Introduction
Burkitt lymphoma (BL) is a highly aggressive,
mature, B-cell lymphoma, and a type of non-Hodgkin lymphoma (NHL),
and is associated with the Epstein-Barr virus (EBV) and oncogene
(MYC) (1). BL is diagnosed in between
30 and 70 children per million and accounts for 82% of all NHLs in
children and adolescents in equatorial African countries (2). The highest incidence rates of BL are
located in tropical African countries, including Malawi, Uganda,
Nigeria and Niger, with other African countries having notably
lower incidence rates, which are similar to those of highly
developed countries (3).
Transcription factors (TFs) and microRNAs (miRNA)
are two types of key gene regulatory factors in multicellular
genomes (4), which regulate gene
expression at the transcriptional and post-transcriptional levels,
respectively. TFs are proteins that bind to specific DNA sequences
in the gene promoter region, activating or repressing gene
expression (5,6). miRNAs are endogenous, small, non-coding
RNA molecules [~22 nucleotides (nt) long], which silences mRNA
expression via base-pairing with complementary sequences (7). They function as either tumor suppressors
or oncogenes, and influence numerous cancer processes, including
proliferation, differentiation and apoptosis (8,9).
MiRNAs and TFs are key gene regulatory factors and
can form certain network motifs (10). These motifs serve potential roles in
numerous biological processes, including the occurrence and
development of a number of diseases. For example, Li et al
(11) identified that certain
miRNA-TF synergistic regulatory motifs in human lung cancer
influence proliferation and cell cycle of non-small cell lung
cancer. Hsieh et al (12)
identified that there are numerous crosstalking motif
interconnections, and hypothesized that certain network motifs
serve a function in cancer formation. Zhang et al (13) summarized the types of feed-back loops
(FBLs) and feed-forward loops (FFLs), and reviewed their behaviors
and functions according to biological processes and diseases. Among
them, two of the most-studied network motifs are FBLs and FFLs. FBL
is a motif where TFs and miRNAs regulate each other; whereas, FFL
is a motif where TFs and miRNAs together regulate a common target
gene, and TFs regulate miRNAs or miRNAs target TFs. FBL and FFL
motifs have positive and negative roles in a number of biological
processes with the FBL motif serving as a switch, for example lsy-6
and miR-273 contribute to a bistable switch in ASE neurons of
Caenorhabdtis elegans (14),
with the FFL motif serving as a buffer, for example miR-7 and Yan
form coherent FFL motifs involved in the specification of
photoreceptor cells in the Drosophila melanogaster eye
(15). These motifs serve important
roles in gene regulation in mammalians (16).
A number of miRNAs are coded from their associated
genes, and the other miRNAs are coded from introns (17). In the transcription process, the
majority of miRNAs are independently transcribed, whereas other
miRNAs are transcribed together with their host gene (17), demonstrating a coupled regulation
between miRNA and the protein-coding gene. A number of studies
determined that intronic miRNA and its host gene are coordinately
expressed, and they frequently function together in biological
processes of diseases (18,19).
Up to now, biological and medical researchers have
discovered numerous TFs, miRNAs and genes associated with BL
through individual experiments. Among them, the dysregulated
miRNAs, TFs and genes serve important roles in various BL
regulation processes processions, such as deregulation of the tumor
protein 53 (TP53) pathway in pediatric BL, and increased expression
of mouse double minute 4 human homolog of P53 binding protein could
deregulate the TP53 pathway in paediatric BL cases (20). Additionally, the expression of miR-143
and miR-145 was consistently low in human BL cell lines, and thus
they may be useful as biomarkers of B-cell malignancies (21). Dysregulated elements functioning
together may result in the occurrence of BL; whereas, other
non-dysregulated elements may serve smaller roles in BL, such as
DUSP16, a novel, epigenetically-regulated determinant of c-Jun
N-terminal kinase activation in BL (22), and miR-146a, the induction of which
serves a role in the induction or maintenance of EBV latency in
type III BL cells (23). Notably,
BL-associated elements include BL-dysregulated elements.
Although these BL-associated elements were
determined one by one, their regulatory mechanisms remain largely
unknown. A previous study indicated that the occurrence of complex
diseases is not the result of a single gene, but due to multiple
genetic variants and environmental risk factors (24). In the present study, the focus was on
BL-associated TFs, miRNAs and genes, as well as their validated
regulatory associations (TFs→miRNAs, TFs→genes, miRNAs→genes and
host gene→miRNA;→represents regulatory relation, meaning that the
former regulates the latter), in order to investigate their
mechanisms in BL. All data were collected from public databases. In
the present study, the dysregulated elements included TFs, miRNAs
and genes that had: Genetic mutations; deletions; abnormal
expression; single nucleotide polymorphisms; inactivation;
overexpression; low expression; downregulation; upregulation; or
differential expression. The associated elements included
dysregulated and non-dysregulated elements. Based on dysregulated
and associated elements, the dysregulated and associated networks
of BL were constructed separately, then the two networks were
systematically investigated to determine the features and key
network motifs (2-nodes FBL motifs, 3-nodes FFL motifs and 4-nodes
motifs). Due to FFL motifs serving important roles in a number of
diseases (25,26), this type of motif was constructed with
the P-match method (27), and it was
combined with the 1,000 nt promoter region sequences of
dysregulated miRNA targets. The workflow of the present study was
depicted in Fig. 1, and further
information was described in the materials and methods. The
understanding of the dysregulated network will improve the
knowledge regarding the pathogenesis of BL, and the understanding
of the associated network will contribute to the prevention and
therapy of BL.
