Introduction
Lung cancer ranks second in incidence and first in
mortality for both males and females, with an estimated 222,500 new
cases and 155,870 deaths in 2017 (1).
Unfortunately, the majority of lung cancer patients are diagnosed
at an advanced stage and are typically not curable. The current
gold standard for lung cancer diagnosis is pathological biopsy,
which is an invasive method. With the development of nondestructive
testing technology, novel non-invasive methods are needed to
monitor and diagnose lung cancer. Though some findings regarding
molecular mechanisms in lung cancer have been reported in recent
years (2–6), the molecular regulatory mechanisms
underlying the initiation and development of lung cancer remain
unclear. Therefore, more studies are needed to explore the
molecular mechanisms in lung cancer.
MicroRNAs (miRNAs) are highly conserved endogenous
small non-coding RNAs approximately 23 nucleotides in length. They
exert their effects through posttranscriptional repression in a
sequence-specific manner (7). miRNAs
have been found to perform crucial biological functions in the
initiation and development of cancer (8). In addition, miRNAs have begun to be used
for the diagnosis and treatment of cancer in recent years (9–11). The
clinical significance of miRNA profiles in diagnosis and prognosis
has also been evaluated in lung cancer (12,13).
Over the past few years, miR-193a-5p has been found
to be involved in multiple cancers, such as esophageal squamous
cell carcinoma (14), bladder cancer
(15), primary bone tumors (16), osteosarcoma (17), colorectal cancer (18) and endometrioid endometrial
adenocarcinoma (19). Some noted
pathways such as the PI3K/AKT signaling pathway, PTEN/AKT signaling
pathway and mTOR signaling pathway have been implicated in the
oncogenesis of different tumors and as therapeutic drug targets
(20–24). Some studies have linked miR-193a-5p to
lung cancer. For example, in Yu's study, miR-193a-5p was found to
block the metastasis of non-small cell lung cancer by inhibiting
the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway (25). Similarly, Chen et al (26)revealed that the
miR-193a-5p-WT1-E-cadherin axis plays a crucial role in the
metastasis of non-small cell lung cancer. However, no study has
reported the diagnostic value of miR-193a-5p in lung cancer.
Thus, we attempted to explore the clinical
diagnostic significance of miR-193a-5p using a microarray
meta-analysis. We also explored the possible molecular mechanisms
using bioinformatics analysis. The results of this study could
provide insights into the clinical diagnosis of lung cancer and
guide future research on the underlying molecular mechanisms.
Materials and methods
GEO dataset retrieval and data
extraction
Lung cancer microarray data published through July
2017 were retrieved from the Gene Expression Omnibus (GEO)
database. The retrieval strategies were as follows: (lung OR
pulmonary OR respiratory OR bronchi OR bronchioles OR alveoli OR
pneumocytes OR ‘air way’) and (cancer OR carcinoma OR tumor OR
neoplas* OR malignan* OR adenocarcinoma) and (MicroRNA OR miRNA OR
‘Micro RNA’ OR ‘Small Temporal RNA’ OR ‘non-coding RNA’ OR ncRNA OR
‘small RNA’). Microarray data meeting the following criteria were
included: i) The subjects in the datasets included lung cancer
patients and corresponding controls; ii) the expression profile of
miR-193a-5p in both lung cancer patients and controls was available
or calculable; and iii) the number of overall subjects was more
than 30. The expression of miR-193a-5p was extracted from the
included datasets and evaluated with means and standard deviations
(SD) using SPSS 22.0 (SPSS, Inc., Chicago, IL, USA).
Meta-analysis of the diagnostic value
of miR-193a-5p in the GEO datasets
A meta-analysis of the included datasets was
conducted to evaluate the diagnostic value of miR-193a-5p using
Stata 12.0 software (StataCorp LP, College Station, TX, USA). The
pooled effect was estimated as the standard mean difference (SMD)
with a 95% confidence interval (CI). The heterogeneity was measured
using a chi-squared test of Q and P values. I2<50% or
P>0.05 indicated no significant heterogeneity. The publication
bias of the datasets was assessed using Begg's funnel plots. A
symmetrical funnel plot indicated no obvious publication bias. In
addition, summary receiver operating characteristic (SROC) analysis
was performed. The pooled diagnostic sensitivity, specificity, odds
ratio (OR), positive likelihood ratio (LR) and negative LR were
calculated to comprehensively evaluate the diagnostic value of
miR-193a-5p.
Identification of miR-193a-5p target
genes
Possible target genes of miR-193a-5p were collected
using miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/mirwalk2) (27), which combines 12 prediction databases.
Genes predicted by at least 2 databases were selected. We also
identified validated target genes of miR-193a-5p using the Tarbase
and MitarBase databases. The predicted genes and validated genes
were further compared to identify the most significant overlapping
miR-193a-5p target genes.
Exploration of the molecular mechanism
of miR-193a-5p using bioinformatics analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID), an online bioinformatics functional
enrichment tool for the analysis of large lists of genes (28,29), was
used to explore the enriched pathways of the overlapping genes.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) analyses were performed using the DAVID online functional
annotation module. Furthermore, to identify hub genes, we used the
STRING v10 database (http://string-db.org/) (30) to construct a protein-protein
interaction (PPI) network. Genes with connection degrees higher
than 3 were considered hub genes.
Validation of hub gene expression
using TCGA data
The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) database is one of the
largest publicly funded project platforms, providing information
regarding cancer-causing genome alterations for more than 30 cancer
types (31). To validate the
expression of the hub genes, we downloaded the RNA-sequencing data
of lung cancer and non-cancerous tissues from the TCGA database.
The hub gene expression data in lung cancer and non-cancerous
tissues were extracted. The expression values were normalized by
log2 transformation and imported into GraphPad Prism 7.0 (GraphPad
Software, Inc., La Jolla, CA, USA). Unpaired Student's t-test and
receiver operating characteristic (ROC) curve analyses were
conducted to assess the expression differences and clinical
significance. Scatter plots were constructed to visualize the
differences in miR-193a-5p expression between lung cancer and
non-cancerous tissues. In addition, the area under the curve (AUC)
was calculated to evaluate the diagnostic capability of
miR-193a-5p. A P-value <0.05 was considered statistically
significant.
Validation of the protein expression
of the hub genes
The Human Protein Atlas database (https://www.proteinatlas.org/) provides abundant
proteome and transcriptome data for tissues, cells and cancers
(32–34). The protein expression of the
upregulated hub genes in lung adenocarcinoma (LUAD) and lung
squamous cell carcinoma (LUSC) specimens was investigated using the
Pathology Altas portal in The Human Protein Atlas database.
Antibody staining was analyzed to reflect the protein expression,
and only samples with a staining quality of over 75% were used.
Pathological images of typical high/medium staining in LUSC cells
were chosen for display in this study.
Validation of the correlation analysis
between miR-193a-5p and hub genes based on TCGA data
The expression data of miR-193a-5p and the six hub
genes in lung cancer were downloaded from the TCGA database. The
expression data of both miR-193a-5p and the hub genes were
normalized by log2 transformation. Spearman's correlation analysis
was further performed using GraphPad Prism 7.0 (GraphPad Software,
Inc.). A Spearman correlation coefficient of r<0 indicated a
negative correlation between miR-193a-5p and its hub genes.
P<0.05 indicated the statistical significance of the correlation
analysis.
Results
Overview of the included datasets
According to the retrieval criteria, a total of 15
datasets published from 2009 to 2016 were selected, including 7
peripheral blood datasets and 8 tissue datasets. The basic
information of the included datasets is provided in Table I (35–49). In
the 7 peripheral blood datasets, 453 lung cancer samples and 306
healthy controls were included. In the 8 tissue datasets, 693 lung
cancer samples and 354 healthy controls were included. The
miR-193a-5p expression data were normalized by log2 transformation
and extracted as the mean and SD.
 | Table I.Characteristics of the GEO datasets
included in the meta-analysis. |
Table I.
Characteristics of the GEO datasets
included in the meta-analysis.
| Study
information | Sample | Array and
annotation information |
|
|---|
|
|
|
|
|---|
| Author | Publication
year | Country | Sample source | Data source | Lung cancer
patients | Healthy
controls | Platform | Refs. |
|---|
| Keller et
al | 2009 | Germany | Peripheral
blood | GSE17681 | 17 | 19 | GPL9040 | (35) |
| Patnaik et
al | 2011 | USA | Peripheral
blood | GSE27486 | 22 | 32 | GPL11432 | (36) |
| Keller et
al | 2011 | Germany | Peripheral
blood | GSE31568 | 32 | 67 | GPL9040 | (37) |
| Patnaik et
al | 2012 | USA | Peripheral
blood | GSE40738 | 82 | 58 | GPL16016 | (38) |
| Keller et
al | 2014 | Germany | Peripheral
blood | GSE61741 | 73 | 94 | GPL9040 | (39) |
| Godrey et
al | 2014 | USA | Peripheral
blood | GSE46729 | 24 | 24 | GPL8786 | (40) |
| Leidinger et
al | 2015 | Germany | Peripheral
blood | GSE68951 | 203 | 12 | GPL16770 | (41) |
| Tan et
al | 2010 | China | SCLC/NSCLC | GSE15008 | 182 | 185 | GPL8176 | (42) |
| Nymark et
al | 2011 | Finland | NSCLC | GSE25508 | 26 | 26 | GPL7731 | (43) |
| Ohba et
al | 2013 | Japan | NSCLC/SCLC | GSE19945 | 55 | 8 | GPL9948 | (44) |
| Bjaanaes et
al | 2014 | Norway | LUAD | GSE48414 | 154 | 20 | GPL16770 | (45) |
| Robles et
al | 2015 | USA | LUAD | GSE63805 | 32 | 30 | GPL18410 | (46) |
| Gasparini et
al | 2015 | Switzerland | NSCLC | GSE72526 | 67 | 18 | GPL20275 | (47) |
| Jin et
al | 2015 | China | SCLC/LUAD/LUSC | GSE74190 | 92 | 44 | GPL19622 | (48) |
| Yoshimoto et
al | 2016 | Japan | SCLC/LUAD | GSE77380 | 85 | 23 | GPL16770 | (49) |
Meta-analysis of the diagnostic value
of peripheral blood miR-193a-5p
The expression of peripheral blood miR-193-5p in the
7 independent peripheral blood datasets was pooled in a forest plot
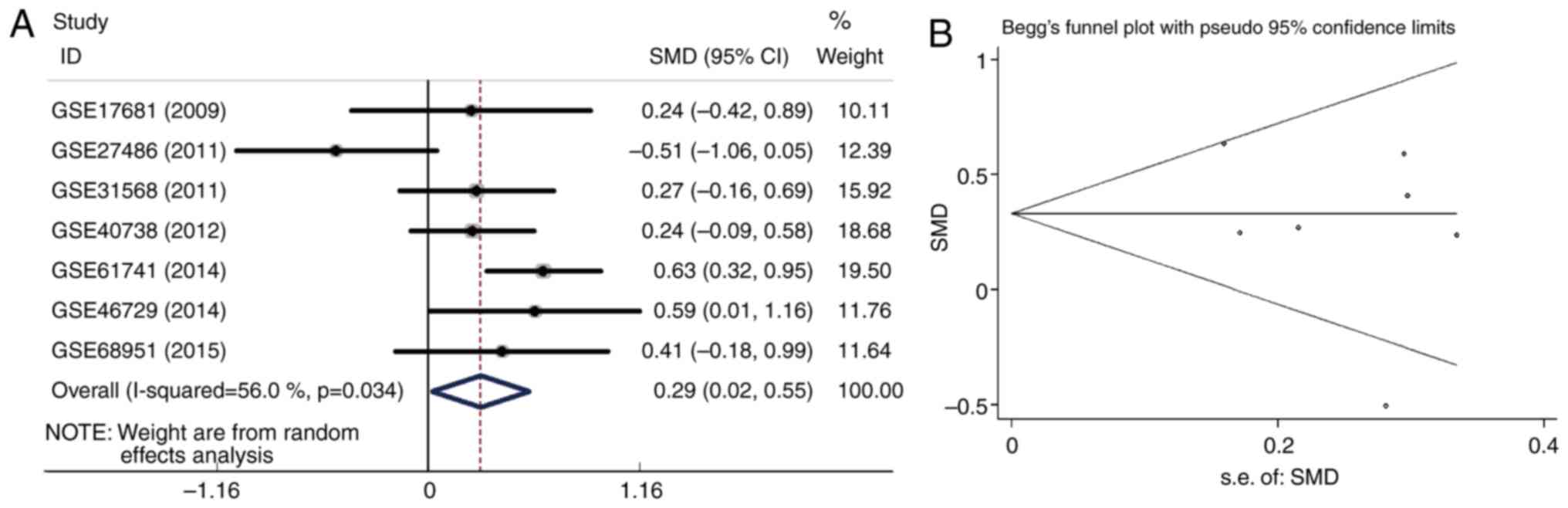
(Fig. 1A). The SMD was 0.29 (95% CI:
0.02 to 0.55; I2=56.0%; P=0.034). The SMD was pooled
using a random-effects model since heterogeneity was observed
(I2>50% or P-value <0.05). Publication bias was
analyzed with a Begg's funnel plot (Fig.
