Introduction
Lung cancer is a highly malignant human cancer in
the world as its mortality rate is highest in contrast to other
types of cancers (1). In addition,
non-small cell lung cancer (NSCLC) accounts for 75 to 80%
pathological types in all lung cancers (2). Although there has been much progress in
the study of NSCLC and its treatment, poor prognosis associated
with lung cancer remains a serious problem for patients (3). Therefore, effective early diagnosis and
prognostic markers can be significant and can enhance clinical
value for NSCLC. The discovery of new anti-tumor molecules, as
potential targets for clinical prevention and treatment of lung
cancer, will create a new approach to tackle lung cancer
treatment.
Long non-coding RNA (lncRNA) is a group of RNA with
>200 nt length and is located in the nucleus or cytoplasm
(4). Although lncRNA does not have
the function of an encoded protein, its finding promotes the
progress of research in the field of non-coding RNA (5). Recent studies found that lncRNA
participates in the process of X chromosome gene silencing, genomic
imprinting, chromatin modification, transcriptional activation and
regulation of gene expression in the nucleus and other
transcription processes (6–8). Studies found that lncRNA is involved in
many biological processes such as apoptosis and cell cycle
(9). Also, lncRNA plays a crucial
role in human disease occurrence and development processes
(10,11). Therefore, the role of lncRNA in tumors
draws considerable attention.
H19 was one of the earliest lncRNA groups identified
by researchers. Naturally, it is a highly conserved lncRNA that is
found in mammals and it is approximately 2.3 kb in length (12). Previous studies confirmed that H19
plays an important role in embryonic development, and it is
abnormally expressed in a variety of tumors including bladder
cancer, gastric cancer and hepatocellular carcinoma through
regulating proliferation, invasion and metastasis of tumors
(13–15). However, the role of H19 in NSCLC and
its underlying mechanisms are unclear. Our study compared the
expression of H19 in NSCLC tissues and adjacent tissues. Also, the
association between differential expression and clinicopathological
parameters of NSCLC was also analyzed. Furthermore, in vitro
experiments investigated the influence of H19 on NSCLC cell
proliferation, epithelial-mesenchymal transition (EMT) impact and
invasion ability. Also, the potential regulation mechanisms of H19
induced EMT process was investigated.
Materials and methods
Clinical features
A total of 76 NSCLC tissue samples were collected
from patients (age 45–78 years; 32 males and 44 females) who
underwent thoracic surgery from 2009 to 2014 in The First
Affiliated Hospital of Zunyi Medical University. Adjacent non-tumor
tissues were obtained at least 2 cm away from the tumor edges (no
observed cancer cells under endoscopic) and used as controls. The
clinical stage and histological classification was evaluated with
the National Comprehensive Cancer Network (NCCN) NSCLC Guidelines
(v.7. 2015). Resection specimens were stored into liquid nitrogen.
The study was approved by the Ethics Committee of Zunyi Medical
University. Informed consent was obtained from each patient prior
to surgery. All experimental procedures were carried out in
accordance with the approved guidelines and were in agreement with
the Declaration of Helsinki.
Cell culture
Normal bronchial epithelial cell line BEAS-2B, NSCLC
adenocarcinoma cell line A549, SPC-A1 and squamous cell SKMES-1
were purchased from the Shanghai Institute of Life Sciences
Institute of Biochemistry and the Cell Biology Institute of Cell
Bank (Shanghai, China). All cell lines were cultured in RPMI-1640
containing 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), 100 U/ml penicillin and 100 mg/ml streptomycin
and were placed in 37°C cell incubation supplied with 5%
CO2. Culture medium was replaced every 1–2 days and
subcultured when cell reached 80 to 90% confluence. Cell
morphological observation was performed with an ordinary optical
microscope (Nikon Corporation, Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues or cells were extracted with
the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's instructions. For
tissue samples, 1 ml TRIzol reagent per 100 mg of tissue sample was
used followed by homogenization of tissue samples with a glass
Teflon homogenizer (Invitrogen; Thermo Fisher Scientific, Inc.).
Then, the homogenized samples were processed to phase separation
following the manufacturer's instructions. PCR primers were
designed and synthesized by the Shenzhen Huada Gene Science and
Technology Services Limited Company, Co., Ltd., (Shenzhen, China).
The specific primers used in qRT-PCR were shown in Table I. Relative expression was calculated
using the 2−ΔΔCq method (16).
 | Table I.Quantitative PCR primer
sequences. |
Table I.
Quantitative PCR primer
sequences.
|
| Forward primer
sequence | Reverse primer
sequence |
|---|
| H19 |
5′-GCCTTGACGTGCTGGATCT-3′ |
5′-TCCGATGCTTTACTCAAGAAGTT-3′ |
| Internal control
U6 |
5′-GACGGACACCCTCACTACTG-3′ |
5′-GACGTTCATGATTCAAGCATGC-3′ |
| miR-203 |
5′-TGCTCTAGAGGCGTCTAAGGCGTCCG-3′ |
5′-CCCAAGCTTCACCTCCCAGCAGCACTTG-3′ |
After transfection with LipofectamineTM2000 plasmid
containing over-expressed H19, A549 cells were collected. cDNA
synthesis was performed with the one-step method using commercial
reverse transcription kit (Fermentas, no. K1633; Wuhan Boster
Biological Technology, Ltd., Wuhan, China). qPCR amplification and
analysis was performed using ABI 7500 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The PCR condition was: 95°C for 10
min, 40 cycles of 95°C for 15 sec, 60°C for 1 min and finally an
elongation step at 72°C for 30 sec. U6 small RNA and β-actin was
chosen as a loading control for normalization and for
quantification of miR-203 and H19 expression.