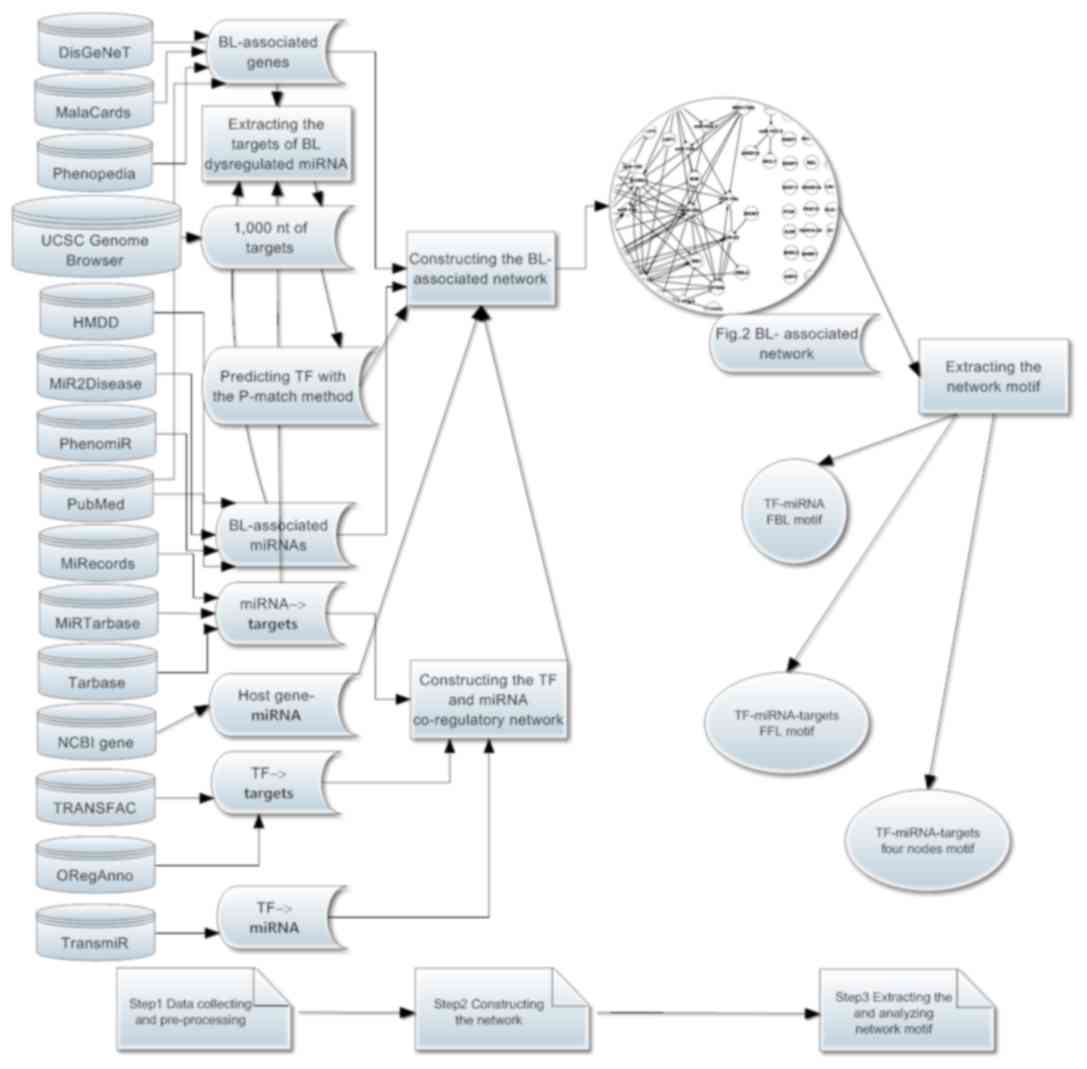 | Figure 1.The workflow of the present study.
(Step 1) Data collection and pre-processing, including
BL-associated TFs, miRNAs, genes and their regulatory associations.
(Step 2) Based on step 1 data, construction of the BL-dysregulated
and BL-associated networks. (Step 3) Extracting and analyzing
network motifs, including FBL, FFL and TF-miRNA-genes 4-nodes
motifs. The arrow represents process flows. BL, Burkitt lymphoma;
targets, target genes; TF, transcription factor; FBL, feed-back
loop; FFL, feed-forward loop; nt, nucleotide; miRNA, microRNA. |
Materials and methods
BL-associated genes
BL-associated genes were collected from the DisGeNET
(version 4.0) (28), MalaCards
(29), Phenopedia (30) and PubMed (http://www.ncbi.nlm.nih.gov/pubmed) databases.
DisGeNET (28) is an open platform
that integrates the information regarding the amount of
gene-disease associations from a number of public data sources and
literature. MalaCards (29) is a
searchable database integrating human maladies and their
annotations. Phenopedia (30) is a
web-based application used to search for summarized information
regarding human genetic associations with genes or diseases from
PubMed. DisGeNET, MalaCards and Phenopedia provided relevant files
regarding disease-genes, which were imported into a spreadsheet
using Microsoft Excel 2010 (Microsoft Corporation, Redmond, WA,
USA). In the PubMed database, the search terms ‘Burkitt lymphoma’
and ‘gene’ were used, and the date was set from 2010 to present. A
large number of study abstracts were reviewed for BL-associated
genes and these genes were also listed in the spreadsheet.
Experimentally-validated data were divided into two types:
Dysregulated and non-dysregulated genes. All symbols of genes were
unified by the official symbols of the NCBI gene database
(https://www.ncbi.nlm.nih.gov/gene).
The unique gene IDs were used as key indexes and duplicated genes
were removed.