1B). The Begg's funnel plot was basically symmetrical,
indicating that no significant publication bias existed among the 7
peripheral blood datasets. SROC analysis was carried out to further
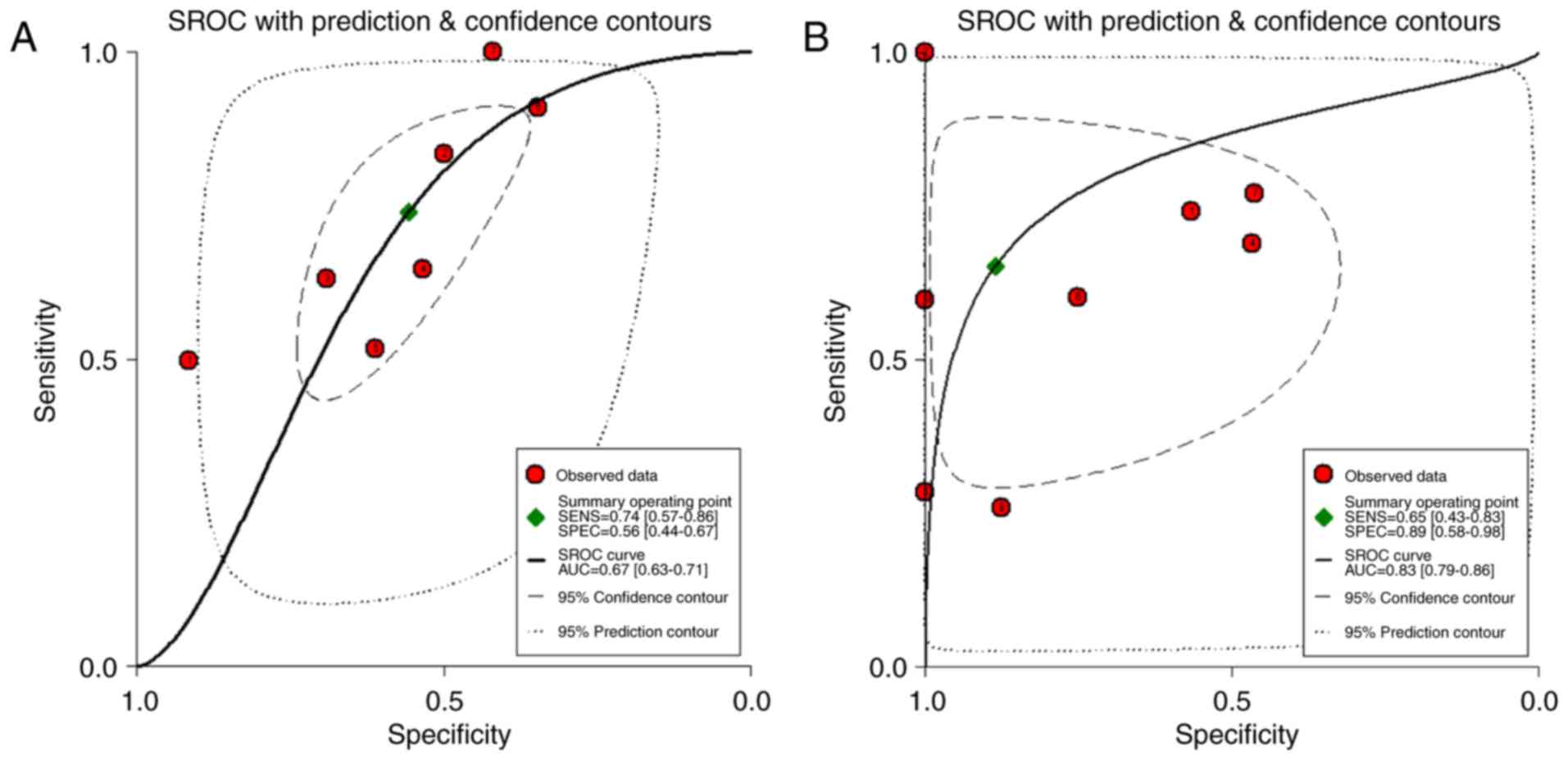
examine the diagnostic value of peripheral blood miR-193a-5p. As
shown in Fig. 2, the AUC was 0.67
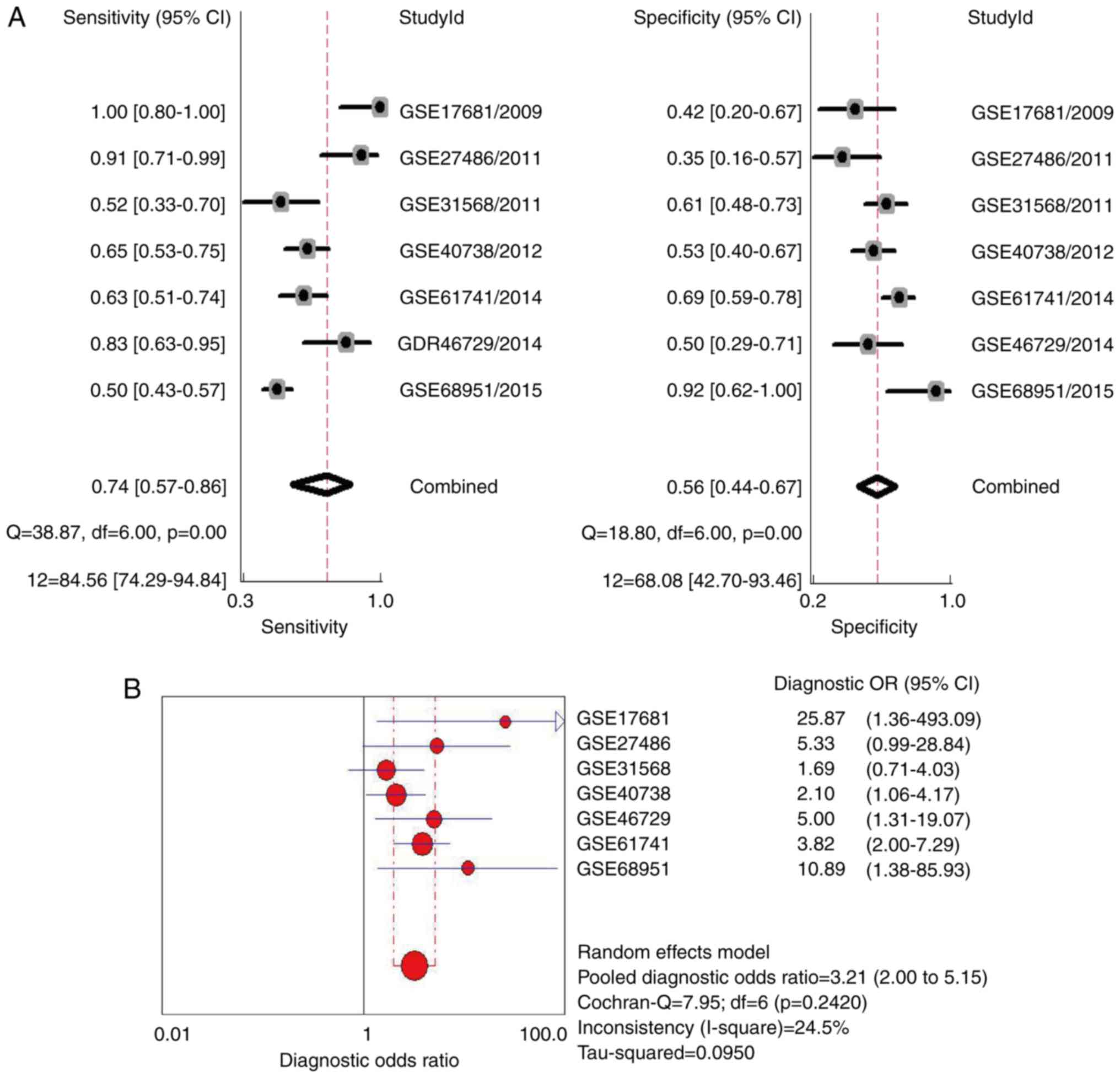
with a 95% CI of 0.63 to 0.71. As shown in Fig. 3A, the combined sensitivity was 0.74
(95% CI: 0.57 to 0.86; I2=84.56%; P=0.00), while the
specificity was 0.56 (95% CI: 0.44 to 0.67; I2=68.08%;
P=0.00). Fig. 3B shows that the
pooled diagnostic odds ratio (OR) was 3.21 (95% CI: 2.00 to 5.15).
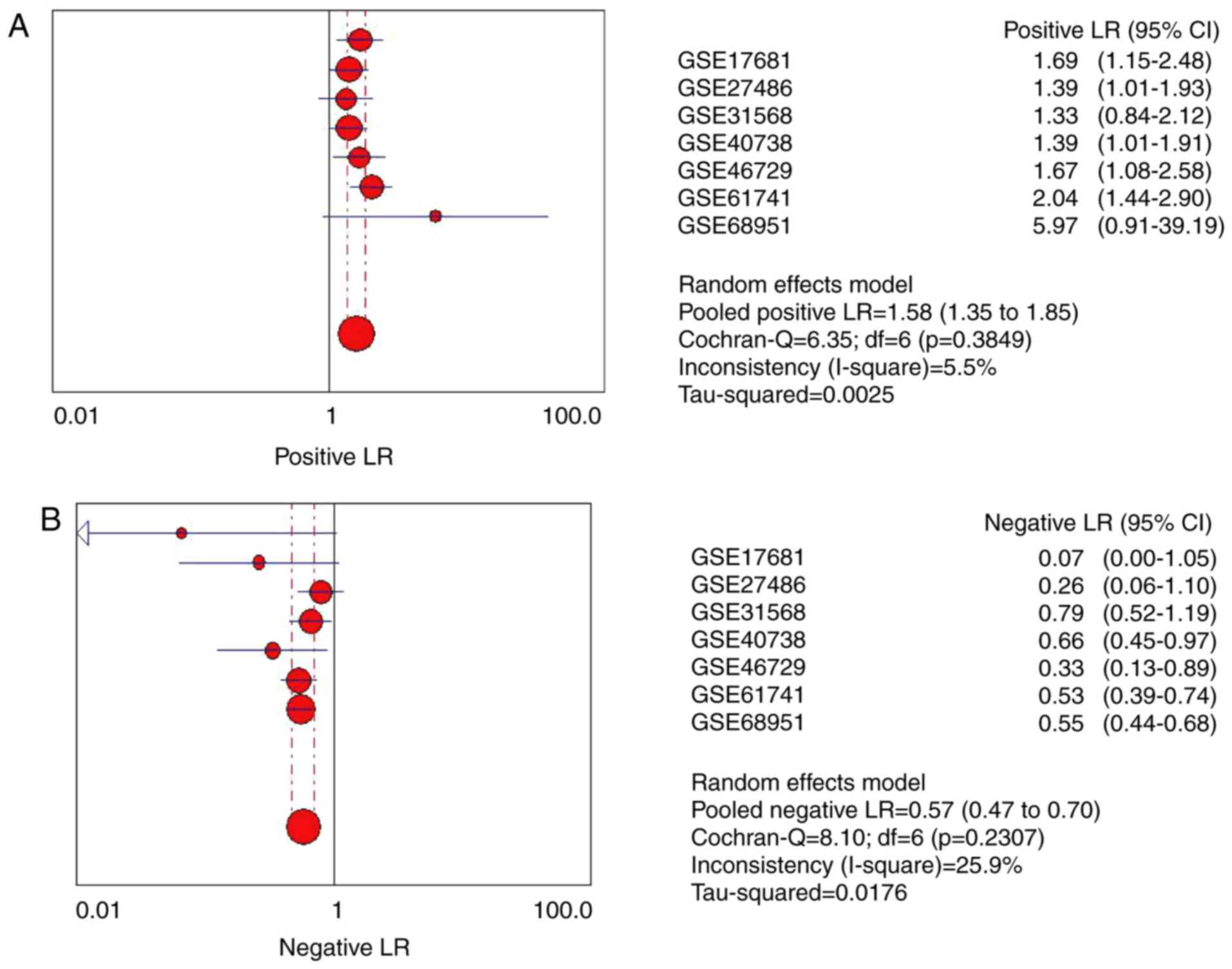
The pooled positive LR and negative LR were also calculated to
evaluate the association between miR-193a-5p and lung cancer
(Fig. 4A and B). The pooled positive
LR was 1.58 (95% CI: 1.35 to 1.85), while the negative LR was 0.57
(95% CI: 0.47 to 0.70).
