Introduction
The promyelocytic leukemia zinc finger (PLZF)
protein is a transcription factor belonging to the Krüppel-like
zinc finger family (1). It regulates
a variety of developmental and physiological pathways, and has been
demonstrated to be involved in leukocyte differentiation and
oncogenic transformation (2,3). The potency and self-renewal abilities of
many stem and early progenitor cells have also been linked to PLZF,
which may maintain these phenotypic characteristics in certain cell
lineages (4,5). Since the identification of the PLZF/RARα
fusion protein in 1998 (6), several
studies have linked the level of PLZF expression with the oncogenic
transformation of numerous types of cancer cells (7,8). Under
normal physiological conditions, PLZF expression is minimally
maintained to regulate both the cell cycle and cell differentiation
potential through several accessory molecules, including cyclin A2,
checkpoint suppressor 1, c-Myc and telomerase reverse transcriptase
(5,9,10). This
low-level expression of PLZF may be further reduced to relieve the
PLZF transcriptional control in cells, which ultimately enables
tumorigenic cell transformation (11). In recent years, PLZF expression has
also been implicated in the innate immune response. PLZF expression
was demonstrated to control the clonal expansion and functional
capacity of cluster of differentiation (CD)1d-restricted natural
killer T cells (12), the induction
of memory-like or innate CD8+ T cells (13) and, most notably, the positive
regulation of a specific subset of interferon-stimulated genes
(ISGs) in the interferon pathway (14). This complex and intriguing
immunomodulatory function of PLZF is achieved through the
stabilization of a histone deacetylase corepressor complex
comprising HDAC3 and the p50 subunit of nuclear factor-κB (15).
The 2–5A system is a well-characterized pathway that
stimulates apoptosis when activated in cells. The system is
triggered by the accumulation of double-stranded RNA in the
cytoplasm caused by a viral infection or non-specific degradation.
This, in turn, activates 2′-5′-oligoadenylate synthetase 1, an
enzyme that converts ATP to short 2′-5′-linked oligoadenylates,
known as 2–5A. 2–5A binds and activates the ribonuclease L enzyme
(RNase L), which then cleaves single-stranded RNA, eventually
leading to the degradation of ribosomal RNA (rRNA) and apoptosis
(16). In a number of cells, the
levels of RNase L are kept under tight spatial and temporal control
during transcriptional and post-transcriptional events. This
ensures that RNase L activity does not interfere with physiological
cellular proliferation (17). RNase L
and PLZF control viral pathogenesis through the induction of
specific ISGs, which, when activated, stimulate various cellular
proteins, including protein kinase R, interferon regulatory factor
7, and signal transducer and activator of transcription 3, to limit
the synthesis of viral DNA and to promote the activation of
apoptotic pathways in virally infected cells (18,19).
The ATP-binding cassette subfamily E member 1
(ABCE1) protein is a member of the ATP-binding cassette (ABC)
transporter family, and was originally described as an RNase L
inhibitor (20), as well as a
PLZF-targeted gene (21). ABCE1
expression was observed to be higher in lung cancer (22), retinoblastoma (23), melanoma (24) and colon cancer (25), compared with in normal cells, thereby
demonstrating an opposite pattern to the expression signature of
PLZF in cancer and in normal cells. Furthermore, the relatively
high expression level of ABCE1 was implicated in the ongoing
survival of these cancerous cells.
The aim of the present study was to explore the
functional association between PLZF and RNase L. Therefore, the
present study investigated whether ABCE1 inhibition, possibly
through PLZF regulation, may induce the anti-proliferative effect
of RNase L in a breast cancer cell line.
Materials and methods
Oncomine database
In order to evaluate the potential difference in
PLZF expression between breast cancer and normal breast tissues,
transcriptomic microarray data (reporter ID: 11-113531123,
ILMN_1750496 and A_23_P104802) from the Oncomine cancer microarray
database (www.oncomine.org) was utilized (26,27). PLZF
was added to the search inquiry, breast cancer was selected as the
cancer type and the filter ‘Cancer vs. Normal Analysis’ was
applied, using the database threshold values of odds ratio >2.0
and P<0.0001. The PLZF mRNA copy number was analyzed in the
following: 1,992 breast carcinoma and 144 normal breast samples
from the Curtis dataset; 532 invasive breast carcinoma, 61 paired
normal breast tissue and 3 paired metastatic samples from the TCGA
dataset; and 786 invasive breast carcinoma, 702 paired
blood-derived normal, 111 paired normal breast tissue and 3 paired
metastatic samples from the TCGA molecular dataset. Table I demonstrates the number of each of
the assessed tissue types observed in the TCGA and Curtis datasets.
The datasets were classified into two main types: Normal tissue vs.
different cancer types or based on the molecular classification of
the tissue samples types [expression status of progesterone (PR),
estrogen (ER) and human epidermal growth factor receptor 2 (HER2)].
The expression data were log-transformed and median-centered per
array, and the standard deviation was normalized to one per array.
Array data were used following adjustment of the threshold to those
genes with a fold-change of 2 and a mean value with a significance
level of P<0.0001, using GraphPad Prism Version 7.00 for Windows
(GraphPad Software, Inc., La Jolla, CA, USA).
 | Table I.Detailed numbers of samples for each
of the TCGA and Curtis datasets obtained from the Oncomine cancer
microarray database. |
Table I.