BL-associated miRNAs
BL-associated miRNAs were collected from the Human
microRNA Disease Database (HMDD; version 2.0) (31), miR2Disease (32), PhenomiR (33) and PubMed databases. HMDD (31) is a database containing
experimentally-validated human miRNA-disease associations from
original papers. miR2Disease (32) is
a manually curated database providing a comprehensive resource
regarding miRNA in various human diseases, by reviewing numerous
published papers. PhenomiR (33) is a
knowledgebase providing miRNA-disease and miRNA-biological
processes associations from a number of studies. HMDD, miR2Disease
and PhenomiR provided relevant data regarding disease-miRNAs which
were imported into a spreadsheet using Microsoft Excel. In the
PubMed database, the search terms ‘Burkitt lymphoma’ and ‘miRNA’
were used, and the date was set from 2010 to present. Numerous
abstracts were read and any BL-associated miRNAs identified were
also added to the spreadsheet. Experimentally-validated data were
divided into dysregulated and non-dysregulated miRNAs. All symbols
of miRNAs were unified by the official symbols of the NCBI gene
database (https://www.ncbi.nlm.nih.gov/gene). The unique miRNA
IDs were used as key indexes and duplicated miRNAs were
removed.
TF-miRNA
TF-miRNA validated associations were collected from
TransmiR (version 1.2) (34), a
database containing 735 TF-miRNA regulatory pairs that were
manually curated from published literature. TransmiR provides
spreadsheet containing TF-miRNAs to the user.
TF-gene
TF-gene validated associations were collected from
TRANScription FACtor (TRANSFAC; version 11.4) (35) and Open REGulatory ANNOtation
(ORegAnno) (36) databases. TRANSFAC
(35) is a database containing
eukaryotic TFs, their genomic binding sites (TFBSs) and DNA-binding
profiles. This database provides details on TFs and their features,
DNA-binding sites and relevant targets, extracted from public
literature. ORegAnno (36) is an open
access database containing regulatory regions, TFBSs, RNA binding
sites, regulatory variants and other regulatory elements. Data from
ORegAnno are extracted from the literature and highly curated
resources. TRANSFAC and ORegAnno provide data regarding TF-genes,
which were imported into a spreadsheet using Microsoft Excel. All
symbols of TFs and genes were unified by the official symbols of
the NCBI gene database (https://www.ncbi.nlm.nih.gov/gene). The unique TF and
gene IDs were used as key indexes and any duplications of
regulatory associations between TFs and genes were removed.
Host gene of miRNA
Host gene-miRNA associations were collected from
National Center for Biotechnology Information (NCBI) gene database
(http://www.ncbi.nlm.nih.gov/gene/), a
comprehensive gene database covering a wide range of species and
including detailed gene information. Each BL-associated miRNA
symbol was input into the search field of the NCBI gene database.
In the genomic context section of each return page, host genes of
miRNAs were extracted and placed in a spreadsheet using Microsoft
Excel. All symbols of genes and miRNAs were unified by the official
symbols of the NCBI gene database (https://www.ncbi.nlm.nih.gov/gene). The unique gene
and miRNA IDs were used as key indexes and any duplications of
association between miRNAs and genes were removed.
miRNA-gene
miRNA-gene validated associations were collected
from Tarbase (version 7.0) (37),
miRTarBase (version 6) (38) and
miRecords (version 4) (39). Tarbase
(37) is a manually curated database
that contains 100,000s of miRNA-gene pairs that have been
experimentally identified. In this database, the focus is on
regulatory pairs extracted by western blot analysis, northern blot
analysis and quantitative PCR (qPCR). miRTarBase (38) also provides experimentally-validated
miRNA-gene pairs. MiRTarBase (version 6.0) contains 7,439
strongly-validated miRNA-gene pairs and 348,007 miRNA-gene pairs
from CLIP-seq. This database contains strongly-validated regulatory
pairs extracted by western blot analysis, northern blot analysis,
qPCR, and other reporter assays. MiRecords (39) is another resource containing animal
miRNA-gene pairs. It contains validated targets from literature
curation and predicted targets from 11 established miRNA target
prediction programs. Tarbase, miRTarBase and miRecords provided
data regarding miRNA-genes which were copied into a spreadsheet
using Microsoft Excel. All symbols including TFs, miRNAs and genes
were unified by the official symbol of the NCBI gene database. The
unique TF, gene and miRNA IDs were used as key indexes and any
duplications of associations between TFs, miRNAs and genes were
removed.
Networks construction
Based on experimentally-validated regulatory data
consisting of: TFs→miRNAs; TFs→genes; miRNAs→genes; and host
gene→miRNAs, the TFs and miRNAs co-regulatory network was
constructed by combining these regulatory pairs. This network was
named the validated regulatory network.
Based on BL-associated data consisting of TFs,
miRNAs and their host gene, genes, and the validated regulatory
network, the BL-associated network was constructed. Firstly, theses
nodes were mapped onto the validated regulatory network. Secondly,
these regulatory data was extracted to construct the BL-associated
regulatory network. Lastly, the BL-associated regulatory network
and other BL-associated data, that did not contain regulatory
associations with other TFs, genes and miRNAs, were combined into a
network that was designated the BL-associated network (Fig. 2).
According to the features of FBLs and FFLs network
motifs described in the introduction, network motifs were manually
extracted from the BL-associated network. A total of 10 types of
motif were identified (Fig. 3).