CCK-8 proliferation assay
A549 cells in each group were collected after
transfection and seeded in 96-well plates at a density of
1×103 cells. A final concentration of 10% of CCK-8
reagent (Wuhan Biological Co., Ltd., Wuhan, China) was added into
cells at 24, 48 and 72 h incubation time. Wavelength value was
detected at 450 nm absorbance using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Cell invasion assay
A549 cells in each group were collected after
transfection and cultured with serum-free RPMI-1640 medium. Cells
was re-suspended and added to the pre-paved Matrigel Transwell
chamber (Corning Incorporated, Corning, NY, USA) at a density of
5×104 per well. The lower chamber contained 600 ml
complete medium. Normal culture was incubated and removed from the
upper chamber following 24 h incubation. Cells ware collected and
fixed with 4% paraformaldehyde. Afterwards, Cells were treated with
0.01% crystal violet staining to determine the cell count. Cell
numbers in the lower chamber were counted from 5 random selected
fields at ×200 optical microscope. Each experiment was repeated
three times.
Western blot analysis
Cells were added with an appropriate amount of RIPA
lysis to obtain total cellular protein solution for protein
extraction. After treatment with an appropriate amount of SDS
buffer at 100°C water bath solution, protein samples underwent
polyacrylamide gel electrophoresis and transferred to PVDF
membranes followed by incubation of primary antibody and secondary
antibody (Abcam, Cambridge, UK). Afterwards, the results were
analyzed using an enhanced chemiluminescence kit (Cell Signaling
Technology, Inc., Danvers, MA, USA) according to the manufacturer's
protocols. GAPDH was used for normalization and quantification of
protein. Quantification of bands was analyzed using the ImageJ
software (National Institutes of Health, Bethesda, MD, USA).
Cell transfection
NSCLC cells were seeded into 6-well plates for 24 h
and grew to 70% confluence for transfection. The LipofecamineTM2000
mixed with H19 mimic, inhibition or negative control and miR-203
mimic, inhibition or negative control (final concentration of 100
nmol/l) were added into cells after cells were incubated for 6 h
and cultured in RPMI-1640 for 24 h. Afterwards, cells were
collected for subsequent experiments.
Luciferase reporter gene assay
A density of 1×105 cells were seeded in
24-well plates until they reached 70% confluence. Cells were
co-transfected with the LipofectamineTM2000 plasmid which contained
the luciferase promoter H19 with overexpressed plasmids. Cells that
were cultured for 48 h were measured and analyzed in accordance
with the dual luciferase reporter gene assay kit (Promega
Corporation, Madison, WI, USA).
Online prediction of potential
target
The miRanda (http://www.microRNA.org) (17), PicTar (https://pictar.mdc-berlin.de/) (18), TargetScan (http://www.targetscan.org) (19) online prediction tools were used to
predict miRNA target genes separately based on previous reports
(20–22). Generally, the default settings were
adopted according to instructions unless otherwise specified.
Taking miRanda for an example, the homo sapiens parameter was
selected. Other important parameters were set to individual values,
e.g. −8.0 for Gap Open Penalty, −2.0 for Gap Extend, 50.0 for Score
Threshold, −20.0 kcal/mol for Energy Threshold, and 2.0 for Scaling
Parameter. For all three software the average minimum free energy
change was measured and used as the reference. Target selection
were based on the following criteria: i) Potential target sites
were determined by at least two different approaches; ii) target
sites must be located in accessible regions and iii) multiple
target sites were prioritized (23).
Statistical analysis
Statistical analysis was performed using SPSS v.12.0
statistical software (SPSS Inc., Chicago, IL, USA). Experimental
data was presented as Mean ± SEM (stand error of mean) or in
percentages (%). Comparison between different experimental groups
was performed with the two-tailed student's T test and multiple
group comparisons used one-way ANOVA followed by Tukey's post hoc
test. Log-rank regression was used to analyze the overall survival
curve. P<0.05 was considered to indicate a statistically
significant difference.