Detailed numbers of samples for each
of the TCGA and Curtis datasets obtained from the Oncomine cancer
microarray database.
| Array | Cells | No. |
|---|
| TCGA type | Normal tissue | 61 |
|
| Invasive breast
carcinoma | 75 |
|
| Mixed lobular +
ductal | 7 |
|
| Invasive ductal
carcinoma | 392 |
|
| Invasive lobular
carcinoma | 36 |
| TCGA molecular | Normal tissue | 61 |
|
| ERBB2 +ve | 73 |
|
| ERBB2 -ve | 228 |
|
| PR -ve | 144 |
|
| PR +ve | 228 |
|
| ER -ve | 94 |
|
| ER +ve | 274 |
|
| Triple -ve | 49 |
| Curtis type | Normal tissue | 144 |
|
| Ductal BRCa in
situ | 10 |
|
| Invasive breast
carcinoma | 21 |
|
| Mixed lobular +
ductal | 89 |
|
| Invasive ductal
carcinoma | 1,556 |
|
| Invasive lobular
carcinoma | 148 |
| Curtis
molecular | Normal tissue | 144 |
|
| ERBB2 +ve | 1743 |
|
| ERBB2 -ve | 249 |
|
| PR -ve | 943 |
|
| PR +ve | 1,049 |
|
| ER -ve | 440 |
|
| ER +ve | 1,552 |
|
| Triple -ve | 250 |
Survival analysis
The hazard ratios for the expression levels of PLZF
and ABCE1 in patients with breast cancer were estimated using the
Kaplan-Meier method, and the log-rank test was used to compare the
survival curves of patients grouped according to median PLZF and
ABCE1 expression levels (low-medium and high-medium expression).
Patient data were obtained from the Kaplan-Meier database, a
comprehensive database containing microarray datasets from the
Curtis and TCGA projects, among others. The distant metastasis-free
survival (DMFS) and relapse-free survival (RFS) rates were
determined as previously described (28). The number of patient samples used to
determine DMFS was 1,769, and the number of samples used to
determine RFS was 3,955. The patient samples included all breast
cancer molecular subtypes, including ER-, PR- and
HER2-positive.
Cell culture
HeLa, 293T, MDA-MB-231, MCF7, MCF10 and MCF12 cells
were obtained from the American Type Culture Collection (Manassas,
VA, USA). The 293, HeLa, MDA-MB-231 and MCF7 cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 4.5
g/l D-glucose, 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), 2 mM L-Glutamine (Sigma-Aldrich; Merck KGaA)
and 100 U/ml penicillin/streptomycin (Sigma-Aldrich; Merck KGaA) in
5% CO2 with 95% air at 37°C. The MCF10 and MCF12 cell
lines were maintained in DMEM/F12 (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum, 1% L-glutamine
(Sigma-Aldrich; Merck KGaA), growth factors from the HuMEC
Supplement kit (Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin under the same conditions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from 293T, MCF-10, MCF-12, MCF7
and MDA-MB-231 cell lines by cell lysis using TRIzol (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Complementary DNA (cDNA) was synthesized using the
SuperScript® III First-Strand Synthesis system (cat no.
18080-051; Invitrogen; Thermo Fisher Scientific, Inc.) by
incubating 5 µg total extracted RNA at 65°C for 5 min with the 10
mM dNTP mixture and Oligo (dT) primers. The cDNA synthesis mix
containing 25 mM MgCl2, 10× RT buffer, 0.1 M DTT and SuperScript
III Reverse Transcriptase enzyme was added to the RNA mix and
incubated at 50°C for 50 min and the reaction was terminated by
heating at 85°C for 5 min. RT-qPCR was performed in triplicate
using a Bio-Rad CFX96™ Real-Time system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) with oligonucleotides for PLZF forward,
5′-AACCACAAGGCTGACGCTGTA-3′ and reverse,
5′-CATAGGTGCTGAAGTCCATGGA-3′; ABCE1 forward,
5′-TTGGTTGTGGGAAGTCGT-3′ and reverse, 5′-GCTTATGTAGTTAATGGGAGGT-3′;
HuR forward, 5′-GAGGCTCCAGTCAAAAACCA-3′ and reverse,
5′-GTTGGCGTCTTTGATCACCT-3′; TTP forward, 5′-CGCTACAAGACTGAGCTAT-3′
and reverse, 5′-GAGGTAGAACTTGTGACAGA-3′. The thermocycler
conditions for the PCR were as follows: 55°C for 2 min, then 95°C
for 10 min, 95°C for 15 sec and 60°C for 1 min for a total of 45
cycles. The levels of target mRNA were normalized to that of 18S
(reference ID: Hs03003631_g1; Thermo Fisher Scientific, Inc.) and
were charted using the 2−ΔΔCq method (29). Results are expressed as the relative
gene expression for each of the target genes, and are presented as
the mean ± standard deviation (SD).
Western blot analysis
In order to assess protein levels, cells were lysed
in radioimmunoprecipitation assay buffer [150 mM NaCl, 50 mM Tris
(pH 8.0), 1.5 mM EDTA, 0.5% Triton-X and 5% glycerol] containing
0.2 M phenylmethylsulfonyl fluoride protease inhibitor (Roche
Diagnostics, Basel, Switzerland). Protein quantification using
Bradford protein assay (cat no. 500-0006; Bio-Rad Laboratories,
Inc., Hercules, CA, USA) was performed as per the manufacture's
recommendations by employing the Ultrospec 3100 pro UV/visible
spectrophotometer Cell lysates for ~30 µg of total protein were
resolved on a 10% gel, blocked with 5% milk in 1X PBS at 37°C for 1
h, transferred to a nitrocellulose membrane (GenHunter Corporation,
Nashville, TN, USA), immunoblotted with primary antibodies (PLZF,
cat no. sc-22839; Santa Cruz Biotechnology Inc., Dallas, TX, USA;
β-actin, cat no. sc-69879; Santa Cruz Biotechnology Inc.; ABCE1,
cat no. ab32270; Abcam, Cambridge, UK), incubated overnight
(dilution, 1:500) at 4°C and detected using a horseradish
peroxidase-conjugated IgG rabbit anti-mouse secondary antibody
(dilution, 1:2,000; cat no. SC-2030; Santa Cruz Biotechnology,
Inc.) at room temperature for 1 h. Blots were developed using an
SuperSignal West Pico Chemiluminescent Substrate (cat no. 34080;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol, and then imaged using a ChemiDoc™ XRS+
system v.2011 (Bio-Rad Laboratories, Inc.).