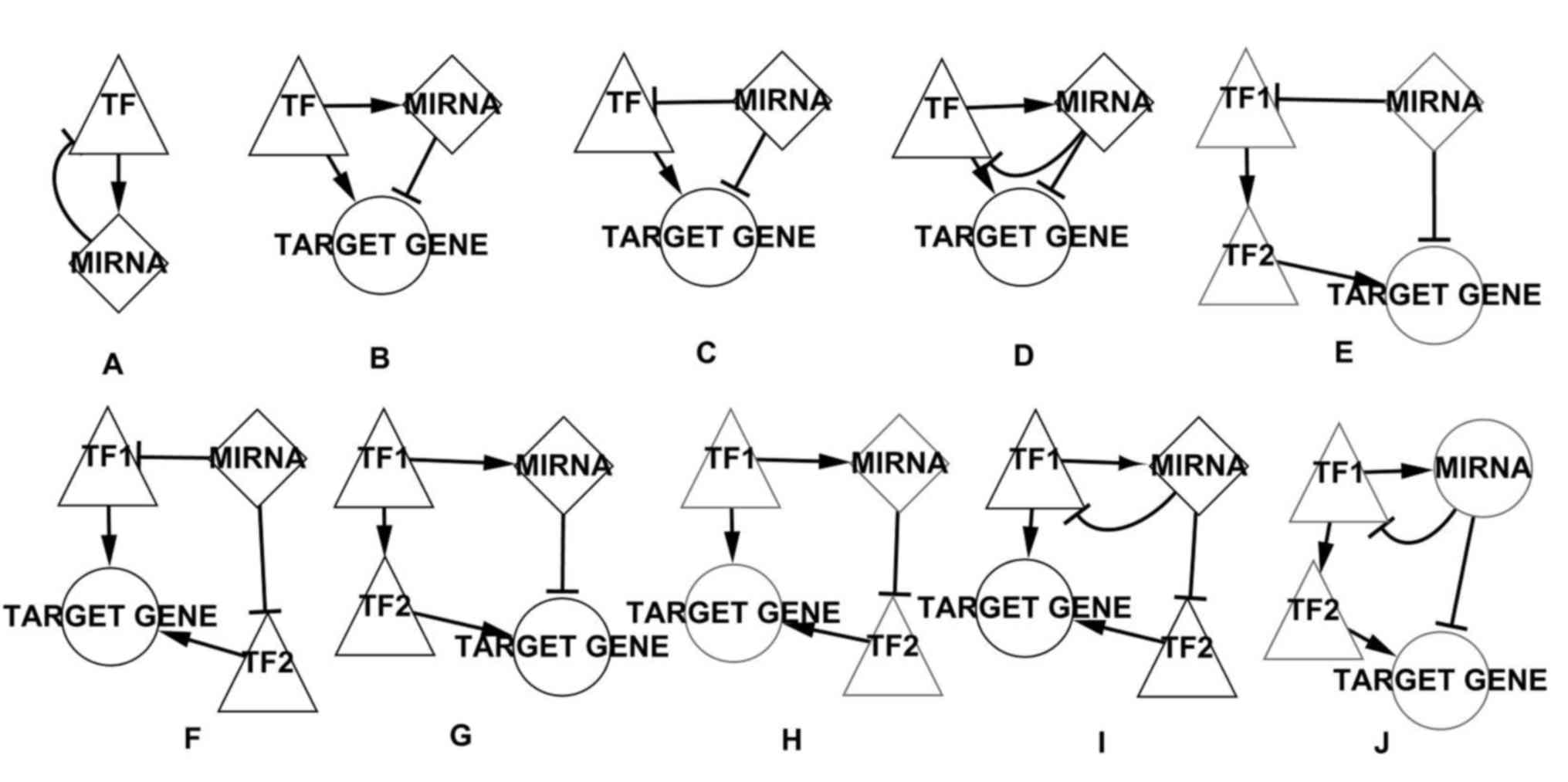 | Figure 3.A number of network motifs in the
BL-associated network. (A) The FBL motif. (B) The TF-regulated FFL
motif. (C) The miRNA-regulated FFL motif. (D) The composite FFL
motif. (E) The 4-nodes miRNA-regulated motif that miRNA and TF
together regulate a target gene, for example miR-17, TCF3, GLI1 and
CCND2 form this type of motif. (F) The 4-nodes miRNA-regulated
motif that two TFs together regulate a target gene, for example
miR-20, TP53, E2F1 and SMO form this type of motif. (G) The 4-nodes
TF-regulated motif that miRNA and TF together regulate a target
gene, for example miR-155, TP53, NFATC2 and ICAM1 form this type of
motif. (H) The 4-nodes TF-regulated motif that two TFs together
regulate a target gene, for example miR-155, TP53, BCL6 and BCL2L1
form this type of motif. (I) The 4-nodes composite motif that two
TFs together regulate a target gene, for example miR-17, E2F1, RBL2
and POU2F2 form this type of motif. (J) The 4-nodes composite motif
that miRNA and TF together regulate a target gene, for example
let-7a, E2F1, RBL2 and CCND2 form this type of motif. Triangles
represent TFs, diamonds represent miRNAs, circles represent target
genes. FBL, feed-back loop; FFL, feed-forward loop; miRNA,
microRNA; BL, Burkitt lymphoma; TF, transcription factor. |
In total, 4 TFs (E2F1, NFKB1, E2F4 and TCF3)
exhibited complicated regulatory relations in the BL-associated
network, with maximum in- and out-degrees, which alongside miRNAs
form network motifs (Fig. 4).