Results
Overexpression of H19 in NSCLC tissues
and cells
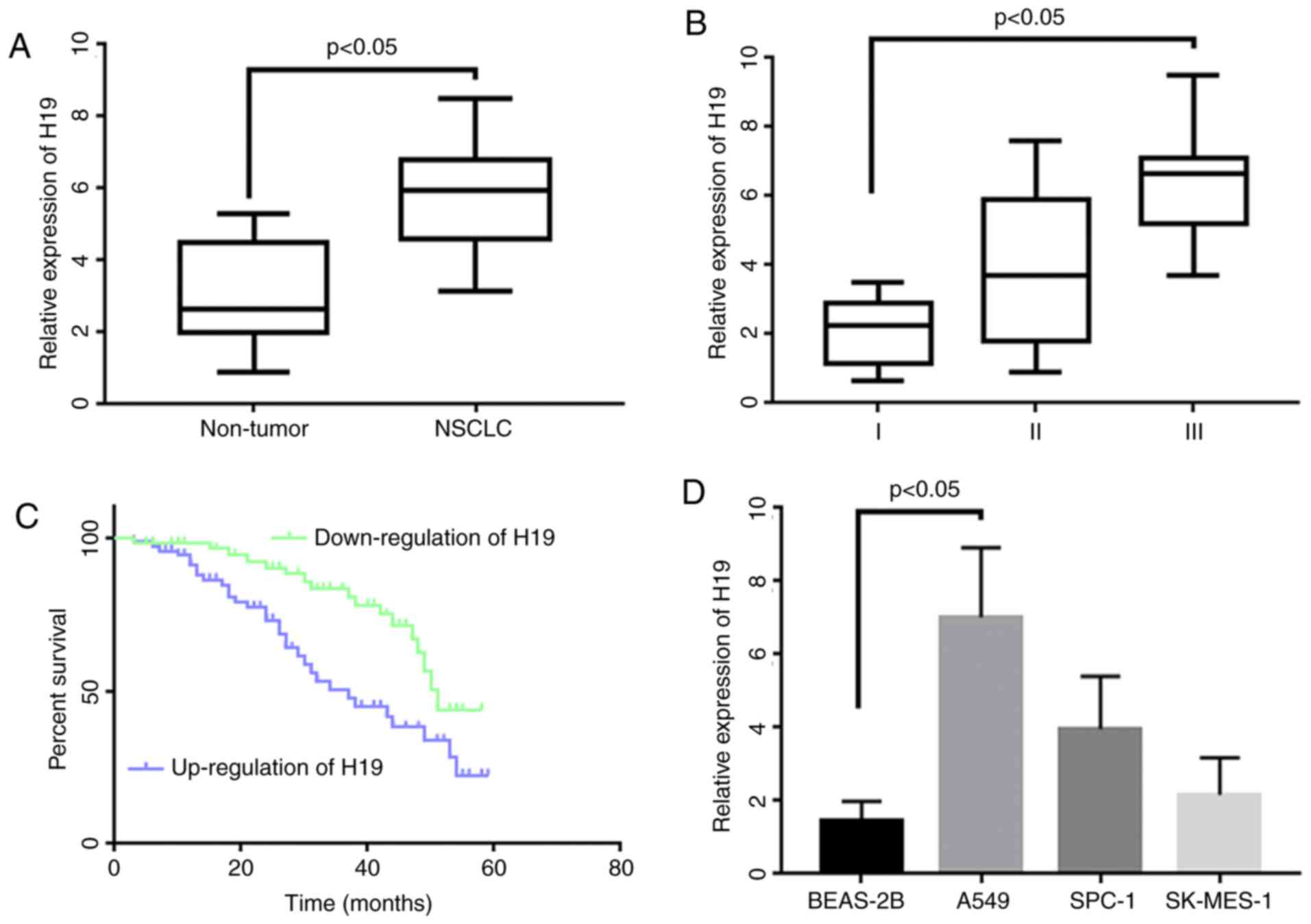
The expression of H19 was significantly increased in
lung cancer tissues compared to adjacent tissues (Fig. 1A). Based on the relative expression
level of H19, patients were divided into a high expression
(tumor/control ≥1.5) and a low expression group (tumor/control
<1.5). It was found that patients in the higher expression group
presented with advanced clinical stages in comparison to the lower
expression group (Table II and
Fig. 1B). The overall survival time
of patients in the higher expression group was also shorter
compared to the lower expression group (Fig. 1C). In addition, the study examined the
H19 expression levels in different NSCLC cells. The results showed
that H19 in A549, SPC-A1 and SK-MES-1 cells increased by 4.2-fold,
3.4-fold and 2.6-fold in contrast to the normal bronchial
epithetical cell line, BEAS (Fig.
1D).
 | Table II.Association of clinicalpathological
features and H19 expression in NSCLC patients. |
Table II.
Association of clinicalpathological
features and H19 expression in NSCLC patients.