Meta-analysis of the diagnostic value
of tissue miR-193-5p
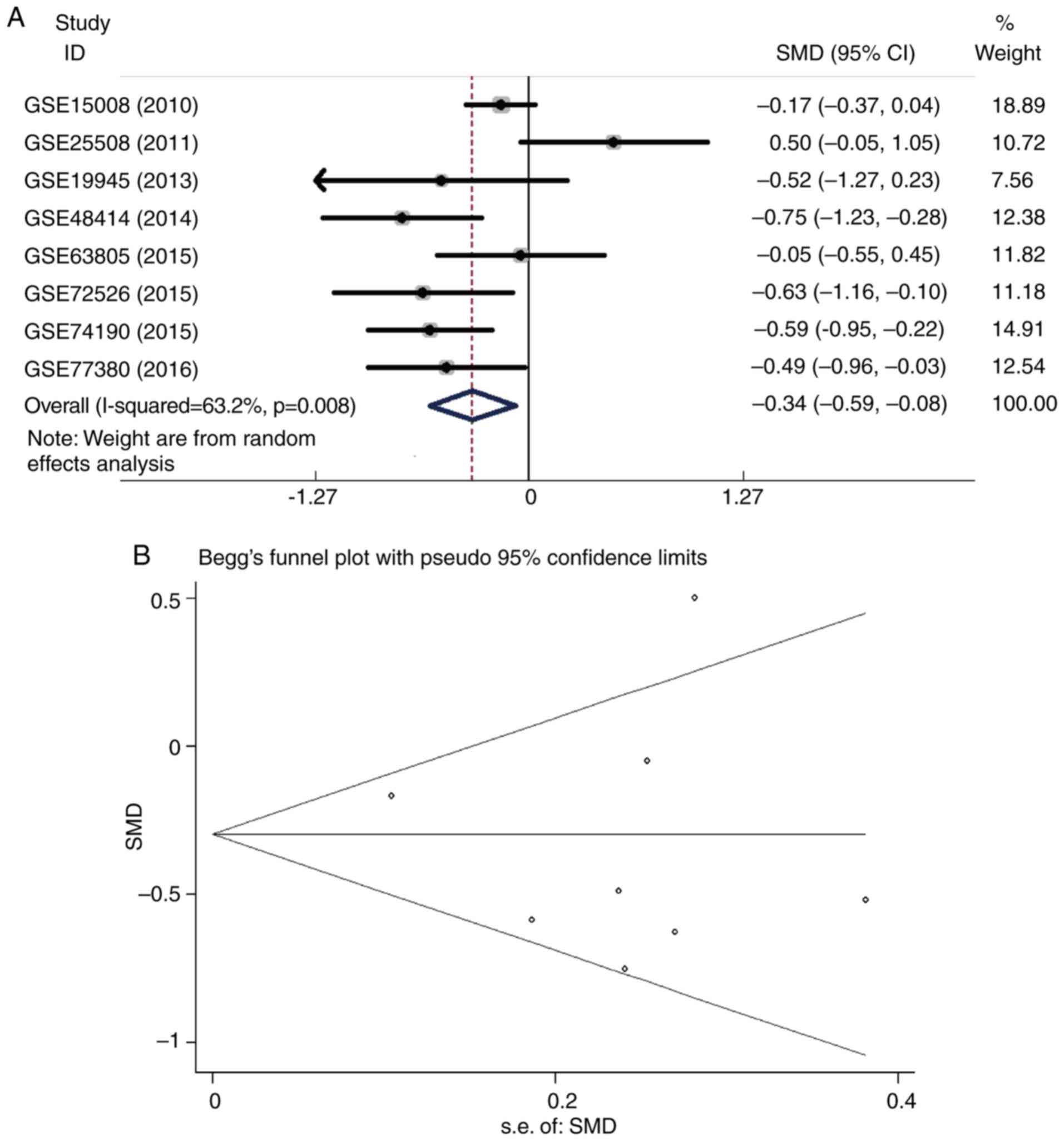
As shown in Fig. 5A,
eight tissue datasets were pooled using a random-effects model, and
the pooled SMD was −0.34 (95% CI: −0.59 to −0.08;
I2=63.2%; P=0.008). A Begg's funnel plot was
constructed, and no publication bias was found, as the funnel plot
was symmetrical (Fig. 5B). In
addition, SROC curve analysis showed satisfactory diagnostic value
of tissue miR-193-5p, with an AUC of 0.83 (95% CI: 0.79 to 0.86)
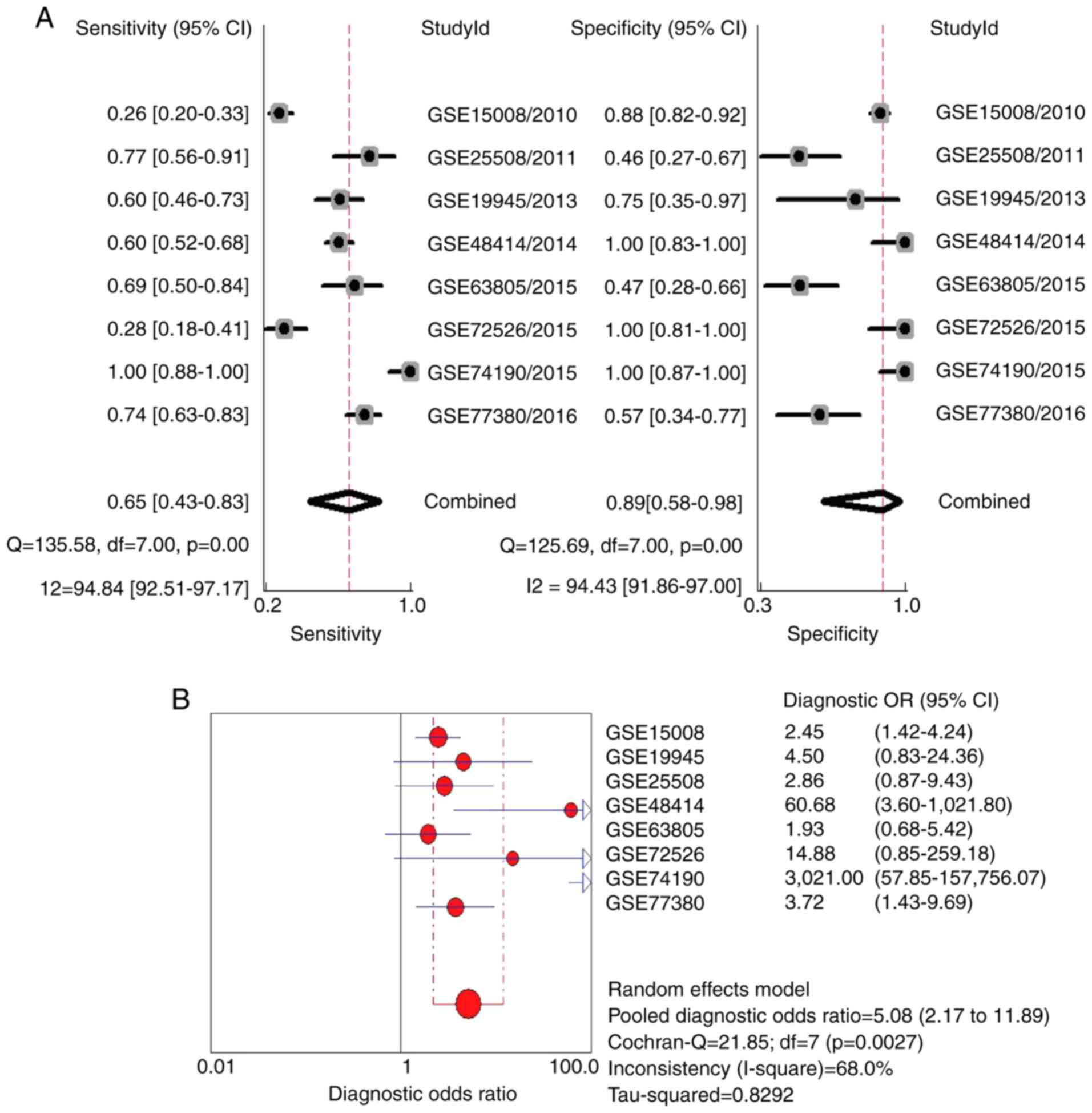
(Fig. 2B). The combined sensitivity
and specificity was 0.65 (95% CI: 0.43–0.83) and 0.89 (95% CI: 0.58
to 0.98), respectively (Fig. 6A).
However, significant heterogeneity was found (I2>50%;
P<0.05). The combined diagnostic OR was 5.08 (95% CI: 2.17 to
11.89), which was calculated using a random-effects model due to
the heterogeneity (Fig. 6B).
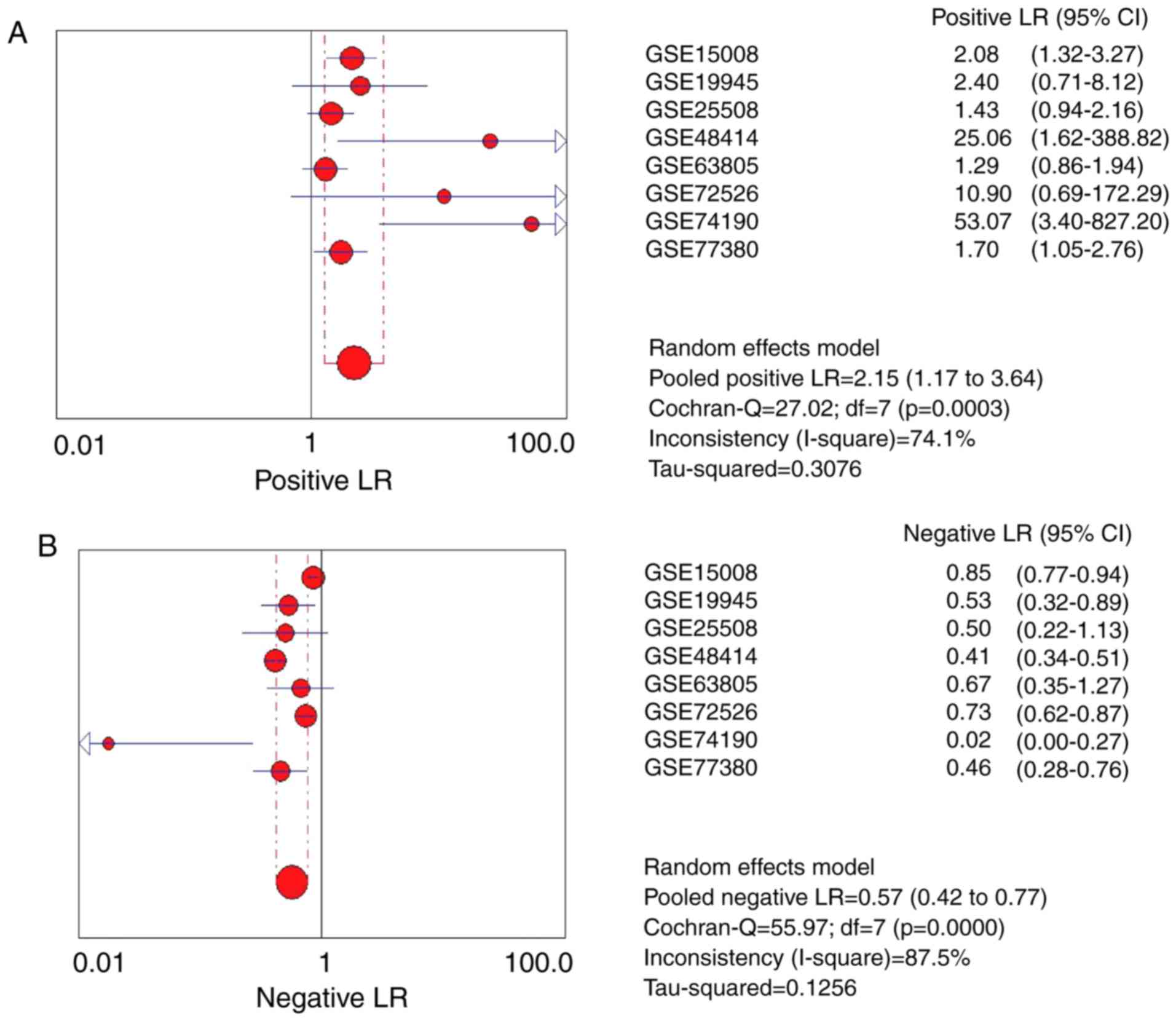
Moreover, the pooled positive LR and negative LR was 2.15 (95% CI:
1.27 to 3.64) and 0.57 (95% CI: 0.42 to 0.77), respectively
(Fig. 7A and B).