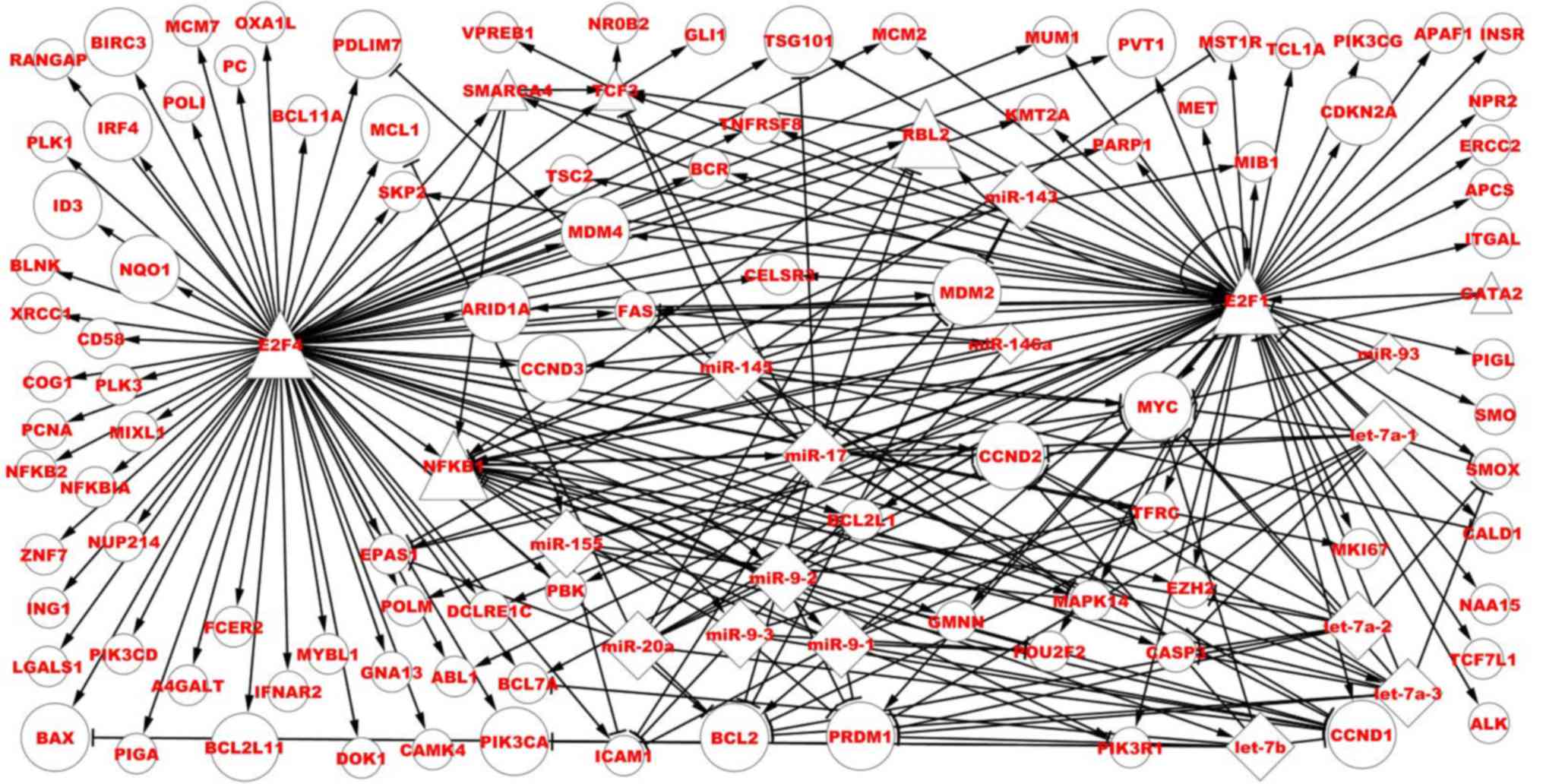 | Figure 4.Direct regulatory associations of
E2F1, NFKB1, E2F4 and TCF3 in the BL-associated network. Bigger
nodes represent dysregulated TFs, miRNAs and targets, smaller nodes
represent associated and non-dysregulated TFs, miRNAs and target
genes. Circles represent targets, triangles represent TFs and
diamonds represent miRNAs. TF, transcription factors; E2F1, E2F
TF1; NFKB1, nuclear factor κB subunit 1; TCF3, TF3; miR, microRNA;
BL, Burkitt lymphoma. |
Using a similar method that was used to construct
the BL-dysregulated network, the dysregulated data (TFs, miRNAs and
genes in BL) was mapped onto the validated regulatory network and
these regulatory data was extracted to construct the
BL-dysregulated regulatory network. Lastly, the network and other
BL-dysregulated data, that did not contain regulatory associations
with other TFs, genes and miRNAs, were combined into the called
BL-dysregulated network. The software (version 2.8.2) (40) was used to visualize the network.
Dysregulated expression was present in three modes: Upregulation,
downregulation and no expression.
Table I presents
statistical data regarding BL-associated network. BL-associated
data include associated genes, miRNAs, TF-miRNA pairs, TF-gene
pairs, host gene-miRNA pairs and miRNA-gene pairs. BL-associated
genes were collected from five data sources and 320 genes were
identified. BL-associated miRNAs were collected from four data
sources and 25 miRNAs were identified. BL-associated TF-miRNA pairs
were collected from TransmiR and 64 pairs were identified.
BL-associated TF-gene pairs were collected from two data sources
and 534 pairs were identified. BL-associated host gene-miRNA pairs
were collected from the NCBI gene database and 24 pairs were
identified. BL-associated miRNA-gene pairs were collected from
three data sources and 256 pairs were identified.
 | Table I.Statistical data regarding
BL-associated network. |
Table I.
Statistical data regarding
BL-associated network.
| BL-associated
data | Data source | Number |
|---|
| BL-associated
genes | DisGeNET,
MalaCards, Phenopedia PubMed, P-match | 320 |
| BL-associated
miRNAs | HMDD, miR2Disease,
PhenomiR, PubMed | 25 |
| BL-associated
TF-miRNA pairs | TransmiR | 64 |
| BL-associated
TF-gene pairs | TransFAC
ORegAnno | 534 |
| BL-associated host
gene-miRNA pairs | NCBI gene
database | 24 |
| BL-associated
miRNA-gene pairs | Tarbase miRTarbase
miRecords | 256 |
According to the type of nodes and type of
regulatory associations, statistical data, including number of
nodes and edges from BL-associated network and BL-dysregulated
network, were manually extracted (Table
II).
 | Table II.Statistical data regarding
dysregulated and associated networks in Burritt lymphoma. |
Table II.