Real-time proliferation assay
Prior to the proliferation assay, MDA-MB-231 and
MCF7 cells (5×105 cells/well) were cultured to ~60%
confluency on 6-cm plates and transfected with 0.5 µg pTRE3G-PLZF +
0.5 µg Tet-On® constructs using 6 µl
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) as
previously described (30). After 24
h, the cells were washed with trypsin and seeded onto a 96-well
plate at a density of 15×103 cells/well in full growth
medium with or without 0.2 µg/ml doxycycline (Dox) for the
proliferation assay. The plates were subsequently inserted into the
xCELLigence® Real-Time Cell Analyzer instrument (ACEA
Biosciences, San Diego, CA, USA), according to the manufacturer's
protocols, in order to monitor cell proliferation.
Statistical analysis
Results were analyzed and graphed using GraphPad
Prism® version 7.02 (GraphPad Software, Inc., La Jolla,
CA, USA). Survival curves were compared between each group using
the log-rank test; the data are reported as the mean mRNA
expression of both PLZF and ABCE1. RT-qPCR results for PLZF mRNA
were compared using one-way analysis of variance and a Holm-Sidak
post-hoc test and are reported as the mean ± standard deviation
(SD). Real-time proliferation assay data are reported as the mean ±
SD. P<0.05 was considered to indicate a statistically
significant difference.
Results
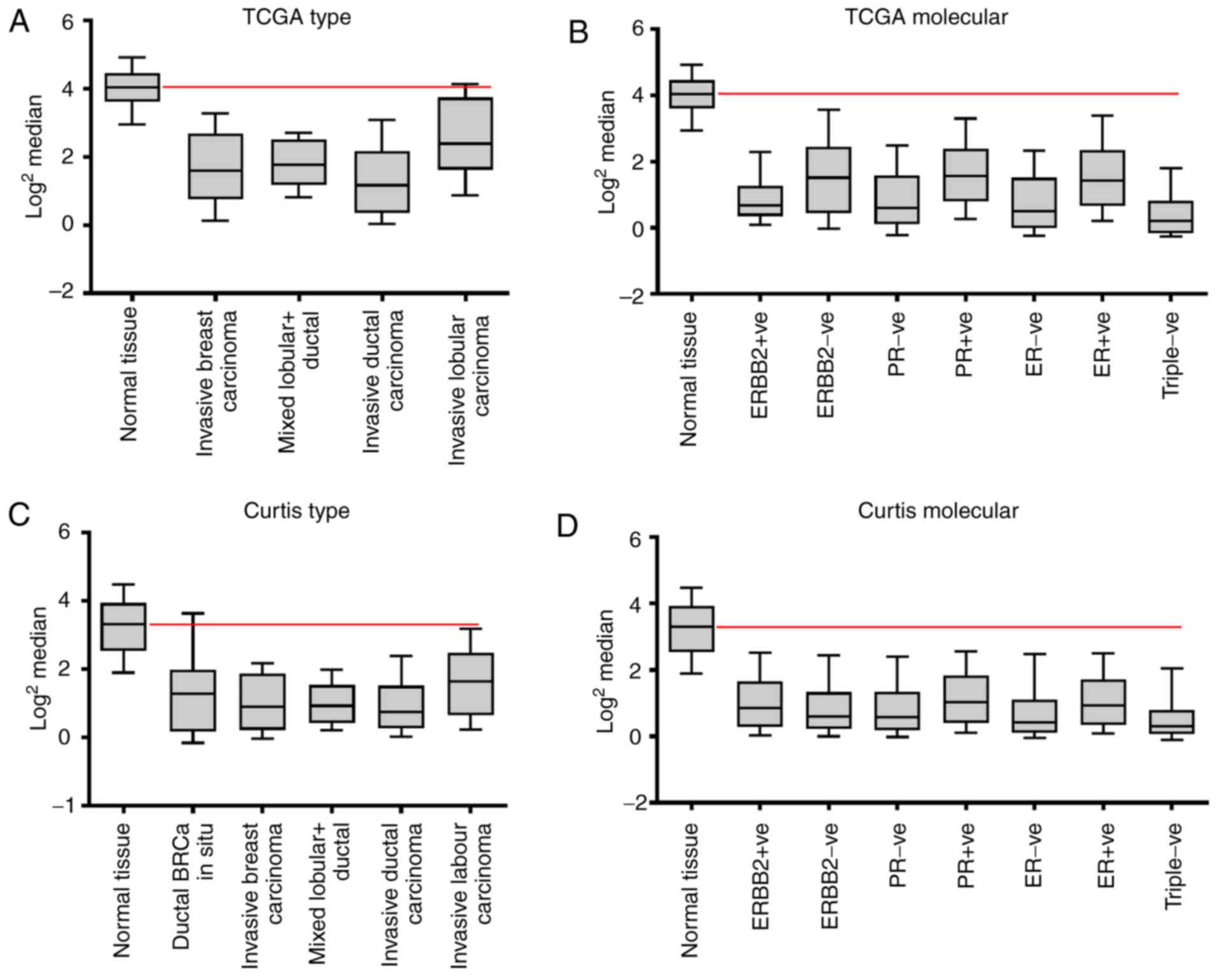
PLZF expression is diminished in
breast cancer tissues compared with in normal breast tissues
The expression of PLZF in breast cancer and normal
tissue samples included in the Oncomine online microarray database
was analyzed. Gene expression data were retrieved from the TCGA and
Curtis databases (Table I) and were
plotted to reveal the mean expression levels of PLZF. The mRNA
levels of PLZF were found to be increased in normal tissues when
compared with various types of adenocarcinoma and lobular carcinoma
of the breast (Fig. 1). These results
are similar to those of previous studies that identified an
involvement of PLZF in the oncogenic transformation of prostate
(31), colon (32) and leukemic cells (6).