Identification of miR-193a-5p
targets
A total of 12666 predicted target genes were
obtained from miRWalk 2.0. Meanwhile, 94 validated target genes
were collected from miRTarbase and Tarbase. Eighty-one overlapping
genes were identified by comparing the predicted and validated
target genes. The 81 overlapping genes are important miR-193a-5p
target genes and were used for further bioinformatics analysis.
Bioinformatics analysis of the
overlapping genes
GO, KEGG and PPI bioinformatics analyses were
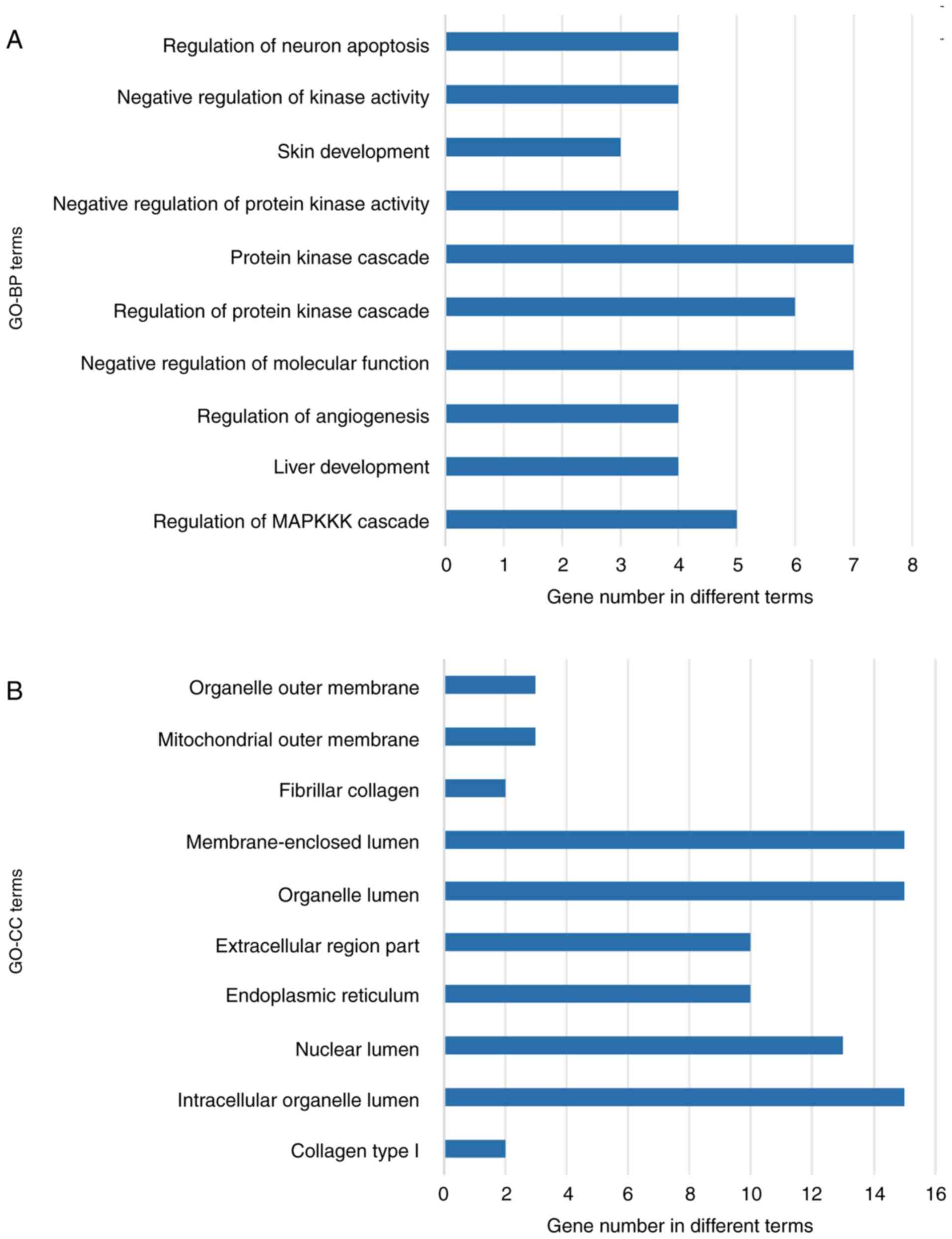
carried out for the overlapping genes (Table II). As shown in Fig. 8A, in biological process (BP), the top
three enriched items were regulation of neuron apoptosis, negative
regulation of kinase activity and skin development. In cellular
component (CC), organelle outer membrane, mitochondrial outer
membrane and fibrillar collagen were the top three enriched terms
(Fig. 8B). For molecular function
(MF), the target genes were mainly enriched in tubulin-tyrosine
ligase activity, protein dimerization activity and platelet-derived
growth factor binding (Fig. 8C). KEGG
analysis showed that the significant terms associated with
miR-193a-5p in lung cancer were pathway in cancer, prostate cancer
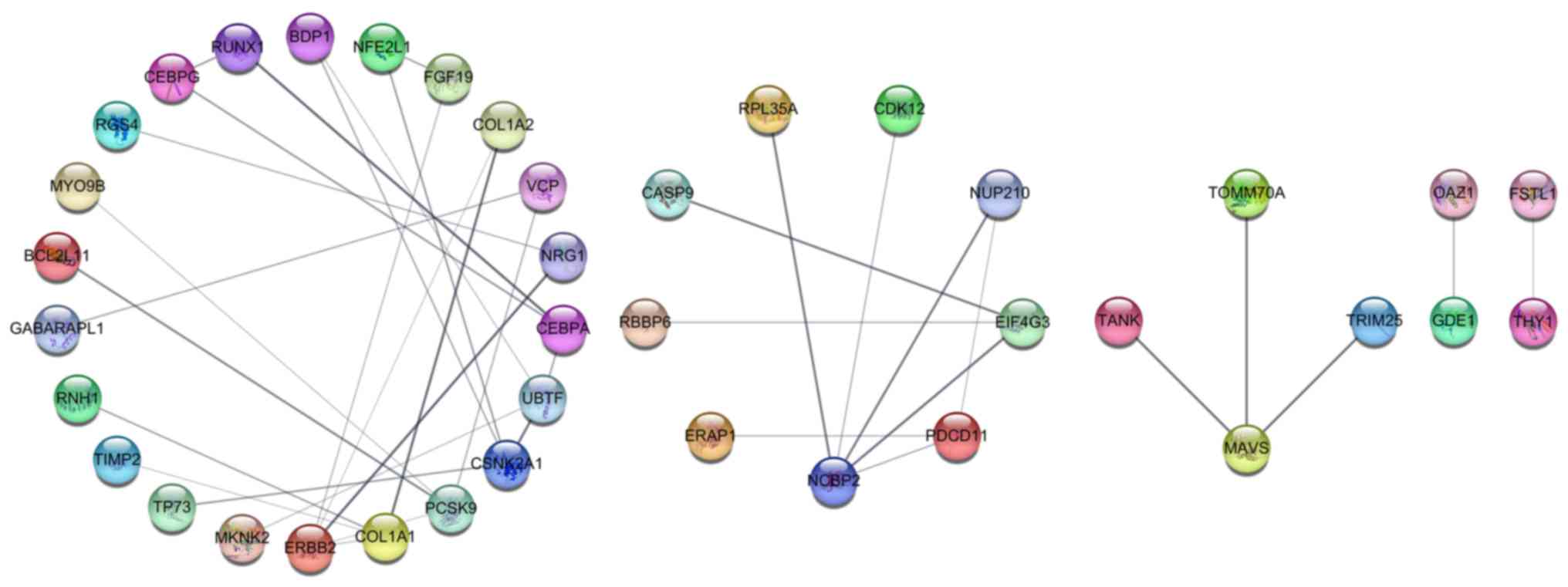
and RIG-I-like receptor signaling pathway (Fig. 8D). By constructing a PPI network
(Fig. 9), we identified 6 hub genes
with a connection degree >3 (ERBB2, NCBP2, COL1A1, PCSK9,
CSNK2A1, and UBTF). These 6 hub genes were the most densely
connected areas in the network and were supported by additional
evidence based on known interactions. These genes were more likely
to be functionally connected, thus showing great potential to be
key target genes of miR-193a-5p in lung cancer.
 | Table II.The significant GO and KEGG enriched
terms of the 81 overlapping genes. |
Table II.
The significant GO and KEGG enriched
terms of the 81 overlapping genes.