|
|
| H19 expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | N=76 | Low | % | High | % | P-value |
|---|
| Age (years) |
| 41 |
| 35 |
| 0.692 |
|
<60 | 28 | 15 | 36.59 | 13 | 37.14 |
|
|
≥60 | 48 | 26 | 63.41 | 22 | 62.86 |
|
| Sex |
|
|
|
|
| 0.577 |
|
Male | 32 | 17 | 41.46 | 15 | 42.86 |
|
|
Female | 44 | 24 | 58.54 | 20 | 57.14 |
|
| Smoking |
|
|
|
|
| 0.107 |
|
Yes | 21 | 11 | 26.83 | 10 | 28.57 |
|
| No | 55 | 30 | 73.17 | 25 | 71.43 |
|
| TNM |
|
|
|
|
| 0.024 |
| I | 42 | 26 | 63.41 | 16 | 45.71 |
|
| II | 24 | 11 | 26.83 | 13 | 31.71 |
|
|
III | 10 | 4 | 9.76 | 6 | 22.58 |
|
| Tumor size
(cm) |
|
|
|
|
| 0.085 |
|
<5 | 49 | 25 | 60.98 | 24 | 68.57 |
|
| ≥5 | 27 | 16 | 39.02 | 11 | 31.43 |
|
| Pathological
histology |
|
|
|
|
| 0.142 |
|
Squamous cell carcinoma | 47 | 26 | 63.41 | 21 | 60 |
|
|
Adenocarcinoma | 19 | 8 | 19.51 | 11 | 26.83 |
|
| Large
cell carcinoma | 10 | 7 | 17.07 | 3 | 13.17 |
|
| Survival time
(months) |
|
|
|
|
|
|
|
<24 | 45 | 20 | 48.78 | 25 | 71.43 | 0.021 |
|
≥24 | 31 | 21 | 51.22 | 10 | 28.57 |
|
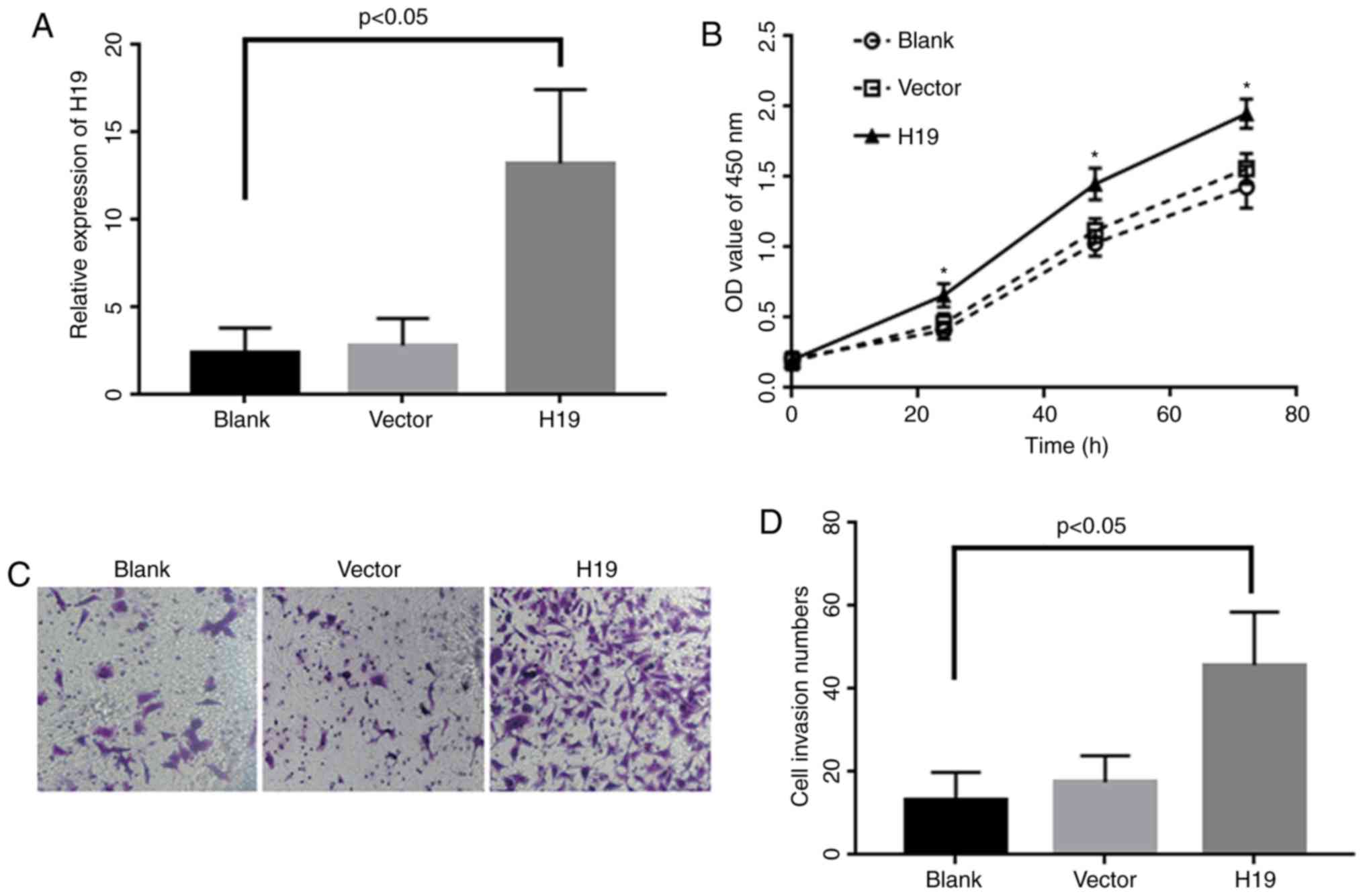
Overexpression of H19 can promote
proliferation and EMT process in A549
Based on the expression results, A549 cell line was
chosen for another functional study. Cells were transfected with
H19 plasmid and validated by quantitative real time polymerase
chain reaction (qRT-PCR). The results showed that the expression
level of H19 in transfected cells increased more than 20-fold than
control cells (P<0.05; Fig. 2A).
Cell proliferation results indicated that cell proliferation became
significantly enhanced in H19 over expressed cells in contrast to
the control group following 72 h incubation period (Fig. 2B). The invasion results demonstrated
that the number of crossing members were significantly increased in
H19 overexpression group when compared to negative control group
(P<0.05; Fig. 2C and D).