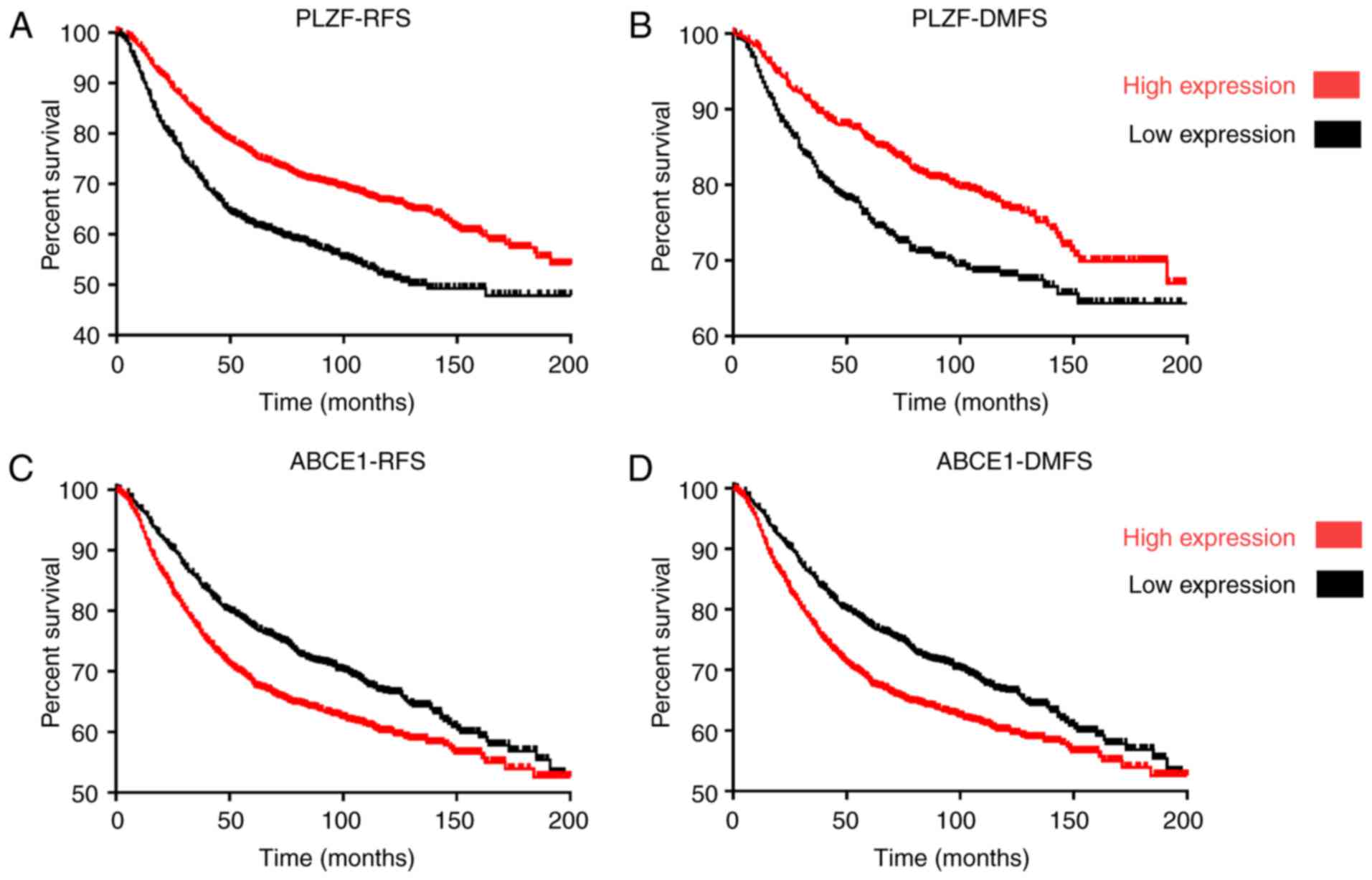
PLZF expression is associated with
survival in patients with breast cancer
The hazard ratio for the expression of PLZF in
patients with breast cancer was calculated using Kaplan-Meier
analysis and the log-rank test (Fig.
2). The results revealed that increased PLZF expression in
patients with breast cancer was associated with prolonged survival
compared with patients with low PLZF expression. The DMFS rates of
patients with low-medium PLZF expression were 78.16 and 69.33%
compared with 87.94 and 79.58% in the patients with high-medium
PLZF at 50 and 100 months, respectively (P=9.1×10−06),
and the RFS rates of patients with low-medium PLZF expression were
lower, at 64.45 and 55.31% compared with 78.69 and 69.39% in
patients with high-medium PLZF at 50 and 100 months, respectively
(P=1×10−16).
Decreased ABCE1 expression in patients with breast
cancer was associated with prolonged survival, as compared with
patients with high ABCE1 expression. In patients with low-medium
ABCE1 expression, the DMFS rates were 79.99 and 70.28% compared
with 71.84 and 62.36% in patients with high-medium ABCE1 at 50 and
100 months, respectively (P=4.4×10−04), while the RFS
rates in patients with low-medium ABCE1 expression were 79.91 and
70.26% compared with 71.14 and 62.37% in patients with high-medium
PLZF at 50 and 100 months, respectively
(P=2.3×10−5).