| Category | ID | Term | Count | P-value | Genes |
|---|
| GOTERM_BP_FAT | GO:0043408 | Regulation of
MAPKKK cascade | 5 | 0.001553 | FGF19, ERBB2, IGF2,
TIMP2, TP73 |
| GOTERM_BP_FAT | GO:0001889 | Liver
development | 4 | 0.001824 | CEBPA, ERBB2,
CEBPG, PCSK9 |
| GOTERM_BP_FAT | GO:0045765 | Regulation of
angiogenesis | 4 | 0.002994 | ERBB2, RNH1, ERAP1,
RUNX1 |
| GOTERM_BP_FAT | GO:0044092 | Negative regulation
of molecular function | 7 | 0.004164 | CEBPG, RGS4, PCSK9,
IGF2, TP73, DHCR24, THY1 |
| GOTERM_BP_FAT | GO:0010627 | Regulation of
protein kinase cascade | 6 | 0.005595 | MAVS, FGF19, ERBB2,
IGF2, TIMP2, TP73 |
| GOTERM_BP_FAT | GO:0007243 | Protein kinase
cascade | 7 | 0.006808 | ERBB2, RGS4, WNK1,
DUSP10, MKNK2, IGF2, TANK |
| GOTERM_BP_FAT | GO:0006469 | Negative regulation
of protein kinase activity | 4 | 0.007393 | RGS4, IGF2, TP73,
THY1 |
| GOTERM_BP_FAT | GO:0043588 | Skin
development | 3 | 0.007749 | COL1A2, COL1A1,
DHCR24 |
| GOTERM_BP_FAT | GO:0033673 | Negative regulation
of kinase activity | 4 | 0.008115 | RGS4, IGF2, TP73,
THY1 |
| GOTERM_BP_FAT | GO:0043523 | Regulation of
neuron apoptosis | 4 | 0.008115 | CASP9, PCSK9, TP73,
DHCR24 |
| GOTERM_CC_FAT | GO:0005584 | Collagen type
I | 2 | 0.008743 | COL1A2, COL1A1 |
| GOTERM_CC_FAT | GO:0070013 | Intracellular
organelle lumen | 15 | 0.018944 | CEBPA, NCBP2,
PDCD11, CEBPG, TRIM25, IGF2, SENP5, RBBP6, CSNK2A1, UBTF, VCP,
INTS4, ANXA11, CDK12, ERAP1 |
| GOTERM_CC_FAT | GO:0031981 | Nuclear lumen | 13 | 0.021220 | NCBP2, CEBPA,
PDCD11, CSNK2A1, UBTF, VCP, INTS4, CEBPG, ANXA11, CDK12, TRIM25,
SENP5, RBBP6 |
| GOTERM_CC_FAT | GO:0005783 | Endoplasmic
reticulum | 10 | 0.022714 | GABARAPL1, CYP1B1,
VCP, ACO1, NUP210, PCSK9, ERAP1, IGF2, DHCR24, THY1 |
| GOTERM_CC_FAT | GO:0044421 | Extracellular
region part | 10 | 0.022714 | HDGF, COL1A2, RNH1,
PCSK9, IGF2, FSTL1, COL1A1, NRG1, TNFSF9, TIMP2 |
| GOTERM_CC_FAT | GO:0043233 | Organelle
lumen | 15 | 0.022719 | CEBPA, NCBP2,
PDCD11, CEBPG, TRIM25, IGF2, SENP5, RBBP6, CSNK2A1, UBTF, VCP,
INTS4, ANXA11, CDK12, ERAP1 |
| GOTERM_CC_FAT | GO:0031974 | Membrane-enclosed
lumen | 15 | 0.026486 | CEBPA, NCBP2,
PDCD11, CEBPG, TRIM25, IGF2, SENP5, RBBP6, CSNK2A1, UBTF, VCP,
INTS4, ANXA11, CDK12, ERAP1 |
| GOTERM_CC_FAT | GO:0005583 | Fibrillar
collagen | 2 | 0.051347 | COL1A2, COL1A1 |
| GOTERM_CC_FAT | GO:0005741 | Mitochondrial outer
membrane | 3 | 0.059102 | MAVS, TOMM70A,
BCL2L11 |
| GOTERM_CC_FAT | GO:0031968 | Organelle outer
membrane | 3 | 0.076066 | MAVS, TOMM70A,
BCL2L11 |
| GOTERM_MF_FAT | GO:0008047 | Enzyme activator
activity | 7 | 0.006987 | CASP9, RGS4,
RANBP1, MYO9B, NRG1, TIMP2, THY1 |
| GOTERM_MF_FAT | GO:0042802 | Identical protein
binding | 9 | 0.015402 | CEBPA, ERBB2,
COL1A2, CLDN1, PCSK9, TPRG1 L, MYO9B, COL1A1, RUNX1 |
| GOTERM_MF_FAT | GO:0019838 | Growth factor
binding | 4 | 0.016275 | ERBB2, COL1A2,
IGF2, COL1A1 |
| GOTERM_MF_FAT | GO:0043125 | ErbB-3 class
receptor binding | 2 | 0.020182 | ERBB2, NRG1 |
| GOTERM_MF_FAT | GO:0000339 | RNA cap
binding | 2 | 0.039963 | NCBP2, EIF4G3 |
| GOTERM_MF_FAT | GO:0008083 | Growth factor
activity | 4 | 0.048576 | FGF19, HDGF, IGF2,
NRG1 |
| GOTERM_MF_FAT | GO:0048407 | Platelet-derived
growth factor binding | 2 | 0.054540 | COL1A2, COL1A1 |
| GOTERM_MF_FAT | GO:0046983 | Protein
dimerization activity | 7 | 0.056783 | CEBPA, ERBB2,
NUP210, CEBPG, NFE2L1, MYO9B, RUNX1 |
| GOTERM_MF_FAT | GO:0,004835 | Tubulin-tyrosine
ligase activity | 2 | 0.068899 | TTLL4, TTLL11 |
| KEGG_PATHWAY | hsa04622 | RIG-I-like receptor
signaling pathway | 3 | 0.053910 | MAVS, TRIM25,
TANK |
| KEGG_PATHWAY | hsa05215 | Prostate
cancer | 3 | 0.080182 | CASP9, ERBB2,
IGF2 |
| KEGG_PATHWAY | hsa05200 | Pathways in
cancer | 5 | 0.092374 | FGF19, CEBPA,
CASP9, ERBB2, RUNX1 |
Hub gene expression validation and
clinical significance
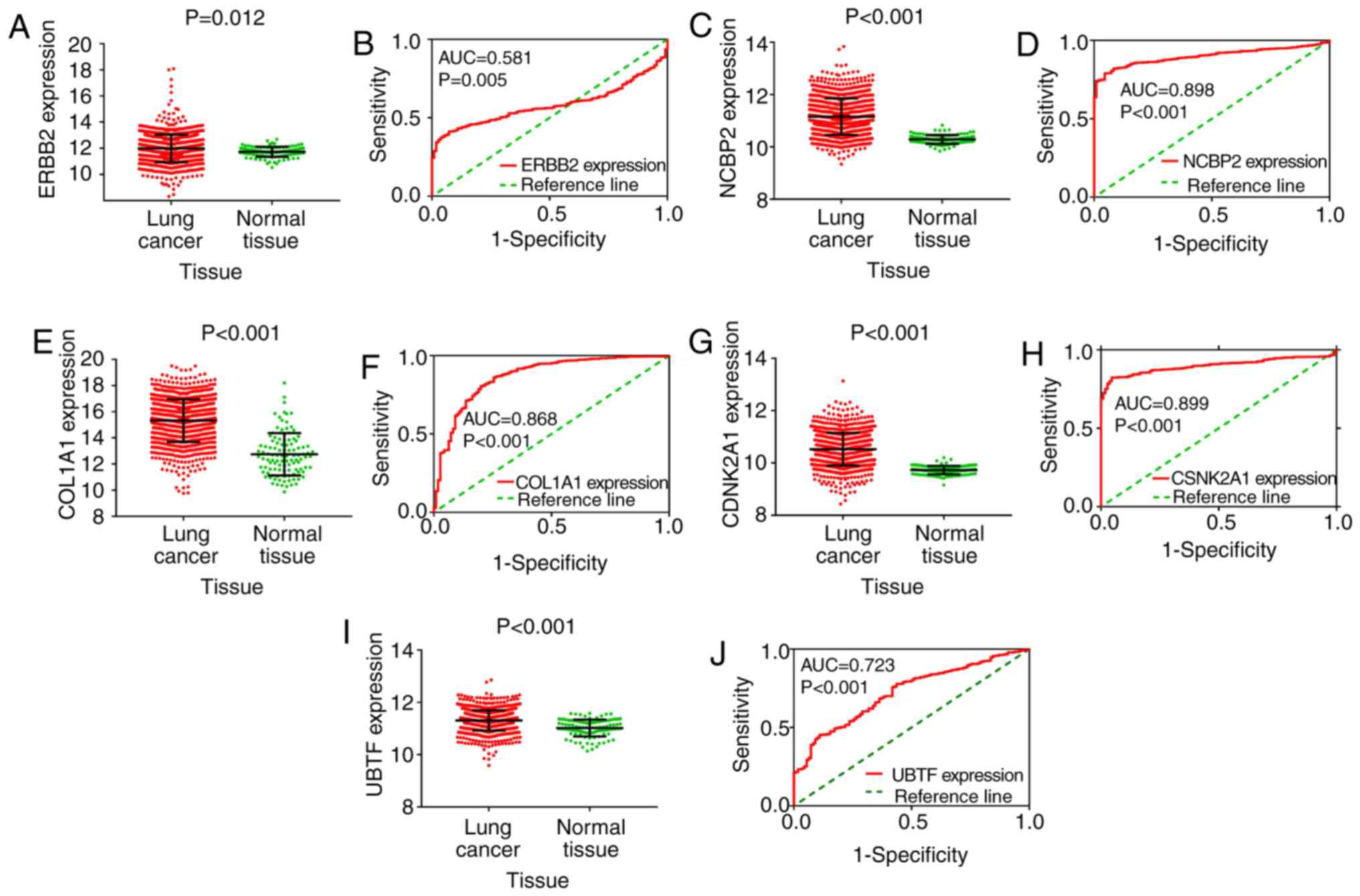
The expression of the six hub genes (ERBB2, NCBP2,
COL1A1, PCSK9, CSNK2A1, UBTF) is shown in scatter plots. ROC curves
were also generated. We found that the expression of five of the
hub genes (ERBB2, NCBP2, COL1A1, CSNK2A1, UBTF) were significantly
upregulated in cancer. In addition, ROC curve analysis also showed
that NCBP2, COL1A1, CSNK2A1 had satisfactory diagnostic value
(Fig. 10). The AUC of ERBB2, NCBP2,
COL1A1, CSNK2A1 and UBTF was 0.581, 0.898, 0.868, 0.899 and 0.723,
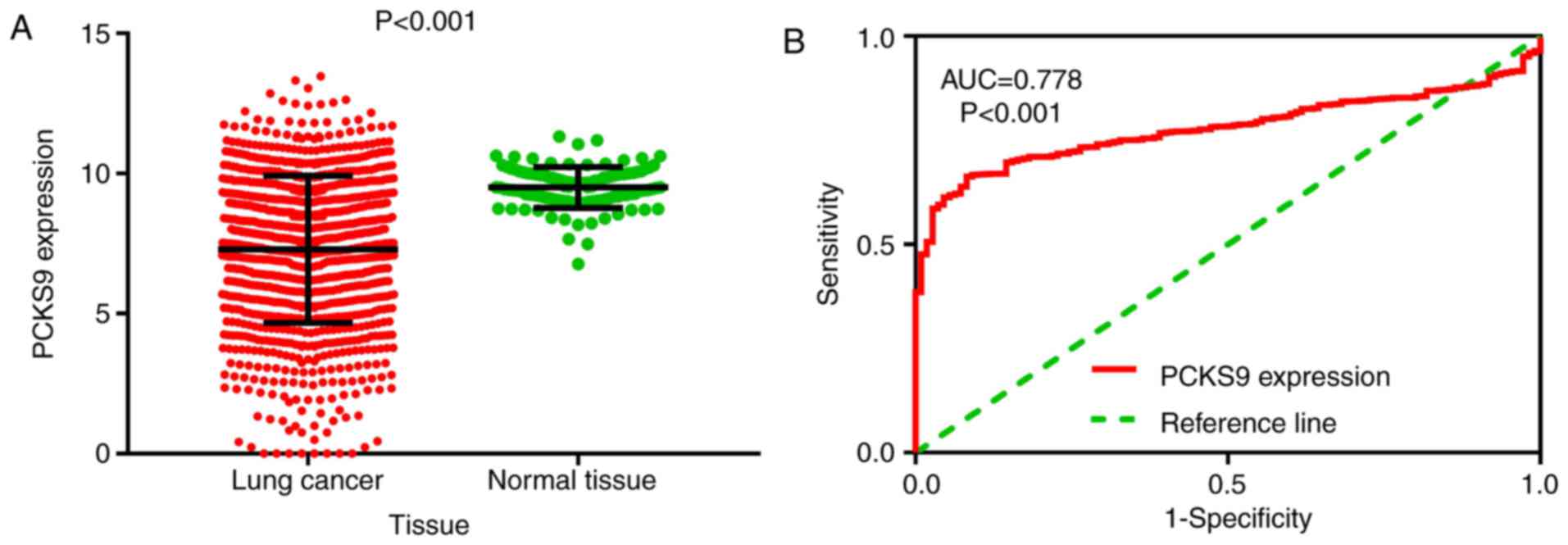
respectively. However, the expression of PCSK9 was significantly
downregulated in cancer, and the ROC curve analysis showed that its
AUC was 0.899 (Fig. 11).