Statistical data regarding
dysregulated and associated networks in Burritt lymphoma.
|
| Number of
nodes | Number of
edges |
|---|
|
|
|
|
|---|
| Network | TF | miRNA | Host gene | Target gene | Single node | TF→TF | TF→target gene | TF→miRNA | miRNA→TF | miRNA→target
gene | Host
gene→miRNA |
|---|
| Dysregulated
network | 15 | 23 | 12 | 47 | 27 | 9 | 21 | 21 | 83 | 70 | 23 |
| Associated
network | 37 | 25 | 14 | 181 | 83 | 63 | 470 | 64 | 81 | 175 | 24 |
Predicted FFL motif
The predicted FFL motifs were constructed with the
following method. Firstly, the 1,000 nt promoter region sequences
of BL-associated genes targeted by dysregulated miRNAs were
downloaded from UCSC (41);
subsequently, the regulatory relations between the aforementioned
dysregulated miRNAs and aforementioned targets in the BL-associated
network were focused on and miRNA-gene pairs were extracted from
the BL-associated network. Secondly, TFs of the aforementioned
targets were predicted with the 1,000 nt promoter region sequences
of the genes and the P-match method, and then the TF-gene pairs
were obtained. The P-Match method is able to identify TFBSs in DNA
sequences, and includes a matrix library as well as sets of known
TFBSs collected from TRANSFAC. In the present study, the vertebrate
matrixes and restricted high quality matrixes were used. The high
quality criterion denotes the following: The threshold rate for a
false negative result was set at 50% (threshold value is 0.5), and
a threshold was also set to differentiate positive from false
positive results. Matrixes which produce the highest number of
false positive results are defined as low quality. Thirdly, the
focus was only on the predicted TFs included in transmiR and
BL-associated miRNAs; subsequently, the corresponding TF-miRNA
pairs were extracted from transmiR, and then the TF-miRNA pairs
were obtained. Lastly, the TF-gene, TF-miRNA and miRNA-gene pairs
were combined, and then FFL motifs were obtained.
According to types of motifs, statistical data,
including number of 3-nodes FFLs, number of 4-nodes motifs and
number of 2-nodes FBLs, were manually extracted from the
BL-associated and BL-dysregulated networks (Table III).
 | Table III.Statistical data of key network
motifs in BL-dysregulated and BL-associated networks. |
Table III.
Statistical data of key network
motifs in BL-dysregulated and BL-associated networks.
| Motif type | Dysregulated
network | Associated
network |
|---|
| Number of 3-nodes
FFLs |
|
|
|
miRNA-regulated FFLs | 18 | 24 |
|
TF-regulated FFLs | 15 | 20 |
|
Composite FFLs | 12 | 31 |
| Number of 4-nodes
motifs |
|
|
|
miRNA-regulated motifs | 4 | 21 |
|
TF-regulated motifs | 0 | 3 |
|
Composite motifs | 11 | 48 |
| Number of 2-nodes
FBLs | 26 | 31 |
Results
BL-associated network
Fig. 2 depicts the
BL-associated network, which includes dysregulated and
non-dysregulated elements. Notably, the BL-associated network
includes the dysregulated network. Table
II displays the statistical data of the two networks. Although
14 host genes are not dysregulated, they may serve a role in the
BL-associated processes. The dysregulated network partially reveals
the pathogenesis of BL. The associated network not only partially
demonstrated the regulatory mechanism underlying the occurrence of
BL, but also determined a number of regulatory mechanisms
associated with the prevention and therapy of BL.
The key network motifs were manually extracted from
the BL-associated network. Fig. 3
depicts 10 types of key network motifs, consisting of TFs, miRNAs
and targets in the BL-associated network. Fig. 3A depicts the FBL motif, a type of key
regulator associated with cancer, in which TFs and miRNAs interact
with each other. Fig. 3B depicts the
TF-regulated FFL motif, where TFs and miRNAs together regulate a
target gene, TFs regulate miRNAs and TFs serve as regulatory
initiators. Fig. 3C depicts the
miRNA-regulated FFL motif, where TFs and miRNAs together regulate a
target gene, miRNAs regulate TFs and miRNAs serve as regulatory
initiators. Fig. 3D depicts the
composite FFL motif, where TFs and miRNAs together regulate a
target gene and miRNAs and TFs regulate each other. Fig. 3E and F depict two types of 4 nodes
miRNA-regulated motifs, in Fig. 3E,
miRNAs regulate a target gene and a TF, which regulates another TF
that regulates the target gene regulated by the miRNAs, in Fig. 3F, miRNAs regulate two TFs, which
together regulate a target gene, and miRNAs serve as regulatory
initiators. Fig. 3G and H depict two
types of 4-nodes TF-regulated motifs, in Fig. 3G, TFs regulate a miRNA and a TF, which
regulates a target gene regulated by the miRNA, in Fig. 3H, TFs regulate a miRNA and a target
gene, which is regulated by a TF regulated by the miRNA, and TFs
serve as regulatory initiators, Fig. 3I
and J depict two types of 4-nodes composite motifs, which are
similar to the aforementioned two types (miRNA-regulated and
TF-regulated) of 4-nodes motif, however, in these types the miRNAs
and TFs regulate each other.
Table III displays
statistical data regarding network motifs in dysregulated and
associated networks of BL. The dysregulated network includes 45
3-nodes FFL motifs, 15 4-nodes motifs and 26 FBL motifs, whilst the
associated network includes 75 3-nodes FFL motifs, 99 4-nodes
motifs and 31 FBL motifs.