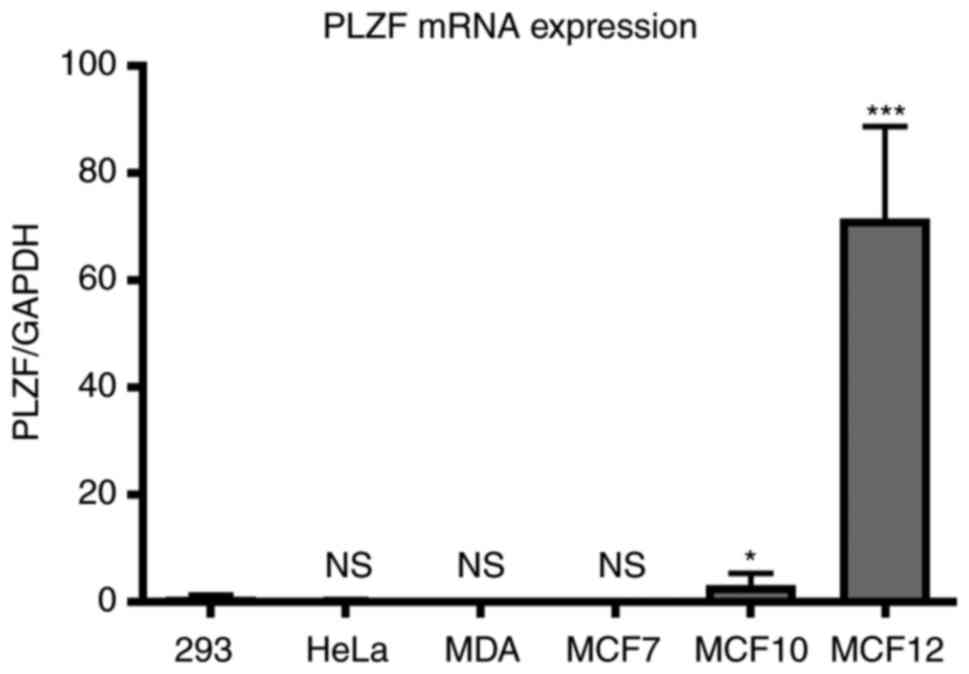
PLZF expression is associated with the
tumorigenicity of breast cancer cell lines in vitro
The mRNA expression of PLZF is detectable in various
cell types during their differentiation process, but this
expression quickly becomes inhibited during the course of oncogenic
transformation. The present study aimed to assess the expression of
PLZF in the breast cancer cell lines MDA-MB-231 and MCF7, and in
the breast basal epithelial cell lines MCF10 and MCF12. Cells were
grown as aforementioned for >48 h prior to total RNA extraction
and cDNA synthesis. The basal expression of PLZF was revealed to be
downregulated in the two cancer cell lines when compared with their
epithelial cell equivalents (Fig. 3).
Our previous study concerning PLZF expression in different cancer
cells indicated that PLZF mRNA expression was higher in 293 cells
compared with in HeLa cells (33).
HeLa and 293 cell lines were included in the RT-qPCR assay of this
study to compare the levels of PLZF mRNA expression in these two
cell lines with corresponding levels in breast cancer cells. The
results of the present study demonstrate that PLZF expression was
markedly higher in MCF10 and MCF12 cells compared with in MCF7 and
MDA-MB-231 cells, the latter of which retain the cytokeratin
profiles of breast luminal cells, fail to form tumors in nude mice
and exhibited no detectable PLZF mRNA expression (34). The fact that PLZF expression was
almost undetectable in MDA-MB-231 and MCF7 cells rendered these
cell lines suitable for overexpression experiments. Although
MDA-MB-231 and MCF7 cells differ in terms of cell type (basal and
luminal, respectively), the two are relatively tumorigenic and
express numerous oncogenic phenotypes (35). These results demonstrated that PLZF
expression is negatively associated with the tumorigenicity of
breast cancer cell lines.
ABCE1 is implicated as a PLZF-targeted
gene
Using an Affymetrix microarray chip, induction of
the PLZF transcript has previously been demonstrated to
downregulate a number of genes, including ABCE1 (21). Similarly, using Oncomine gene
expression signatures, the present study compared the gene
expression profiles of normal breast tissues with those of
different breast cancer types and observed that ABCE1 was
significantly overexpressed in cancer tissues when compared with in
normal tissues (Table II). Notably,
ABCE1 was revealed to be in the top 10% of overexpressed genes in
both the invasive breast ductal carcinoma (TCGA Breast 2) and the
invasive ductal carcinoma (Curtis Breast) datasets, and in the top
5% of overexpressed genes in the invasive ductal carcinoma (TCGA
Breast) dataset. This was in agreement with our previous
observation that the apparent loss of PLZF expression in cancer
cells may cause an increase in ABCE1 expression in cancer
tissues.
 | Table II.Comparison of ABCE1 expression in
breast cancer vs. normal tissues samples in the TCGA and Curtis
datasets. |
Table II.
Comparison of ABCE1 expression in
breast cancer vs. normal tissues samples in the TCGA and Curtis
datasets.