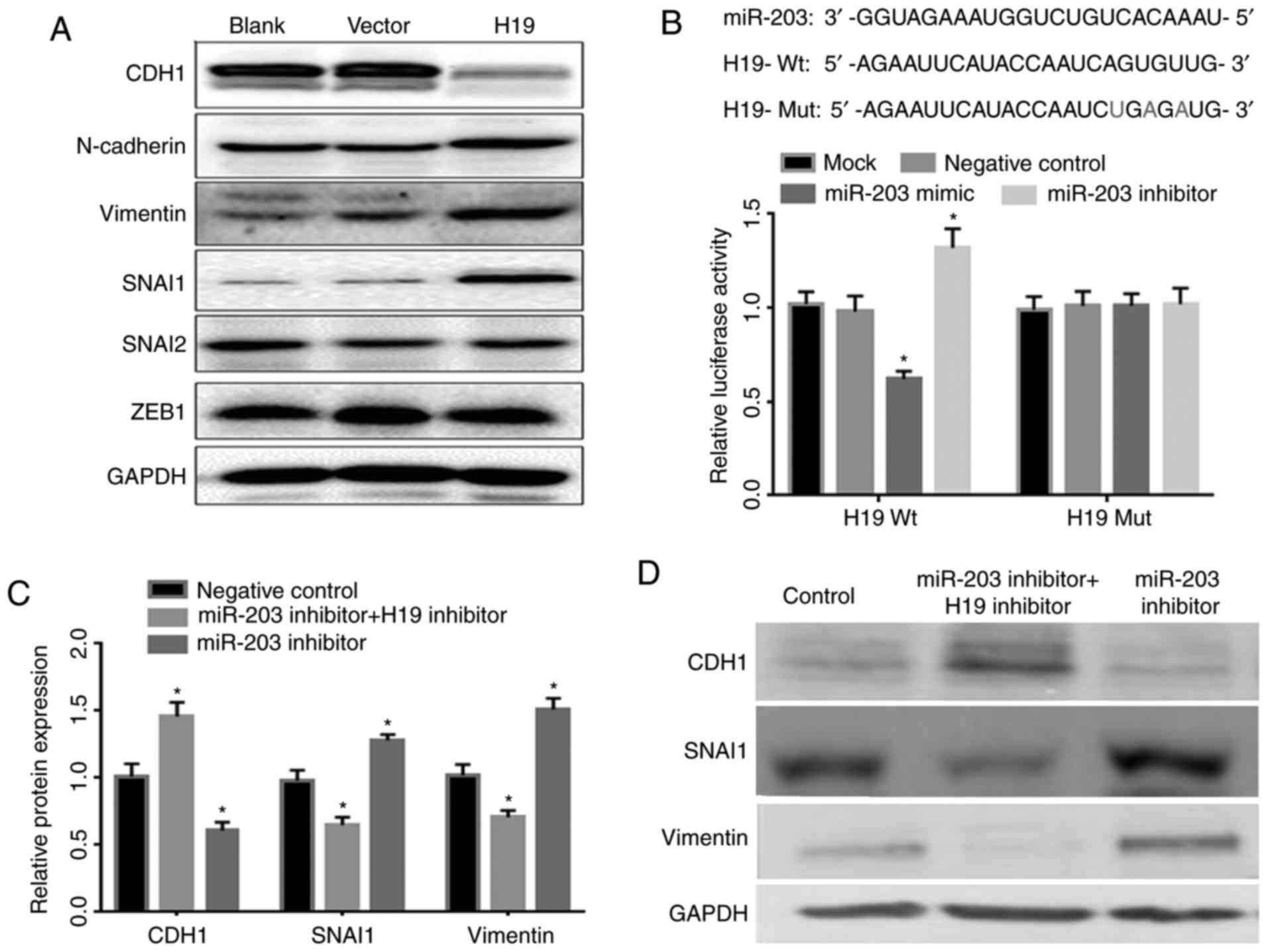
Association of miR-203 with H19 and
EMT expression
EMT is a well-documented event in the progression of
tumor invasion. Afterwards, western blot assays were used to detect
the expression of EMT markers (CDH1, N-cadherin and Vimentin,
SNAI1, SNAI2, and ZEB1) between H19 overexpressed cells and control
cells. Western blot testing found that epithelial marker CDH1 was
significantly reduced after H19 overexpression. In contrast, the
expressions of mesenchymal markers which included SNAI1 and
Vimentin, were increased (Fig.
3A).
Accumulative studies have revealed that lncRNA H19
mediate EMT by functioning as a sponge for the miRNAs in a variety
of cancers (24–27). Recently, miR-203 was reported to play
a crucial role in the EMT core network, which functions as a switch
controlling epithelial cell plasticity during cancer progression
(28). We hypothesized that H19 could
possibly promote EMT by acting as a miRNA sponge and hijack miR-203
since miR-203 is known to attenuate EMT in many types of cancers.
In order to search for potential targets of miR-203, three online
prediction tools were used such as, miRanda, PicTar and TargetScan,
and we identified putative miR-203 binding site in 3′-UTR of H19 as
a potential target. We also referred to the previously predicted
miRNA targets for H19 by others, e.g. RGD (https://rgd.mcw.edu/) (29) which results are in agreement with our
prediction.
Afterwards, a luciferase reporter was constructed
containing wild type and mutant H19 3′-UTR binding sequence
(Fig. 3B). After transfection,
miR-203 overexpressed plasmid and H19 gene promoter were detected
by luciferase activity and the results are shown in Fig. 3B. Additionally, the luciferase
activity was significantly inhibited, as its expression decreased
approximately by 60%. miR-203 inhibitor significantly increased the
luciferase activity of H19 in compared with the control cells
(P<0.05). Furthermore, we observed that miR-203 inhibitor
suppressed the expression of epithelial marker CDH1 and promoted
the expression of mesenchymal markers, SNAI1 and Vimentin. However,
co-transfection of miR-203 inhibitor and H19 inhibitor reversed the
effect (Fig. 3C and D).