Regulatory network of predicted TF in
the BL-associated network
Finally, 38 predicted TFs were obtained. Among these
TFs, E2F TF1 (E2F1), nuclear factor κB subunit 1 (NFKB1) and E2F4
are dysregulated genes, and TF3 (TCF3) is an associated and
non-dysregulated gene. Fig. 4 depicts
the direct regulatory associations of four TFs in the BL-associated
network. E2F1 regulates 57 targets, and among them, 13 genes are
dysregulated, with the others being non-dysregulated. E2F1
regulates 10 miRNAs, and among them, nine miRNAs are dysregulated
and one miRNA is non-dysregulated, with five TFs regulating E2F1
and six miRNAs targeting E2F1. E2F1, its targets and these miRNAs
form 38 FFLs and six FBLs. E2F4 regulates 79 targets, and among
them, 21 genes are dysregulated, with the others being
non-dysregulated. NFKB1 regulates eight miRNAs, which are all
dysregulated, five TFs [E2F1, E2F4, GATA binding protein 2, RB
transcriptional corepressor-like 2 (RBL2) and SMARCA4] regulate
NFKB1, nine miRNAs target NFKB1 and six miRNAs form six FBLs. TCF3
regulates three targets (GLI family zinc finger 1, V-set pre-B cell
surrogate light chain 1 and nuclear receptor subfamily 0 group B
member 2), which are all non-dysregulated, four TFs (E2F1, E2F4,
RBL2 and SMARCA4) regulate TCF3 and two miRNAs (miR-145 and miR-17)
target TCF3.
Discussion
In the present study, the BL-dysregulated and
BL-associated networks were constructed. These networks determined
a portion of the regulatory mechanism of genes and miRNAs on BL. In
the dysregulated network, all of the nodes had dysregulated
expression, except a number of host genes that did not have
dysregulated expression. The biological network does not exhibit a
dysregulated status for healthy people. When an individual becomes
unwell, their biological network demonstrates a dysregulated
status. If the dysregulated status is regulated to a normal status
by a number of medical methods, the diseases may be treated
successfully. BL-dysregulated networks serve important roles in
pathogenesis mechanisms underlying BL, and the regulation of the
dysregulated network may be an effective strategy that contributes
to gene therapy for BL.
In the dysregulated network, a number of single
nodes, that do not have regulatory associations with other nodes,
serve important roles in portions of the BL progression process,
including apoptosis regulator BHRF1 which functions as a survival
factor in Wp-restricted BL and rescued the cells from the apoptosis
induced by dnEBNA1 expression (42),
is deleted in lung and esophageal cancer 1, and is a modulator of
proliferation, apoptosis and migration in BL (43). The mechanism underlying their
influence on the progression of BL requires further research. A
number of dysregulatory-expressed nodes not only formed
dysregulated regulatory pathways, such as E2F1→MYC,
TP53→miR-20a→B-cell lymphoma 2 and E2F4→MYC→miR-19b-1→cyclin D1,
but also form a number of key network motifs, such as FBL motifs
(phosphatase and tensin homolog↔miR-19a and MYB↔miR-155), 3-nodes
FFL motif (E2F1→BCL2 and E2F1→miR-18a→BCL2) and 4-nodes motif
(miR-17→RBL2→MYC and miR-17→TP53→MYC). The mechanisms underlying
the dysregulated regulatory pathways and network motifs influencing
the occurrence, development and therapy of BL require further
research in order to determine them.
Acknowledgements
Not applicable.
Funding
The present study was supported by the 13th Five
Year Plan of Science and Technology Research by Education
Department of Jilin Province of China (grant no. 2016352), the 13th
Five Year Plan of Science and Technology Research by Education
Department of Jilin Province of China (grant no. JJKH20181369KJ),
Scientific Research Project of Shenzhen Institute of Information
Technology (grant no. QN201716) and the Scientific Research Staring
Foundation of Changchun University of Science and Technology (grant
no. 2016420).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KW wrote and revised the manuscript. KW, CLC, CM and
CX conceived and designed the study and methodology. ZC and YY
collected and preprocessed the data. JW and CT analyzed the
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
TFs
|
transcription factors
|
|
targets
|
target genes
|
|
BL
|
Burkitt lymphoma
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
TFBSs
|
transcription factor binding sites
|
References
|
1
|
Molyneux EM, Rochford R, Griffin B, Newton
R, Jackson G, Menon G, Harrison CJ, Israels T and Bailey S:
Burkitt's lymphoma. Lancet. 379:1234–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Naresh KN, Raphael M, Ayers L, Hurwitz N,
Calbi V, Rogena E, Sayed S, Sherman O, Ibrahim HA, Lazzi S, et al:
Lymphomas in sub-Saharan Africa-what can we learn and how can we
help in improving diagnosis, managing patients and fostering
translational research? Br J Haematol. 154:696–703. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stefan C, Bray F, Ferlay J, Liu B and
Parkin Maxwell D: Cancer of childhood in sub-Saharan Africa.
Ecancermedicalscience. 11:7552017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Latchman DS: Transcription factors: An
overview. Int J Biochem Cell Biol. 29:1305–1312. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karin M: Too many transcription factors:
Positive and negative interactions. New Biol. 2:126–131.
1990.PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ha TY: MicroRNAs in human diseases: From
cancer to cardiovascular disease. Immune Netw. 11:135–154. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su N, Wang Y, Qian M and Deng M:
Combinatorial regulation of transcription factors and microRNAs.