Protein expression of the upregulated
hub genes
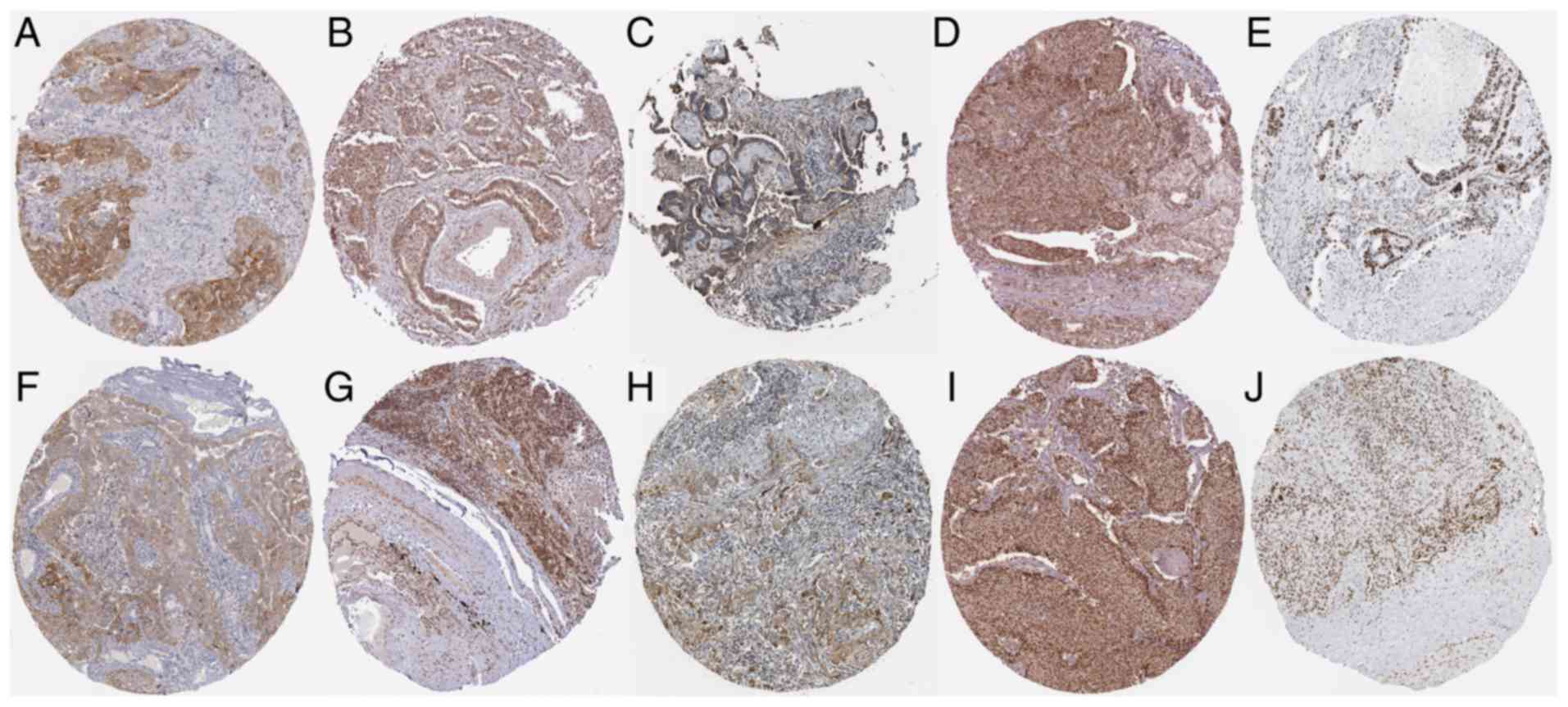
Antibody staining was performed (Fig. 12) and among the five upregulated hub
genes, ERBB2, NCBP2, COL1A1, CSNK2A1 and UBTF, we discovered that
the antibody staining of NCBP2 and UBTF was high (Fig. 12B and E), while that of ERBB2, COL1A1
and CSNK2A1 (Fig. 12A, C and D) was
moderate in pathological LUAD sections. In pathological LUSC
sections, the antibody staining of NCBP2, CSNK2A1 and UBTF was high
(Fig. 12G, I and J), while that of
ERBB2 and COL1A1 was moderate (Fig. 12F
and H). The protein expression of ERBB2, NCBP2, COL1A1, CSNK2A1
and UBTF was upregulated in both LUAD and LUSC pathological
sections. ERBB2 and COL1A1 were localized in the cytoplasm and cell
membrane in both LUAD and LUSC pathological sections. CSNK2A1 and
UBTF staining was predominantly nuclear in both LUAD and LUSC
pathological sections. In LUAD sections, NCBP2 staining was both
cytoplasmic/membranous and nuclear, while in LUSC sections, it was
observed only in the nucleus.
Correlation between miR-193a-5p and
hub genes
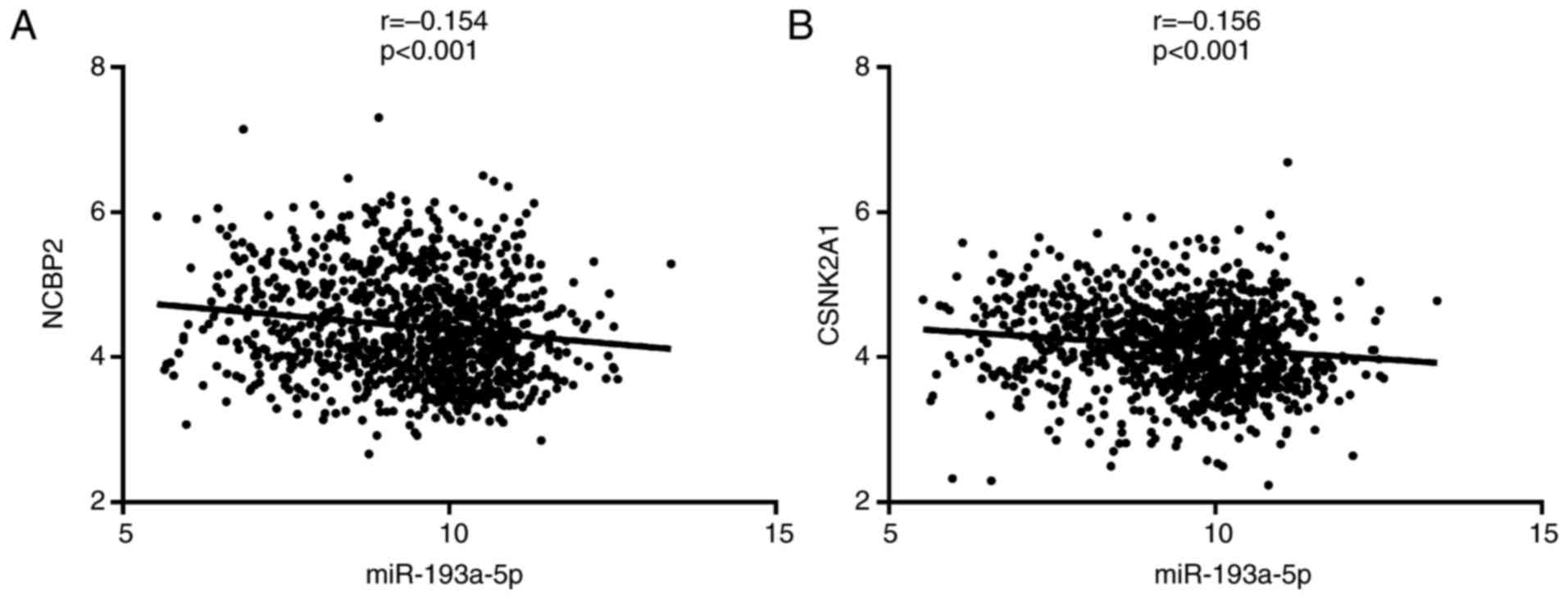
We analyzed a total of 1046 lung cancer samples from
the TCGA database, including 533 LUAD and 513 LUSC samples. Among
the five upregulated hub genes, we found that the expression of
NCBP2 and CSNK2A1 was negatively correlated with that of
miR-193a-5p (P<0.05). The correlation coefficient r was −0.154
(CI: −0.214 to −0.092) and −0.156 (CI: −0.216 to −0.094),
respectively (Fig. 13). This finding
further suggested that NCBP2 and CSNK2A1 are target genes of
miR-193a-5p. However, there was no significant negative correlation
between ERBB2, COL1A1 or UBTF and miR-193a-5p. Thus, based on our
correlation analysis, we were unable to verify that ERBB2, COL1A1,
and UBTF are miR-193a-5p target genes.
Discussion
In this study, we aimed to examine the diagnostic
value of peripheral blood and tissue miR-193a-5p expression. We
also attempted to elucidate the molecular regulatory mechanism
underlying miR-193a-5p in lung cancer. We selected eligible
microarray datasets and conducted a meta-analysis to explore the
clinical diagnostic significance of miR-193a-5p. We then used
bioinformatics analysis to explore the potential molecular
mechanism involved. We compared predicted and validated target
genes of miR-193a-5p and identified the overlapping genes. GO, KEGG
and PPI network analyses were further performed for the overlapping
genes. This study therefore provides a foundation for future
research on miR-193a-5p.
Over the past few decades, the identification of
diagnostic biomarkers in lung cancer has been an important research
focus. Some classic and useful biomarkers such as TP53 (50,51),
neuron-specific enolase (NSE) (52,53),
carcinoembryonic antigen (CEA) (54,55), and
the cytokeratin 19 fragment (CYFRA21-1) (56,57) have
been identified and applied in clinical diagnosis. Nevertheless,
there is still a need to identify diagnostic markers of lung
cancer. Increasingly, lncRNAs and miRNAs have been found to act as
diagnostic markers in lung cancer (58–62). The
combination of multiple diagnostic biomarkers would greatly improve
the sensitivity and specificity of lung cancer diagnosis (56,63,64). Thus,
each finding is likely to contribute to the future clinical
diagnosis of lung cancer. No previous studies have elucidated the
diagnostic value of miR-193a-5p through a comprehensive
meta-analysis. In this study, we used a microarray meta-analysis to
explore the diagnostic value of miR-193a-5p in lung cancer. We
found that the expression of peripheral blood miR-193a-5p was
significantly higher in lung cancer samples than in normal samples.
The pooled AUC was 0.67, with a sensitivity of 0.74 and specificity
of 0.56. The pooled diagnostic OR was 3.21, the positive likelihood
ratio was 1.58 and the negative likelihood ratio was 0.57. Thus,
peripheral blood miR-193a-5p displayed moderate diagnostic value.
It may be useful to combine peripheral blood miR-193a-5p with other
diagnostic markers to improve its clinical diagnostic efficacy.
Surprisingly, in tissue samples, the expression of miR-193a-5p in
lung cancer tissue was evidently reduced compared with normal
tissue. The pooled AUC was 0.83, with a sensitivity of 0.65 and
specificity of 0.89. The diagnostic OR was 5.08, the positive
likelihood ratio was 2.15, and the negative likelihood ratio was
0.57. Thus, tissue miR-193a-5p expression exhibited satisfactory
performance in diagnosing lung cancer and might be a promising
clinical diagnostic biomarker. Interestingly, the expression of
peripheral blood miR-193a-5p was not consistent with the expression
observed in lung cancer tissue samples. The reasons for this
discrepancy between peripheral blood and tissue miRNA expression
remain unclear. The origin of peripheral blood miRNAs remains
controversial. Some researchers have suggested that miRNAs are
secreted through microvesicles/exosomes, and some miRNAs are
expressed at a higher level in microvesicles than inside tumor
cells (65–67). In another study, Pigati et al
(68) found that miRNAs were
selectively released from malignant cells and that the level of
miRNAs released did not necessarily reflect the original abundance
of miRNAs in cells. In Hu's study, it was suggested that the
clinical role of serum miRNAs was independent from that in tissue
samples (69). These findings may
indicate why the expression profiles of miR-193a-5p were opposite
in peripheral blood and tissue specimens in our study. Further
studies are needed to determine the exact mechanism that causes the
difference in expression. Still, peripheral blood and tissue
miR-193a-5p could facilitate the diagnosis of lung cancer to some
extent.
Since miR-193a-5p exerts its regulatory effects by
specifically targeting certain genes, we identified potential
miR-193a-5p target genes and further uncovered the underlying
regulatory pathways. We found that the top enriched GO terms in BP,
CC and MF were regulation of neuron apoptosis, organelle outer
membrane and tubulin-tyrosine ligase activity, respectively.