Down-regulation of miR-203 in tissue
samples and NSCLC cell lines
As shown in Fig. 4A, a
decreased expression of miR-203 was observed in NSCLC tissue
samples compared with paired normal tissue samples (Fig. 4A). Further analysis revealed a
negative association of miR-203 expression level and TNM stage
(Fig. 4B). Next, we determined the
miR-203 expression levels in three NSCLC cell lines (A549, SPC-A1,
SKMES-1) and in the normal bronchial epithelial cell line BEAS-2B.
Our results found that miR-203 was significantly down-regulated in
all cell lines compared to BEAS-2B (Fig.
4C). Based on the observed negative relation between H19 and
miR-203, our results suggested an association of miR-203 expression
with H19 expression in NSCLC samples (Fig. 4D).
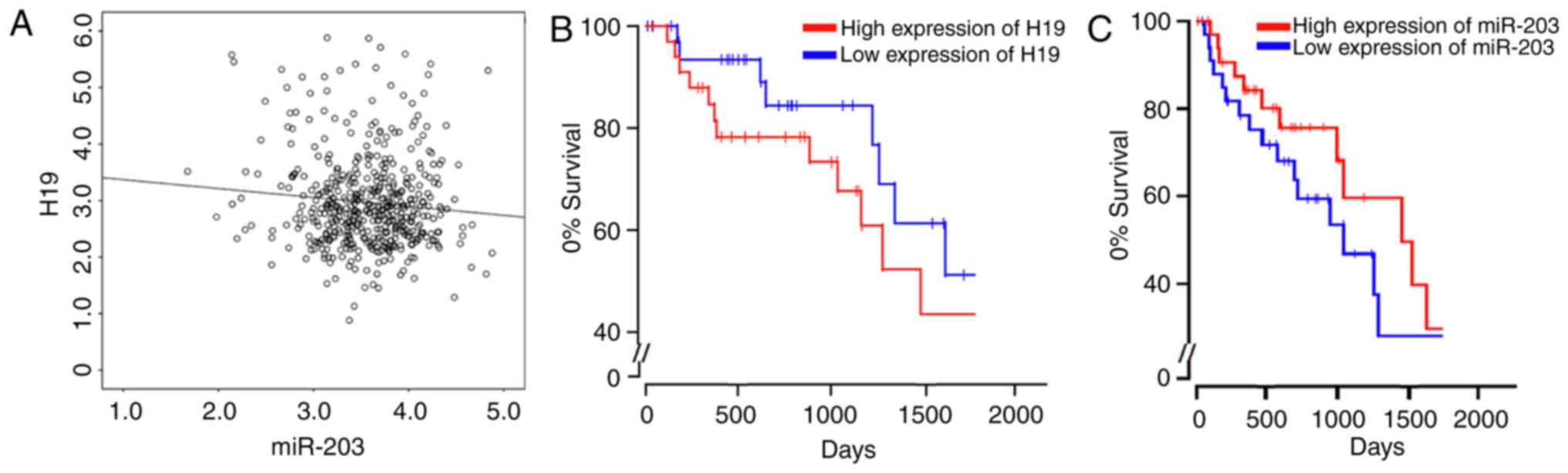
Given the limited number of samples, we also turned
to the public dataset of TCGA (https://cancergenome.nih.gov/) and studied the
expression profiles of both H19 and miR-203 in lung adenocarcinoma
(LUAD), one largest subtype of NSCLC which accounts for 40% of all
total lung cancer cases (30). Our
results from analysis are consistent with the above findings,
indicating a negative correlation between the expressions of H19
and miR-203, which was proved with statistical significance
(P<0.05; Fig. 5A). Moreover, we
also determined the Kaplan-Meier plots for both high and low
expressions of H19 and miR-203. It was shown that high expression
of H19 results in the significant reduction of cumulative survival
among LUAD patients (Fig. 5B), while
high expression of miR-203 largely improves the cumulative survival
(Fig. 5C). All these results provide
a convincing support for the current work.
Discussion
With the completion of the Human Genome Project, it
was discovered that in addition to approximately 20,000
protein-coding genes that account for 2% of the whole genome, there
are a large number of non-coding RNAs (ncRNAs) containing microRNA
and lncRNA.16 Also, ncRNAs are reported to participate in a variety
of functional cell processes on multiple levels including protein
translation and protein degradation. Despite the evidence
supporting the function of ncRNA in carcinogenesis, their role and
potential application in lung cancer is still in its early stages
(31–34).
As a transcriptional regulatory lncRNA, H19 was
originally found in embryonic development related research
(12). In recent years, a series of
studies have found that H19 abnormally over-expressed in many
tumors, such as bladder cancer (35),
gastric cancer (36), hepatocellular
carcinoma (37), and NSCLC (38,39). Also,
an in-vitro study revealed that H19 overexpression promotes
the proliferation of tumor, increases migration and invasion of
tumor cells and reduces sensitivity to chemotherapy, thus
suggesting an important role of H19 in tumorigenesis and cancer
development (40). However, the
expression of H19 and its function in NSCLC is not yet clear.