| Comparison | P-value | Q-value | Odds ratio | Rank (%) |
|---|
| Invasive BRCa
stroma vs. normal |
7.16×10−70 |
9.63×10−67 | 3.1 | Lowest 10 |
| Invasive ductal
BRCa vs. normal |
1.48×10−10 |
1.11×10−7 | 10.1 | Lowest 10 |
| Invasive BRCa
stroma vs. normal |
1.30×10−8 |
5.37×10−6 | 10.4 | Lowest 5 |
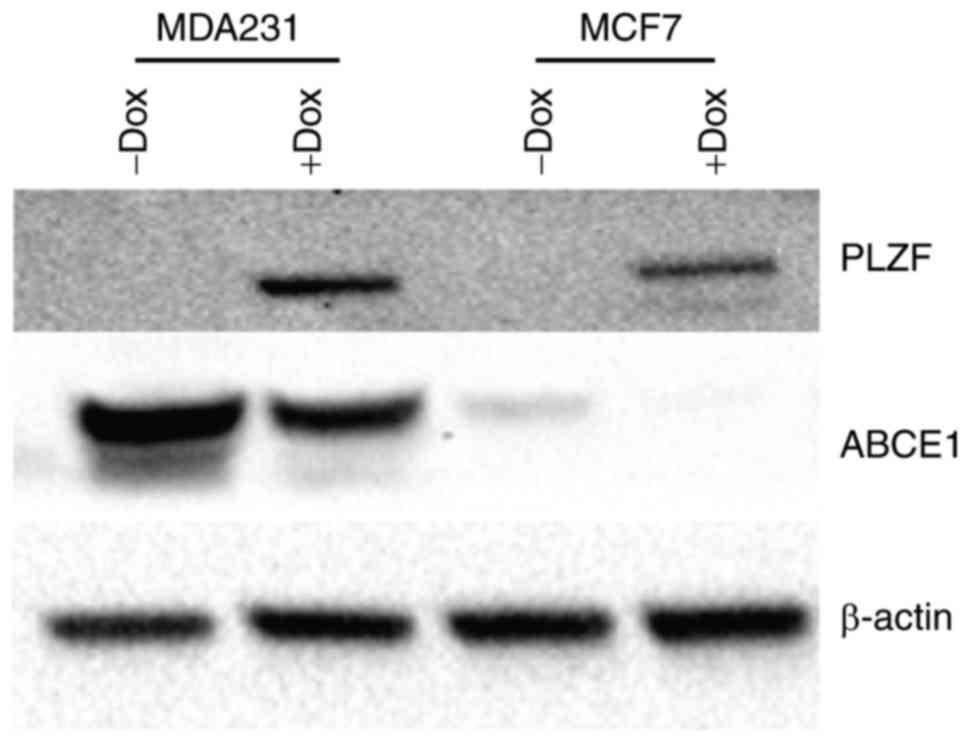
PLZF inhibits the expression of
ABCE1
PLZF is a transcriptional repressor that acts on a
number of different signaling pathways and usually exerts its
inhibitory effect through accessory molecules. To confirm that PLZF
exerts the same transcriptional control over ABCE1, a PLZF
Tet-On® system was used, as described previously
(30), to induce PLZF expression in
the cancer MDA-MB-231 and MCF7 cell lines. MDA-MB-231 cells lacked
PLZF expression (both at the transcriptional and protein levels),
as demonstrated in Figs. 3 and
4. It was observed that PLZF
expression, induced by overnight treatment with 0.2 µg/ml Dox,
markedly downregulated ABCE1 protein expression (Fig. 4).
PLZF expression induces an
ABCE1-mediated inhibition of cellular proliferation in breast
cancer cells
Although Dox treatment has been reported to alter
the genetic signature of a number of common cancer cell lines
(36), its effect on the
proliferation capacity of these cells is less significant than the
marked effect observed in the present study. Furthermore, the
addition of 0.2 µg/ml Dox to the culture medium in a study using
Dox in gene-regulated assays demonstrated that there was no
significant effect compared with the vehicle control (37).
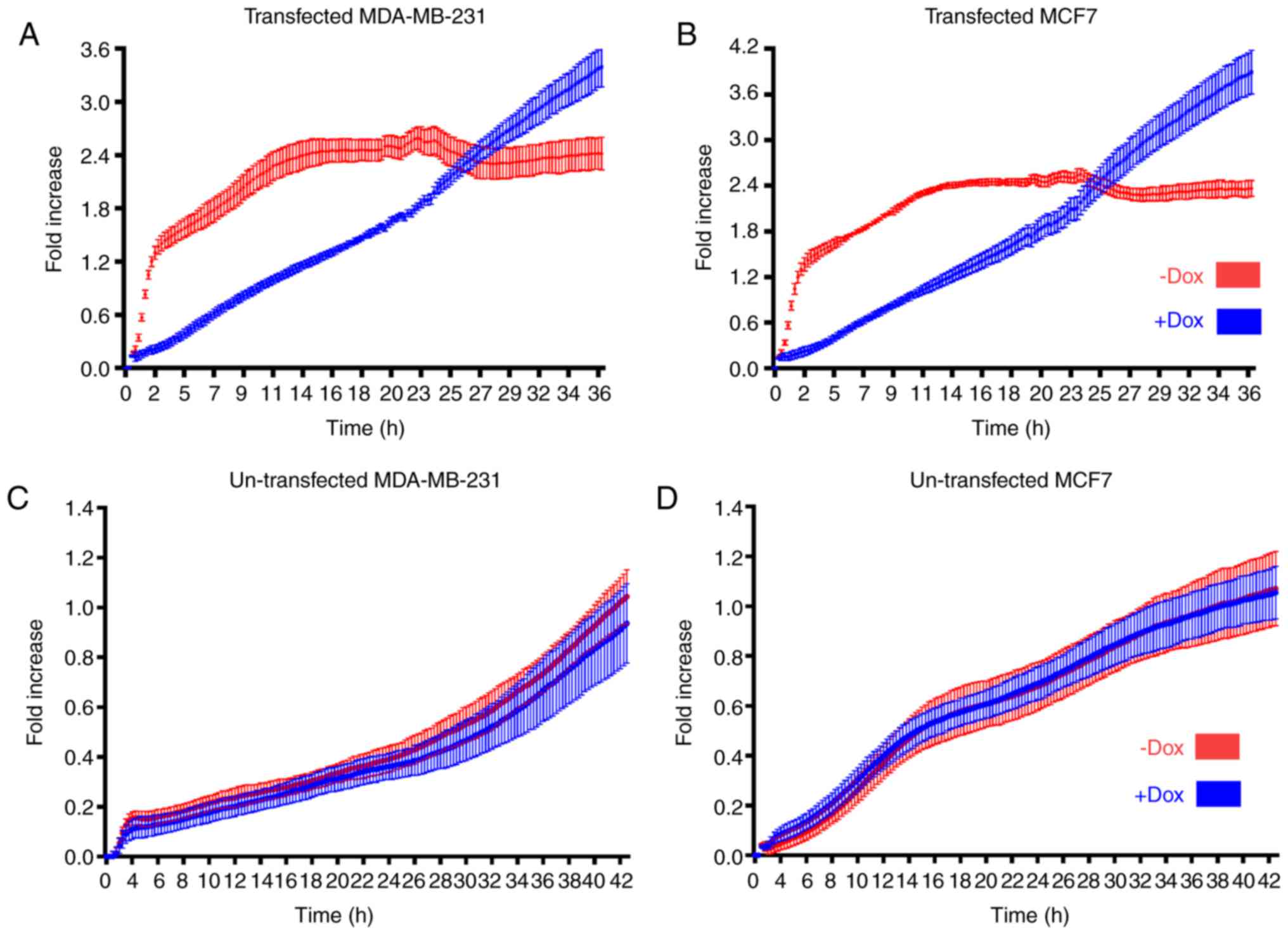
Through real-time cell analysis using an
xCELLigence® RTCA instrument, a significant decrease in
the proliferation capacity of MDA-MB-231 and MCF-7 cells, which
were previously transfected with the PLZF tet-on system, was
observed following treatment with 0.2 µg/ml Dox. Dox-induced PLZF
expression markedly diminished the cellular proliferation
capability within 36 h of treatment (Fig.