BMC Syst Biol. 4:1502010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li K, Li Z, Zhao N and Xu Y, Liu Y, Zhou
Y, Shang D, Qiu F, Zhang R, Chang Z and Xu Y: Functional analysis
of microRNA and transcription factor synergistic regulatory network
based on identifying regulatory motifs in non-small cell lung
cancer. BMC Syst Biol. 7:1222013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hsieh WT, Tzeng KR, Ciou JS, Tsai JJ,
Kurubanjerdjit N, Huang CH and Ng KL: Transcription factor and
microRNA-regulated network motifs for cancer and signal
transduction networks. BMC Syst Biol. 9 Suppl 1:S52015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang HM, Kuang S, Xiong X, Gao T, Liu C
and Guo AY: Transcription factor and microRNA co-regulatory loops:
Important regulatory motifs in biological processes and diseases.
Brief Bioinform. 16:45–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hobert O: Architecture of a
microRNA-controlled gene regulatory network that diversifies
neuronal cell fates. Cold Spring Harb Symp Quant Biol. 71:181–188.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Cassidy JJ, Reinke CA, Fischboeck S
and Carthew RW: A microRNA imparts robustness against environmental
fluctuation during development. Cell. 137:273–282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsang J, Zhu J and van Oudenaarden A:
MicroRNA-mediated feedback and feedforward loops are recurrent
network motifs in mammals. Mol Cell. 26:753–767. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, Li J, Li S, Chen L, Ding H, et al: Intronic miR-301 feedback
regulates its host gene, ska2, in A549 cells by targeting MEOX2 to
affect ERK/CREB pathways. Biochem Biophys Res Commun. 396:978–982.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leventaki V, Rodic V, Tripp SR, Bayerl MG,
Perkins SL, Barnette P, Schiffman JD and Miles RR: TP53 pathway
analysis in paediatric Burkitt lymphoma reveals increased MDM4
expression as the only TP53 pathway abnormality detected in a
subset of cases. Br J Haematol. 158:763–771. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Akao Y, Nakagawa Y, Kitade Y, Kinoshita T
and Naoe T: Downregulation of microRNAs-143 and −145 in B-cell
malignancies. Cancer Sci. 98:1914–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee S, Syed N, Taylor J, Smith P, Griffin
B, Baens M, Bai M, Bourantas K, Stebbing J, Naresh K, et al: DUSP16
isan epigenetically regulated determinant of JNK signalling in
Burkitt's lymphoma. Br J Cancer. 103:265–274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Motsch N, Pfuhl T, Mrazek J, Barth S and
Grässer FA: Epstein-Barr virus-encoded latent membrane protein 1
(LMP1) induces the expression of the cellular microRNA miR-146a.
RNA Biol. 4:131–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirschhorn JN and Daly MJ: Genome-wide
association studies for common diseases and complex traits. Nat Rev
Genet. 6:95–108. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qin S, Ma F and Chen L: Gene regulatory
networks by transcription factors and microRNAs in breast cancer.
Bioinformatics. 31:76–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin Y, Zhang Q, Zhang HM, Liu W, Liu CJ,
Li Q and Guo AY: Transcription factor and miRNA co-regulatory
network reveals shared and specific regulators in the development
of B cell and T cell. Sci Rep. 5:152152015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chekmenev DS, Haid C and Kel AE: P-Match:
Transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:(Web Server issue).
W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piñero J, Queralt-Rosinach N, Bravo À,
Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F and Furlong LI:
DisGeNET: A discovery platform for the dynamical exploration of
human diseases and their genes. Database (Oxford): bav028.
2015.
|
|
29
|
Rappaport N, Nativ N, Stelzer G, Twik M,
Guan-Golan Y, Stein TI, Bahir I, Belinky F, Morrey CP, Safran M and
Lancet D: MalaCards: An integrated compendium for diseases and
their annotation. Database (Oxford): bat018. 2013. View Article : Google Scholar
|
|
30
|
Yu W, Clyne M, Khoury MJ and Gwinn M:
Phenopedia and genopedia: Disease-centered and gene-centered views
of the evolving knowledge of human genetic associations.
Bioinformatics. 26:145–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T
and Cui Q: HMDD v2.0: A database for experimentally supported human
microRNA and disease associations. Nucleic Acids Res. 42:(Database
Issue). D1070–D1074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: A manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res. 37:(Database Issue). D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ruepp A, Kowarsch A and Theis F: PhenomiR:
microRNAs in human diseases and biological processes. Methods Mol
Biol. 822:249–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:(Database Issue). D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wingender E, Chen X, Hehl R, Karas H,
Liebich I, Matys V, Meinhardt T, Prüss M, Reuter I and Schacherer
F: TRANSFAC: An integrated system for gene expression regulation.
Nucleic Acids Res. 28:316–319. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lesurf R, Cotto KC, Wang G, Griffith M,
Kasaian K, Jones SJ, Montgomery SB and Griffith OL: Open Regulatory
Annotation Consortium: ORegAnno 3.0: A community-driven resource
for curated regulatory annotation. Nucleic Acids Res. 44:(D1).
D126–D132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vlachos IS, Paraskevopoulou MD, Karagkouni
D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL,
Maniou S, Karathanou K, Kalfakakou D, et al: DIANA-TarBase v7.0:
Indexing more than half a million experimentally supported
miRNA:mRNA interactions. Nucleic Acids Res. 43:(Database Issue).
D153–D159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database Issue). D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC Genome Browser database: Update 2011. Nucleic Acids
Res. 39:(Database Issue). D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Watanabe A, Maruo S, Ito T, Ito M,
Katsumura KR and Takada K: Epstein-Barr virus-encoded Bcl-2
homologue functions as a survival factor in Wp-restricted Burkitt
lymphoma cell line P3HR-1. J Virol. 84:2893–2901. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng M, Huang B, Du Z, Xu X and Chen Z:
DLC-1 as a modulator of proliferation, apoptosis and migration in
Burkitt's lymphoma cells. Mol Biol Rep. 38:1915–1920. 2011.
View Article : Google Scholar : PubMed/NCBI
|


