The results of our study indicated that compared
with normal tissues, H19 was significantly up regulated in NSCLC
tissues, suggesting its oncogenic role in NSCLC cancer occurrence
and development. Further analysis demonstrated that H19
over-expression is closely associated with an advanced clinical
stage and an aggressive lymph node metastasis in patients,
suggesting that a high expression of H19 may be involved in
malignant lung cancer proliferation. Overall survival analysis
found that high expression is related to poor prognosis in
patients, suggesting a potential application of H19 as a prognostic
biomarker in NSCLC.
Our results showed that overexpression of H19
increased cell numbers of crossing basement members which suggested
an enhanced cell invasiveness ability. The present study also found
that A549 cells following an overexpression of H19 demonstrated
significant morphological changes with a shuttle-shaped oval
deformation growth, increased pseudopodia variable length and a
larger cell gap. All these changes indicated involvement of the EMT
process. It is well known that EMT is a complex and an important
biological process that participates in a variety of
pathophysiological processes including cancer invasion and
transformation (41,42). When tumor cells undergo EMT, they lose
their polarity and increase migration and invasion ability.
Therefore, we analyzed the changes of EMT related proteins to
confirm whether EMT stimulates the effect of H19 on cell invasion.
The results showed a decreased expression of epithelial marker CDH1
and increased expression of mesenchymal markers, SNAI1 and Vimentin
along with an over expression of H19 suggesting a mediatory role of
EMT in enhanced cell invasion.
Previous studies provided evidence of a decreased
expression of miR-203 in a variety of lung cancer tissue samples in
contrast to the expression of normal human bronchial epithelial
cells or normal lung tissues, respectively (43,44). Also,
reduced expression of miR-203 was found to be associated with
metastatic tumors (45).
Additionally, over-expression of miR-203 in various cancer cell
lines led to inhibition of EMT processes, such as cell
proliferation, migration, invasion and tumor metastasis (46). Furthermore, previous studies
demonstrated the association of H19 with miRNA ribonucleoprotein
complexes, thus acting as a natural molecular sponge for various
miRNAs (47–49). Also, studies showed lncRNAs have the
ability to modulate downstream targets of miRNA by acting as a
decoy to sequester miRNAs (50–52). Given
that EMT is attenuated by miR-203 in many cancer types, we
hypothesized that H19 acts as a miRNA sponge and hijacks miR-203
hence promotes EMT in the process.
Finding of our study revealed that H19 expression
was elevated in NSCLC tissues along with a decreased miR-203
expression level. It was also found that patients in advanced
clinical stages had a higher H19 and a lower miR-203 expression
compared normal tissues. The overall survival time of patients in
higher H19 expression group was shorter in contrast to the lower
H19 expression group. Up-regulation of H19 enhanced cell
proliferation and promoted invasion. Over expression of H19
stimulated the EMT process in lung cancer cells and presented with
typical morphological characteristics of EMT. The level of
mesenchymal marker proteins such as Vimentin and SNAI1 increased;
while CDH1 protein level decreased. Also, H19 negatively regulated
miR-203. Inhibition of H19 attenuated miR-203 induced EMT process.
Our study also reported a negative correlation between miR-203 and
H19 expression level in tissue samples and cell lines. In addition,
a decreased miR-203 expression was associated with the TNM stage.
Furthermore, inhibition of H19 significantly reversed the EMT
promotion effect of miR-203 on lung cancer cells through
down-regulation of epithelial markers and an up-regulation of
mesenchymal markers. In short, H19 may promote invasion in NSCLC by
influencing the EMT process, which can affect regulation of
specific miRNAs.
In summary, H19 overexpression can induce the
occurrence of EMT and promote invasion in lung cancer cells. The
direct binding effect of miR-203 and H19 on EMT process suggested
that miRNA-lncRNA acts as a potential regulatory network in
H19-mediated EMT changes. Therefore, our study provides evidence
that H19, which is involved in the tumorigenesis of NSCLC, can
serve as a potential target for new drugs and clinical
biomarkers.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Nature
Science Foundation of China (grant no. 81560487).
Availability of data and materials
The data used and analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
XJG contributed to the present study design and
performed the majority of the experiments. LMZ and ZXF contributed
to the data analysis and data interpretation. LL, YJZ, MYL and JYJ
contributed to collection of the clinical samples.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Zunyi Medical University. Informed consent was
obtained from each patient prior to surgery. All experimental
procedures were carried out in accordance with the approved
guidelines and were in agreement with the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rosell R and Karachaliou N: Lung cancer in
2014: Optimizing lung cancer treatment approaches. Nat Rev Clin
Oncol. 12:75–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pérez-Soler R: Individualized therapy in
non-small-cell lung cancer: Future versus current clinical
practice. Oncogene. 28 Suppl 1:S38–S45. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNA in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brannan CI, Dees EC, Ingram RS and
Tilghman SM: The product of the H19 gene may function as an RNA.
Mol Cell Biol. 10:28–36. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vennin C, Spruyt N, Dahmani F, Julien S,
Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X and
Adriaenssens E: H19 non coding RNA-derived miR-675 enhances
tumorigenesis and metastasis of breast cancer cells by
downregulating c-Cbl and Cbl-b. Oncotarget. 6:29209–29223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matouk IJ, Halle D, Gilon M and Hochberg
A: The non-coding RNAs of the H19-IGF2 imprinted loci: A focus on
biological roles and therapeutic potential in Lung Cancer. J Transl
Med. 13:1132015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hariharan M, Scaria V, Pillai B and
Brahmachari SK: Targets for human encoded microRNAs in HIV genes.