5A and B). MDA-MB-231 and MCF-7 cells that were not treated
with Dox continued to proliferate in a logarithmic manner
consistent with normal physiological proliferation behavior in
vitro. The same number of non-transfected MDA-MB-231 and MCF-7
cells were then cultured with or without 0.2 µg/ml Dox. No
significant difference was observed in the two proliferation curves
over 48 h (Fig. 5C and D). The
results of the present study demonstrated that PLZF expression
causes an ABCE1-mediated inhibition of the proliferation of breast
cancer cell lines.
Discussion
Transcription factors serve an essential role in the
molecular regulation of protein expression. Numerous types of
cancer cells reach their oncogenic state through alterations in
their protein signature (4,5). This is commonly achieved by the
transcriptional inhibition of safeguard proteins (which safeguard
against cell cycle override; for example, tumor protein p53), which
would otherwise prevent the cell from undergoing continuous
division and from evading apoptosis (5,9,10). PLZF has been previously demonstrated
to be a tumor suppressor gene in certain types of cancer (11). However, the exact manner of its
inhibitory effect in different cellular environments has yet to be
elucidated.
Using a number of microarray datasets, the mRNA
expression of PLZF was found to be associated with the survival
rate of patients with breast cancer. Higher PLZF expression not
only corresponded with a better survival rate in these patients, it
was also negatively associated with the tumorigenicity of breast
cancer cell lines commonly used in cancer research. PLZF, like
other transcription factors, exerts its biological effects through
numerous downstream targets. In turn, these targets participate in
a number of other signaling pathways, which further complicates our
understanding of how each molecule or protein is involved in the
oncogenic transformation of specific cell types.
The RNase L inhibitor ABCE1 has previously been
identified as a PLZF-targeted gene (18,19). Using
western blot analysis to quantify protein expression, it was
observed that PLZF downregulated ABCE1 expression in the breast
cancer cell lines used in the present study. This inhibition of
ABCE1 likely caused RNase L upregulation in the breast cancer
cells, leading to the diminished proliferation of cells. PLZF
exerts a chromatin-stabilization effect that enables a basal
activity state of early response genes to be established, alongside
its ability to control the inflammatory reaction (14). Therefore, PLZF may be a useful
modulator capable of differentially targeting certain signaling
pathways that are yet to be investigated in an oncological context,
but which may ultimately change the overall phenotype of tumors.
The findings of the present study are important for understanding
how PLZF exerts its final inhibitory actions in breast cancer
cells, and potentially in other solid tumors, through the
modulation of immunological pathways. Furthermore, these results
may pave the way for further studies into the targeting of PLZF as
an approach to limiting the oncogenic transformation and
aggressiveness of associated cancer types.
Acknowledgements
The authors would like to thank Professor Khalid Abu
Khabar (King Faisal Specialist Hospital and Research Centre) for
supplying the required reagents and for providing support with the
databases.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
BAS and SAY designed the research, conducted the
experiments, analyzed the data, and drafted the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen Z, Brand NJ, Chen A, Chen SJ, Tong
JH, Wang ZY, Waxman S and Zelent A: Fusion between a novel
Krüppel-like zinc finger gene and the retinoic acid receptor-alpha
locus due to a variant t(11;17) translocation associated with acute
promyelocytic leukaemia. EMBO J. 12:1161–1167. 1993.PubMed/NCBI
|
|
2
|
Li X, Peng H, Schultz DC, Lopez-Guisa JM,
Rauscher FJ III and Marmorstein R: Structure-function studies of
the BTB/POZ transcriptional repression domain from the
promyelocytic leukemia zinc finger oncoprotein. Cancer Res.
59:5275–5282. 1999.PubMed/NCBI
|
|
3
|
Rho SB, Park YG, Park K, Lee SH and Lee
JH: A novel cervical cancer suppressor 3 (CCS-3) interacts with the
BTB domain of PLZF and inhibits the cell growth by inducing
apoptosis. FEBS Lett. 580:4073–4080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park I, Qian D, Kiel M, Becker MW, Pihalja
M, Weissman IL, Morrison SJ and Clarke MF: Bmi-1 is required for
maintenance of adult self-renewing haematopoietic stem cells.
Nature. 423:302–305. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Costoya JA, Hobbs RM, Barna M, Cattoretti
G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ and Pandolfi PP:
Essential role of Plzf in maintenance of spermatogonial stem cells.
Nat Genet. 36:653–659. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeyati PL, Shaknovich R, Boterashvili S,
Li J, Ball HJ, Waxman S, Nason-Burchenal K, Dmitrovsky E, Zelent A
and Licht JD: Leukemia translocation protein PLZF inhibits cell
growth and expression of cyclin A. Oncogene. 18:925–934. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Felicetti F, Errico MC, Bottero L,
Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini
M, Colombo MP, et al: The promyelocytic leukemia zinc
finger-microRNA-221/-222 pathway controls melanoma progression
through multiple oncogenic mechanisms. Cancer Res. 68:2745–2754.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bernardo MV, Yelo E, Gimeno L, Campillo JA
and Parrado A: Identification of apoptosis-related PLZF target
genes. Biochem Biophys Res Commun. 359:317–322. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheung M, Pei J, Pei Y, Jhanwar SC, Pass
HI and Testa JR: The promyelocytic leukemia zinc-finger gene, PLZF,
is frequently downregulated in malignant mesothelioma cells and
contributes to cell survival. Oncogene. 29:1633–1640. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suliman BA, Xu D and Williams BR: The
promyelocytic leukemia zinc finger protein: Two decades of
molecular oncology. Front Oncol. 2:742012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Savage AK, Constantinides MG, Han J,
Picard D, Martin E, Li B, Lantz O and Bendelac A: The transcription
factor PLZF directs the effector program of the NKT cell lineage.
