaguchi T, Arita T, Shiozaki A,
Fujiwara H, et al: Prognostic impact of circulating miR-21 and
miR-375 in plasma of patients with esophageal squamous cell
carcinoma. Expert Opin Biol Ther. 12 Suppl 1:S53–S59. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Avissar M, Christensen BC, Kelsey KT and
Marsit CJ: MicroRNA expression ratio is predictive of head and neck
squamous cell carcinoma. Clin Cancer Res. 15:2850–2855. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Mathé EA, Nguyen GH, Bowman ED, Zhao Y,
Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: Associations with survival. Clin
Cancer Res. 15:6192–6200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Miko E, Czimmerer Z, Csánky E, Boros G,
Buslig J, Dezso B and Scholtz B: Differentially expressed microRNAs
in small cell lung cancer. Exp Lung Res. 35:646–664. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H,
Wang W, Zhang L, Zhang X, Tang Q, et al: MicroRNA-25 functions as a
potential tumor suppressor in colon cancer by targeting Smad7.
Cancer Lett. 335:168–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Esposito F, Tornincasa M, Pallante P,
Federico A, Borbone E, Pierantoni GM and Fusco A: Down-regulation
of the miR-25 and miR-30d contributes to the development of
anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J
Clin Endocrinol Metab. 97:E710–E718. 2012. View Article : Google Scholar : PubMed/NCBI
|