Biochem Biophys Res Commun. 337:1214–1218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grün D, Wang YL, Langenberger D, Gunsalus
KC and Rajewsky N: microRNA target predictions across seven
Drosophila species and comparison to mammalian targets. PLoS Comput
Biol. 1:e132005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reid JG, Nagaraja AK, Lynn FC, Drabek RB,
Muzny DM, Shaw CA, Weiss MK, Naghavi AO, Khan M, Zhu H, et al:
Mouse let-7 miRNA populations exhibit RNA editing that is
constrained in the 5′-seed/cleavage/anchor regions and stabilize
predicted mmu-let-7a:mRNA duplexes. Genome Res. 18:1571–1581. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan WL, Huang HD and Chang JG: lncRNAMap:
A map of putative regulatory functions in the long non-coding
transcriptome. Comput Biol Chem. 50:41–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal. 10:pii:
eaak9557. 2017. View Article : Google Scholar
|
|
25
|
Zhang Q, Li X, Li X, Li X and Chen Z:
LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by
targeting miR-484 in human lung cancer cells. J Cell Biochem.
119:4447–4457. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b-3p as competing
endogenous RNA. Biochim Biophys Acta. 1864:1887–1899. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moes M, Le Béchec A, Crespo I, Laurini C,
Halavatyi A, Vetter G, Del Sol A and Friederich E: A novel network
integrating a miRNA-203/SNAI1 feedback loop which regulates
epithelial to mesenchymal transition. PLoS One. 7:e354402012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shimoyama M, De Pons J, Hayman GT,
Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang
SJ, et al: The Rat Genome Database 2015: Genomic, phenotypic and
environmental variations and disease. Nucleic Acids Res.
43:(Database Issue). D743–D750. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang JT, Lee YM and Huang RS: The impact
of the Cancer Genome Atlas on lung cancer. Transl Res. 166:568–585.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao Y, Gao F, Ma JL, Sun WZ and Song LP:
The potential clinical applications and prospects of microRNAs in
lung cancer. Onco Targets Ther. 7:901–906. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ricciuti B, Mecca C, Crinò L, Baglivo S,
Cenci M and Metro G: Non-coding RNAs in lung cancer. Oncoscience.
1:674–705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ariel I, Sughayer M, Fellig Y, Pizov G,
Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T, Tykocinski ML, et
al: The imprinted H19 gene is a marker of early recurrence in human
bladder carcinoma. Mol Pathol. 53:320–323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang E, Li W, Yin D, De W, Zhu L, Sun S
and Han L: c-Myc-regulated long non-coding RNA H19 indicates a poor
prognosis and affects cell proliferation in non-small-cell lung
cancer. Tumour Biol. 37:4007–4015. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui JD, Mo J, Luo M, Yu Q, Zhou S, Li T,
Zhang Y and Luo W: c-Myc-activated long non-coding RNA H19
downregulates miR-107 and promotes cell cycle progression of
non-small cell lung cancer. Int J Clin Exp Pathol. 8:12400–12409.
2015.PubMed/NCBI
|
|
40
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin CW, Lin PY and Yang PC: Noncoding RNAs
in tumor epithelial-to-mesenchymal transition. Stem Cells Int.
2016:27327052016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Heerboth S, Housman G, Leary M, Longacre
M, Byler S, Lapinska K, Willbanks A and Sarkar S: EMT and tumor
metastasis. Clin Transl Med. 4:62015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin J, Deng J, Wang F, Xia X, Qiu T, Lu W,
Li X, Zhang H, Gu X, Liu Y, et al: The expression and function of
microRNA-203 in lung cancer. Tumour Biol. 34:349–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tang R, Zhong T, Dang Y, Zhang X, Li P and
Chen G: Association between downexpression of MiR-203 and poor
prognosis in non-small cell lung cancer patients. Clin Transl
Oncol. 18:360–368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Funamizu N, Lacy CR, Kamada M, Yanaga K
and Manome Y: MicroRNA-203 induces apoptosis by upregulating Puma
expression in colon and lung cancer cells. Int J Oncol.
47:1981–1986. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ding X, Park SI, McCauley LK and Wang CY:
Signaling between transforming growth factor β (TGF-β) and
transcription factor SNAI2 represses expression of microRNA miR-203
to promote epithelial-mesenchymal transition and tumor metastasis.
J Biol Chem. 288:10241–10253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The interaction between MiR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhuang M, Gao W, Xu J, Wang P and Shu Y:
The long non-coding RNA H19-derived miR-675 modulates human gastric
cancer cell proliferation by targeting tumor suppressor RUNX1.
Biochem Biophys Res Commun. 448:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Imig J, Brunschweiger A, Brümmer A,
Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M
and Hall J: miR-CLIP capture of a miRNA targetome uncovers a
lincRNA H19-miR-106a interaction. Nat Chem Biol. 11:107–114. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cai X and Cullen BR: The imprinted H19
noncoding RNA is a primary microRNA precursor. RNA. 13:313–316.
2007. View Article : Google Scholar : PubMed/NCBI
|