Immunity. 29:391–403. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weinreich MA, Odumade OA, Jameson SC and
Hogquist KA: PLZF+ T cells regulate memory-like CD8+ T cell
development. Nat Immunol. 11:709–716. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu D, Holko M, Sadler AJ, Scott B,
Higashiyama S, Berkofsky-Fessler W, McConnell MJ, Pandolfi PP,
Licht JD and Williams BR: Promyelocytic leukemia zinc finger
protein regulates interferon-mediated innate immunity. Immunity.
30:802–816. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sadler AJ, Suliman BA, Yu L, Yuan X, Wang
D, Irving AT, Sarvestani ST, Banerjee A, Mansell AS, Liu JP, et al:
The acetyltransferase HAT1 moderates the NF-κB response by
regulating the transcription factor PLZF. Nat Commun. 6:67952015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li XL, Blackford JA, Judge CS, Liu M, Xiao
W, Kalvakolanu DV and Hassel BA: RNase-L-dependent Destabilization
of Interferon-induced mRNAs. A role for the 2–5A system in
attenuation of the interferon response. J Biol Chem. 275:8880–8888.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou A, Paranjape J, Brown TL, Nie H, Naik
S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C and
Silverman RH: Interferon action and apoptosis are defective in mice
devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J.
16:6355–6363. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fernandes J: Oncogenes: The passport for
viral oncolysis through PKR inhibition. Biomark Cancer. 8:101–110.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He Z, Jiang J, Kokkinaki M, Tang L, Zeng
W, Gallicano I, Dobrinski I and Dym M: MiRNA-20 and mirna-106a
regulate spermatogonial stem cell renewal at the
post-transcriptional level via targeting STAT3 and Ccnd1. Stem
cells. 31:2205–2217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bisbal C, Martinand C, Silhol M, Lebleu B
and Salehzada T: Cloning and characterization of a RNAse L
inhibitor. A new component of the interferon-regulated 2–5A
pathway. J Biol Chem. 270:13308–13317. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McConnell MJ, Chevallier N,
Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM and Licht
JD: Growth suppression by acute promyelocytic leukemia-associated
protein PLZF is mediated by repression of c-myc expression. Mol
Cell Biol. 23:9375–9388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ren Y, Li Y and Tian D: Role of the ABCE1
gene in human lung adenocarcinoma. Oncol Rep. 27:965–970. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hendig D, Langmann T, Zarbock R, Schmitz
G, Kleesiek K and Götting C: Characterization of the ATP-binding
cassette transporter gene expression profile in Y79: A
retinoblastoma cell line. Mol Cell Biochem. 328:85–92. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heimerl S, Bosserhoff AK, Langmann T,
Ecker J and Schmitz G: Mapping ATP-binding cassette transporter
gene expression profiles in melanocytes and melanoma cells.
Melanoma Res. 17:265–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hlavata I, Mohelnikova-Duchonova B,
Vaclavikova R, Liska V, Pitule P, Novak P, Bruha J, Vycital O,
Holubec L, Treska V, et al: The role of ABC transporters in
progression and clinical outcome of colorectal cancer. Mutagenesis.
27:187–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Glass D, Viñuela A, Davies MN, Ramasamy A,
Parts L, Knowles D, Brown AA, Hedman AK, Small KS, Buil A, et al:
Gene expression changes with age in skin, adipose tissue, blood and
brain. Genome Biol. 14:R752013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bewick V, Cheek L and Ball J: Statistics
review 12: Survival analysis. Critical Care. 8:389–394. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Suliman BA: The generation of a
ZBTB16-inducible expression system in the ACHN adenocarcinoma cell
line. J Taibah Univ Medical Sci. 10:359–364. 2015.
|
|
31
|
Kikugawa T, Kinugasa Y, Shiraishi K, Nanba
D, Nakashiro KI, Tanji N, Yokoyama M and Higashiyama S: PLZF
regulates Pbx1 transcription and Pbx1-HoxC8 complex leads to
androgen-independent prostate cancer proliferation. Prostate.
66:1092–1099. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mariani F, Sena P, Magnani G, Mancini S,
Palumbo C, de Leon Ponz M and Roncucci L: PLZF expression during
colorectal cancer development and in normal colorectal mucosa
according to body size, as marker of colorectal cancer risk.
ScientificWorldJournal. 2013:6308692013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ali Bandar S: The role of PLZF in the
TLR-mediated inflammatory response and the oncogenic
characteristics of renal cell carcinoma. 2017.
|
|
34
|
Paine TM, Soule HD, Pauley RJ and Dawson
PJ: Characterization of epithelial phenotypes in mortal and
immortal human breast cells. Int J Cancer. 50:463–473. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nagaraja G, Othman M, Fox B, Alsaber R,
Pellegrino C, Zeng Y, Khanna R, Tamburini P, Swaroop A and Kandpal
RP: Gene expression signatures and biomarkers of noninvasive and
invasive breast cancer cells: Comprehensive profiles by
representational difference analysis, microarrays and proteomics.
Oncogene. 25:2328–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie J, Nair A and Hermiston TW: A
comparative study examining the cytotoxicity of inducible gene
expression system ligands in different cell types. Toxicology In
Vitro. 22:261–266. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Onoda T, Ono T, Dhar DK, Yamanoi A, Fujii
T and Nagasue N: Doxycycline inhibits cell proliferation and
invasive potential: Combination therapy with cyclooxygenase-2
inhibitor in human colorectal cancer cells. J Lab Clin Med.
143:207–216. 2004. View Article : Google Scholar : PubMed/NCBI
|