Mitochondria are important organelles in most eukaryotes.
Mitochondrial outer membrane permeabilization has been reported to
be involved in cancer and may be a promising therapeutic target
(70,71). Therefore, miR-193a-5p might also
participate in lung cancer through a similar mechanism. Recently,
widespread loss of tubulin tyrosine ligase has been found during
tumor growth, suggesting that tubulin tyrosine ligase activity
might be involved in the regulation of tumor cells (72). Tubulin tyrosine ligase activity may be
associated with miR-193a-5p, which might exert effects in lung
cancer. However, more studies are needed to confirm this. In our
KEGG analysis, the terms pathways in cancer, prostate cancer and
RIG-I-like receptor signaling pathway were determined to be
important. However, no studies have yet determined the relationship
between miR-193a-5p and these pathways. Thus, more studies are
urgently required. Through PPI network construction, six hub genes
were identified (ERBB2, NCBP2, COL1A1, PCSK9, CSNK2A1, UBTF). By
analyzing TCGA data, we discovered that among the six hub genes,
five (ERBB2, NCBP2, COL1A1, CSNK2A1, UBTF) were significantly
upregulated in lung cancer tissue. Since the expression of
miR-193a-5p in lung cancer tissue was downregulated, the five
upregulated hub genes (ERBB2, NCBP2, COL1A1, CSNK2A1, UBTF) are
likely to be target genes of miR-193a-5p in lung cancer. However,
more studies are needed to investigate the relationship between the
significantly downregulated hub gene, PCSK9, and miR-193a-5p in
lung cancer. Interestingly, we further verified that the protein
expression of ERBB2, NCBP2, COL1A1, CSNK2A1 and UBTF was also
upregulated according to antibody staining in LUAD and LUSC
pathological sections. These results further demonstrated the
upregulation of ERBB2, NCBP2, COL1A1, CSNK2A1 and UBTF in lung
cancer, indicating that they are likely to be targets of
miR-193a-5p. Of the five upregulated hub genes, NCBP2 and CSNK2A1
were found to be negatively correlated with miR-193a-5p.
Consequently, we focused on NCBP2 and CSNK2A1 in the detailed
discussion below.
NCBP2 (nuclear cap binding protein subunit 2) is
also known as CBC2. Its protein product is a component of the
nuclear cap-binding protein complex. NCBP2 has been reported as a
key target gene in ovarian carcinoma (73). However, we did not find any studies on
NCBP2 and lung cancer. In our study, we found that the protein
expression of NCBP2 was increased in both LUAD and LUSC samples. In
addition, miR-193a-5p was found to be negatively correlated with
NCBP2 (r=−0.154). These findings provided more evidence suggesting
that NCBP2 may be a target of miR-193a-5p in lung cancer.
Nevertheless, more relevant studies are required to further
validate this hypothesis.
CSNK2A1 (casein kinase 2 alpha 1), also known as
CKII, is a serine/threonine protein kinase that participates in
various cellular processes, including the cell cycle, apoptosis,
and circadian rhythm. Over the past few years, CSNK2A1 has been
found to play a significant role in the survival of cancer
patients, suggesting that it may be a promising therapeutic target
(74–76). For example, in Bae JS's study, CSNK2A1
was found to participate in the progression of breast carcinoma and
to indicate poorer patient survival (77). In addition, Rabjerg et al
(78) identified CSNK2A1 as a
promising novel prognostic biomarker in clear cell renal carcinoma.
Furthermore, CSNK2A1 has also been found to be involved in ovarian
cancer (79), oral cancer (80), prostate cancer (81,82) and
pancreatic cancer (83). A CK2
inhibitor has been implicated in the apoptosis, migration, and
metastasis of lung cancer cells (84–87).
Increased protein expression of CSNK2A1 was observed in both LUAD
and LUSC samples. We also observed a negative correlation between
miR-193a-5p and CSNK2A1 according to an analysis of TCGA data
(r=−0.156). Thus, CSNK2A1 could be a promising therapeutic target
in lung cancer. Still, further experimental studies are needed.
In conclusion, in this study we discovered that
peripheral blood and tissue miR-193a-5p could be a promising
diagnostic biomarker for the clinical diagnosis of lung cancer.
Several pathways regulated by miR-193a-5p and its targets,
including pathways in cancer, prostate cancer and the RIG-I-like
receptor signaling pathway, might be important in lung cancer. In
addition, six hub genes, ERBB2, COL1A1, PCSK9, UBTF, and
particularly NCBP2 and CSNK2A1, were identified as key target genes
of miR-193a-5p. However, there were some limitations in this study.
The analysis was based on online databases, and we only performed
bioinformatics analyses. More experiments are needed to validate
the findings. Luciferase assays would be a powerful method to
validate the findings in the current study. Clinicall, this study
could offer possible insights into lung cancer diagnosis and
provide a basis for future research on the molecular mechanisms
involved.
Acknowledgements
Not applicable.
Funding
The present study was supported the Fund of the
Natural Science Foundation of Guangxi, China (grant no.
2016GXNSFAA380255) and Future Academic Star of Guangxi Medical
University (grant no. WLXSZX17042). The funders were not involved
in the study design, data collection and analysis, the decision to
publish or preparation of the manuscript.
Availability of data and materials
All the data and materials used in the current study
are freely accessible in GEO (https://www.ncbi.nlm.nih.gov/geo/), TCGA (https://cancergenome.nih.gov/), miRWalk 2.0
(http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/),
Tarbase (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index),
MitarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php),
DAVID (https://david.ncifcrf.gov/), STRING
(https://string-db.org/) and the Human Protein
Atlas databases (https://www.proteinatlas.org/).
Authors' contributions
ZYL and GC designed the study and revised the
manuscript. ZCX, RXT, XG, QNX, and JYL contributed to the
collection and analysis of the data, as well as the writing of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ERBB2
|
erb-b2 receptor tyrosine kinase 2
|
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
NCBP2
|
nuclear cap binding protein subunit
2
|
|
COL1A1
|
collagen type I alpha 1 chain
|
|
PCSK9
|
proprotein convertase subtilisin/kexin
type 9
|
|
CSNK2A1
|
casein kinase 2 α1
|
|
UBTF
|
upstream binding transcription factor,
RNA polymerase I
|
|
PIK3R3
|
phosphoinositide-3-kinase regulatory
subunit
|
|
mTOR
|
mechanistic target of rapamycin
kinase
|
|
S6K2
|
serine/threonine protein kinase 2
|
|
SROC
|
summary receiver operating
characteristic
|
|
LR
|
likelihood ratio
|
|
DAVID
|
The Database for Annotation,
Visualization and Integrated Discovery
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ROC
|
receiver operating characteristic
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA A Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar
|
|
2
|
Lu C, Shan Z, Hong J and Yang L:
MicroRNA-92a promotes epithelial-mesenchymal transition through
activation of PTEN/PI3K/AKT signaling pathway in non-small cell
lung cancer metastasis. Int J Oncol. 51:235–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Armas-Lopez L, Piña-Sánchez P, Arrieta O,
de Alba EG, Ortiz-Quintero B, Santillán-Doherty P, Christiani DC,
Zúñiga J and Ávila-Moreno F: Epigenomic study identifies a novel
mesenchyme homeobox2-GLI1 transcription axis involved in cancer
drug resistance, overall survival and therapy prognosis in lung
cancer patients. Oncotarget. 8:67056–67081. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bai M, Li W, Yu N, Zhang H, Long F and
Zeng A: The crosstalk between β-catenin signaling and type I, type
II and type III interferons in lung cancer cells. Am J Transl Res.
9:2788–2797. 2017.PubMed/NCBI
|
|
5
|
Hu T and Lu YR: BCYRN1, a c-MYC-activated
long non-coding RNA, regulates cell metastasis of non-small-cell
lung cancer. Cancer Cell Int. 15:362015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pang L, Han S, Jiao Y, Jiang S, He X and
Li P: Bu Fei Decoction attenuates the tumor associated macrophage
stimulated proliferation, migration, invasion and immunosuppression
of non-small cell lung cancer, partially via IL-10 and PD-L1
regulation. Int J Oncol. 51:25–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Romero-Cordoba SL, Salido-Guadarrama I,
Rodriguez-Dorantes M and Hidalgo-Miranda A: miRNA biogenesis:
Biological impact in the development of cancer. Cancer Biol Ther.
15:1444–1455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naidu S, Magee P and Garofalo M:
miRNA-based therapeutic intervention of cancer. J Hematol Oncol.
8:682015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ganju A, Khan S, Hafeez BB, Behrman SW,
Yallapu MM, Chauhan SC and Jaggi M: miRNA nanotherapeutics for
cancer. Drug Discov Today. 22:424–432. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mishra S, Yadav T and Rani V: Exploring
miRNA based approaches in cancer diagnostics and therapeutics. Crit
Rev Oncol Hematol. 98:12–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao C, Lu F and Chen H, Zhao F, Zhu Z,
Zhao X and Chen H: Clinical significance of circulating miRNA
detection in lung cancer. Med Oncol. 33:412016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang C, Hu X, Alattar M and Zhao H: miRNA
expression profiles associated with diagnosis and prognosis in lung
cancer. Expert Rev Anticancer Ther. 14:453–461. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin CH, Tsai CH, Yeh CT, Liang JL, Hung
WC, Lin FC, Chang WL, Li HY, Yao YC, Hsu TI, et al:
miR-193a-5p/ERBB2 act as concurrent chemoradiation therapy response
indicator of esophageal squamous cell carcinoma. Oncotarget.
7:39680–39693. 2016.PubMed/NCBI
|
|
15
|
Zhou J, Duan H, Xie Y, Ning Y, Zhang X,
Hui N, Wang C, Zhang J and Zhou J: miR-193a-5p targets the coding
region of AP-2a mRNA and induces cisplatin resistance in bladder
cancers. J Cancer. 7:1740–1746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jacques C, Calleja LR, Baud'huin M,
Quillard T, Heymann D, Lamoureux F and Ory B: miRNA-193a-5p
repression of p73 controls Cisplatin chemoresistance in primary
bone tumors. Oncotarget. 7:54503–54514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pu Y, Zhao F, Cai W, Meng X, Li Y and Cai
S: miR-193a-3p and miR-193a-5p suppress the metastasis of human
osteosarcoma cells by down-regulating Rab27B and SRR, respectively.
Clin Exp Metastasis. 33:359–372. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang P, Ji DB, Han HB, Shi YF, Du CZ and
Gu J: Downregulation of miR-193a-5p correlates with lymph node
metastasis and poor prognosis in colorectal cancer. World J
Gastroenterol. 20:12241–12248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang
P, Chan LK, Zhang T, Yu J, Kwong J, et al: A novel
miR-193a-5p-YY1-APC regulatory axis in human endometrioid
endometrial adenocarcinoma. Oncogene. 32:3432–3442. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Toren P and Zoubeidi A: Targeting the
PI3K/Akt pathway in prostate cancer: Challenges and opportunities
(review). Int J Oncol. 45:1793–1801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Georgi B, Korzeniewski N, Hadaschik B,
Grüllich C, Roth W, Sültmann H, Pahernik S, Hohenfellner M and
Duensing S: Evolving therapeutic concepts in prostate cancer based
on genome-wide analyses (review). Int J Oncol. 45:1337–1344. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie Y, Naizabekov S, Chen Z and Tokay T:
Power of PTEN/AKT: Molecular switch between tumor suppressors and
oncogenes. Oncol Lett. 12:375–378. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steelman LS, Martelli AM, Cocco L, Libra
M, Nicoletti F, Abrams SL and McCubrey JA: The therapeutic
potential of mTOR inhibitors in breast cancer. Br J Clin Pharmacol.
82:1189–1212. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nicoletti F, Fagone P, Meroni P, McCubrey
J and Bendtzen K: mTOR as a multifunctional therapeutic target in
HIV infection. Drug Discov Today. 16:715–721. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen J, Gao S, Wang C, Wang Z, Zhang H,
Huang K, Zhou B, Li H, Yu Z, Wu J and Chen C: Pathologically
decreased expression of miR-193a contributes to metastasis by
targeting WT1-E-cadherin axis in non-small cell lung cancers. J Exp
Clin Cancer Res. 35:1732016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tomczak K, Czerwinska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
32
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thul PJ, Akesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:eaal33212017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keller A, Leidinger P, Borries A,
Wendschlag A, Wucherpfennig F, Scheffler M, Huwer H, Lenhof HP and
Meese E: miRNAs in lung cancer - studying complex fingerprints in
patient's blood cells by microarray experiments. BMC cancer.
9:3532009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patnaik SK, Yendamuri S, Kannisto E,
Kucharczuk JC, Singhal S and Vachani A: MicroRNA expression
profiles of whole blood in lung adenocarcinoma. PloS One.
7:e460452012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Keller A, Leidinger P, Bauer A, Elsharawy
A, Haas J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, et al:
Toward the blood-borne miRNome of human diseases. Nat Methods.
8:841–843. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Patnaik SK, Kannisto ED, Mallick R,
Vachani A and Yendamuri S: Whole blood microRNA expression may not
be useful for screening non-small cell lung cancer. PloS One.
12:e01819262017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Keller A, Leidinger P, Vogel B, Backes C,
ElSharawy A, Galata V, Mueller SC, Marquart S, Schrauder MG, Strick
R, et al: miRNAs can be generally associated with human pathologies
as exemplified for miR-144. BMC Med. 12:2242014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46729January
8–2017
|
|
41
|
Leidinger P, Galata V, Backes C, Stähler
C, Rheinheimer S, Huwer H, Meese E and Keller A: Longitudinal study
on circulating miRNAs in patients after lung cancer resection.
Oncotarget. 6:16674–16685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang
C, Wan J, Zhou F, Shao K, Sun Y, et al: A 5-microRNA signature for
lung squamous cell carcinoma diagnosis and hsa-miR-31 for
prognosis. Clin Cancer Res. 17:6802–6811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nymark P, Guled M, Borze I, Faisal A,
Lahti L, Salmenkivi K, Kettunen E, Anttila S and Knuutila S:
Integrative analysis of microRNA, mRNA and aCGH data reveals
asbestos- and histology-related changes in lung cancer. Genes
Chromosomes Cancer. 50:585–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19945January
8–2017
|
|
45
|
Bjaanaes MM, Halvorsen AR, Solberg S,
Jørgensen L, Dragani TA, Galvan A, Colombo F, Anderlini M,
Pastorino U, Kure E, et al: Unique microRNA-profiles in
EGFR-mutated lung adenocarcinomas. Int J Cancer. 135:1812–1821.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Robles AI, Arai E, Mathé EA, Okayama H,
Schetter AJ, Brown D, Petersen D, Bowman ED, Noro R, Welsh JA, et
al: An integrated prognostic classifier for stage I lung
adenocarcinoma based on mRNA, microRNA, and DNA methylation
Biomarkers. J Thorac Oncol. 10:1037–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gasparini P, Cascione L, Landi L, Carasi
S, Lovat F, Tibaldi C, Alì G, D'Incecco A, Minuti G, Chella A, et
al: microRNA classifiers are powerful diagnostic/prognostic tools
in ALK-, EGFR-, and KRAS-driven lung cancers. Proc Natl Acad Sci
USA. 112:pp. 14924–14929. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
48
|
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74190January
8–2017
|
|
49
|
Yoshimoto TI, Motoi N, Yamamoto N, Nagano
H, Ushijima M, Matsuura M, Okumura S, Yamaguchi T, Fukayama M and
Ishikawa Y: Pulmonary carcinoids and low-grade gastrointestinal
neuroendocrine tumors show common microRNA expression profiles,
different from adenocarcinomas and small cell carcinomas.
Neuroendocrinology. 106:47–57. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Halvorsen AR, Silwal-Pandit L, Meza-Zepeda
LA, Vodak D, Vu P, Sagerup C, Hovig E, Myklebost O, Børresen-Dale
AL, Brustugun OT and Helland Å: TP53 mutation spectrum in smokers
and never smoking lung cancer patients. Front Genet. 7:852016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mogi A and Kuwano H: TP53 mutations in
nonsmall cell lung cancer. J Biomed Biotechnol. 2011:5839292011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang L, Zhou JG, Yao WX, Tian X, Lv SP,
Zhang TY, Jin SH, Bai YJ and Ma H: Systematic review and
meta-analysis of the efficacy of serum neuron-specific enolase for
early small cell lung cancer screening. Oncotarget. 8:64358–64372.
2017.PubMed/NCBI
|
|
53
|
Isgro MA, Bottoni P and Scatena R:
Neuron-specific enolase as a biomarker: Biochemical and clinical
aspects. Adv Exp Med Biol. 867:125–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zou Y, Wang L, Zhao C, Hu Y, Xu S, Ying K,
Wang P and Chen X: CEA, SCC and NSE levels in exhaled breath
condensate-possible markers for early detection of lung cancer. J
Breath Res. 7:0471012013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Grunnet M and Sorensen JB:
Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung
Cancer. 76:138–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang B, He YJ, Tian YX, Yang RN, Zhu YR
and Qiu H: Clinical utility of haptoglobin in combination with CEA,
NSE and CYFRA21-1 for diagnosis of lung cancer. Asian Pac J Cancer
Prev. 15:9611–9614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Okamura K, Takayama K, Izumi M, Harada T,
Furuyama K and Nakanishi Y: Diagnostic value of CEA and CYFRA 21-1
tumor markers in primary lung cancer. Lung Cancer. 80:45–49. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M
and Tu J: Identification of circulating long noncoding RNA HOTAIR
as a novel biomarker for diagnosis and monitoring of non-small cell
lung cancer. Technol Cancer Res Treat. Jan 1–2017.(Epub ahead of
print). View Article : Google Scholar
|
|
59
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC and Li DJ: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50:2017.https://doi.org/10.1111/cpr.12394simple10.1111/cpr.12394
View Article : Google Scholar
|
|
60
|
Inamura K: Diagnostic and therapeutic
potential of MicroRNAs in lung cancer. Cancers. 9:E492017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ricciuti B, Mencaroni C, Paglialunga L,
Paciullo F, Crinò L, Chiari R and Metro G: Long noncoding RNAs: New
insights into non-small cell lung cancer biology, diagnosis and
therapy. Med Oncol. 33:182016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen J, Wang R, Zhang K and Chen LB: Long
non-coding RNAs in non-small cell lung cancer as biomarkers and
therapeutic targets. J Cell Mol Med. 18:2425–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Pan YW, Zhou ZG, Wang M, Dong JQ, Du KP,
Li S, Liu YL, Lv PJ and Gao JB: Combination of IL-6, IL-10 and
MCP-1 with traditional serum tumor markers in lung cancer diagnosis
and prognosis. Genet Mol Res. 15:2016.https://doi.org/10.4238/gmr15048949simple10.4238/gmr15048949
View Article : Google Scholar
|
|
64
|
Wang WJ, Tao Z, Gu W and Sun LH: Clinical
observations on the association between diagnosis of lung cancer
and serum tumor markers in combination. Asian Pac J Cancer Prev.
14:4369–4371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA. 105:pp.
10513–10518. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Skog J, Wurdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Valadi H, Ekstrom K, Bossios A, Sjostrand
M, Lee JJ and Lotvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Pigati L, Yaddanapudi SC, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM, et al: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C and Shen H: Serum microRNA
signatures identified in a genome-wide serum microRNA expression
profiling predict survival of non-small-cell lung cancer. J Clin
Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Farsinejad S, Gheisary Z, Ebrahimi Samani
S and Alizadeh AM: Mitochondrial targeted peptides for cancer
therapy. Tumour Biol. 36:5715–5725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fulda S, Galluzzi L and Kroemer G:
Targeting mitochondria for cancer therapy. Nat Rev Drug Discov.
9:447–464. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lafanechere L, Courtay-Cahen C, Kawakami
T, Jacrot M, Rüdiger M, Wehland J, Job D and Margolis RL:
Suppression of tubulin tyrosine ligase during tumor growth. J Cell
Sci. 111:171–181. 1998.PubMed/NCBI
|
|
73
|
Wei S, Wang Y, Xu H and Kuang Y: Screening
of potential biomarkers for chemoresistant ovarian carcinoma with
miRNA expression profiling data by bioinformatics approach. Oncol
Lett. 10:2427–2431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chua MM, Ortega CE, Sheikh A, Lee M,
Abdul-Rassoul H, Hartshorn KL and Dominguez I: CK2 in cancer:
Cellular and biochemical mechanisms and potential therapeutic
target. Pharmaceuticals (Basel). 10:E182017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ortega CE, Seidner Y and Dominguez I:
Mining CK2 in cancer. PLoS One. 9:e1156092014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Abdel-Magid AF: Inhibition of CK2: An
attractive therapeutic target for cancer treatment. ACS Med Chem
Lett. 4:1131–1132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bae JS, Park SH, Jamiyandorj U, Kim KM,
Noh SJ, Kim JR, Park HJ, Kwon KS, Jung SH, Park HS, et al:
CK2α/CSNK2A1 phosphorylates SIRT6 and is involved in the
progression of breast carcinoma and predicts shorter survival of
diagnosed patients. Am J Pathol. 186:3297–3315. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Rabjerg M, Bjerregaard H, Halekoh U,
Jensen BL, Walter S and Marcussen N: Molecular characterization of
clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1
as promising novel prognostic markers. APMIS. 124:372–383. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kulbe H, Iorio F, Chakravarty P, Milagre
CS, Moore R, Thompson RG, Everitt G, Canosa M, Montoya A, Drygin D,
et al: Integrated transcriptomic and proteomic analysis identifies
protein kinase CK2 as a key signaling node in an inflammatory
cytokine network in ovarian cancer cells. Oncotarget.
7:15648–15661. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Srivastava R, Akthar S, Sharma R and
Mishra S: Identification of Ellagic acid analogues as potent
inhibitor of protein Kinase CK2: A chemopreventive role in oral
Cancer. Bioinformation. 11:21–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chatterjee A, Chatterjee U and Ghosh MK:
Activation of protein kinase CK2 attenuates FOXO3a functioning in a
PML-dependent manner: Implications in human prostate cancer. Cell
Death Dis. 4:e5432013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Schneider CC, Kartarius S, Montenarh M,
Orzeszko A and Kazimierczuk Z: Modified tetrahalogenated
benzimidazoles with CK2 inhibitory activity are active against
human prostate cancer cells LNCaP in vitro. Bioorg Med Chem.
20:4390–4396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Nelson N, Szekeres K, Iclozan C, Rivera
IO, McGill A, Johnson G, Nwogu O and Ghansah T: Apigenin: Selective
CK2 inhibitor increases Ikaros expression and improves T cell
homeostasis and function in murine pancreatic cancer. PLoS One.
12:e01701972017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Benavent Acero F, Capobianco CS, Garona J,
Cirigliano SM, Perera Y, Urtreger AJ, Perea SE, Alonso DF and
Farina HG: CIGB-300, an anti-CK2 peptide, inhibits angiogenesis,
tumor cell invasion and metastasis in lung cancer models. Lung
cancer. 107:14–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ku MJ, Park JW, Ryu BJ, Son YJ, Kim SH and
Lee SY: CK2 inhibitor CX4945 induces sequential inactivation of
proteins in the signaling pathways related with cell migration and
suppresses metastasis of A549 human lung cancer cells. Bioor Med
Chem Lett. 23:5609–5613. 2013. View Article : Google Scholar
|
|
86
|
Zhou Y, Li K, Zhang S, Li Q, Li Z, Zhou F,
Dong X, Liu L, Wu G and Meng R: Quinalizarin, a specific CK2
inhibitor, reduces cell viability and suppresses migration and
accelerates apoptosis in different human lung cancer cell lines.
Indian J Cancer. 52 Suppl 2:e119–e124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
So KS, Rho JK, Choi YJ, Kim SY, Choi CM,
Chun YJ and Lee JC: AKT/mTOR down-regulation by CX-4945, a CK2
inhibitor, promotes apoptosis in chemorefractory non-small cell
lung cancer cells. Anticancer Res. 35:1537–1542. 2015.PubMed/NCBI
|