Introduction
Non-small cell lung cancer (NSCLC) is one of the
most common types of human cancer, and makes up >80% of all lung
cancer cases (1,2). Data analyses indicate that the 5-year
survival rate is <5% for the majority of patients with NSCLC who
have suffered from distant metastasis (3). NSCLC includes adenocarcinoma, large cell
carcinoma and squamous cell carcinoma (4–6). Although
various strategies exist for the treatment of NSCLC, the survival
rate remains poor for affected patients (7); therefore an early diagnosis of NSCLC is
beneficial for patients with cancer, so that they can receive the
most appropriate therapeutic regimens.
An early diagnosis enables early treatment of lung
cancer (8–10). Detection of carcinoembryonic antigen
(CEA) in the serum is a sensitive panel for the potential molecular
diagnosis of NSCLC (11). Expression
of cancer antigen 125 (CA125) is associated with cisplatin
susceptibility in human lung cancer cells, which may aid the
diagnostic and therapeutic approaches (12). A previous study indicated that tissue
polypeptide antigen (TPA) and cytokeratin fragment 21-1 (Cyfra21-1)
are serum markers in patients with NSCLC that may be beneficial for
the recognition of tumor relapse (13). In addition, pro-gastrin-releasing
peptide (ProGRB) is a valuable marker with a high specificity for
the diagnosis of lung cancer, with a similar diagnostic accuracy to
pathological diagnosis (14).
Furthermore, neuron-specific enolase (NSE) is a potential target in
the diagnosis and treatment of lung cancer (15). These data indicate that expression
levels of CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE are associated
with the progression of lung cancer.
The purpose of the present study was to analyze the
efficacy of marker gene detection and computed tomography (CT) in
diagnosing human lung cancer. The association between lung cancer
marker genes (CEA, CA125, TPA, ProGRB and cCyfra21-1) and tumor
size in patients with lung cancer was also analyzed.
Materials and methods
Ethics statement
The clinical study design was approved by the Ethics
Committee of the China Japan Union Hospital (Changchun, China;
approval no. 20130701XA) and all patients provided written informed
consent prior to the examinations.
Patients and healthy individuals
A total of 328 patients with NSCLC (186 males and
142 females; median age, 57 years old) and 328 healthy individuals
(168 males and 160 females; median age, 57 years old) were enrolled
between July 2013 and May 2017 at the China Japan Union Hospital.
All patients with NSCLC who received any chemotherapy or
radiotherapy prior to surgery were excluded from the study. All
subjects in the present study underwent CT diagnosis (16). All subjects in the present study
underwent imaging with a trimodality CT system. The characteristics
of the patients with NSCLC are summarized in Table I.
 | Table I.Characteristics of patients with
NSCLC. |
Table I.
Characteristics of patients with
NSCLC.
| Characteristics | NSCLC patients | Healthy patients |
|---|
| Total no., n | 328 | 328 |
| Males, n | 186 | 168 |
| Females, n | 664 | 160 |
| Age range (mean),
years | 36-68 (57) | 36-68 (57) |
CT scan protocol
The CT system consisted of a full-ring
time-of-flight 64-slice positron emission tomography (PET)/CT
scanner (Discovery PET/CT 690 VCT; GE Healthcare, Chicago, IL,
USA). The CT diagnosis system was used to analyze NSCLC tumor size
using the preprogrammed setting. The preprogrammed setting was
optimized, as described previously (17) to achieve the best image formation. The
lungs in all patients underwent contrast-enhanced CT according to
the manufacturer's protocols (Discovery PET/CT 690 VCT). The
Tumor-Node-Metastasis (TNM) stage (seventh edition) (18) was assigned based on intraboard
consensus decision.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cancer tissues using an
RNAeasy Mini kit (Qiagen GmbH, Hilden, Germany). RNA was reverse
transcribed to cDNA using SuperScript™ III One-Step
RT-PCR System with Platinum™ Taq High Fidelity DNA
Polymerase (Thermo Fisher Scientific, Inc.), according to
manufacturer's protocol. All forward and reverse primers were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.; Table II). The thermocycling conditions were
as follows: Initial denaturation at 95°C for 1 min, followed by 45
cycles of 95°C for 30 sec, 56°C for 30 sec and 72°C for 15 sec. The
reaction volume (50 µl) contained 50 ng genomic cDNA, 200 µM dNTPs,
200 µM primers, 2.5 U Taq DNA polymerase (Thermo Fisher Scientific,
Inc.) and 2.5 U SYBR®-Green (Thermo Fisher Scientific,
Inc.). Relative mRNA expression levels were calculated by the
2−ΔΔCq method (19).
 | Table II.Primer sequences for RT-qPCR. |
Table II.
Primer sequences for RT-qPCR.
|
| Sequence |
|---|
|
|
|
|---|
| Gene Name | Reverse | Forward |
|---|
| CEA |
5′-CTTATTACTGCCAGCAAAGGAGTAGTT-3′ |
5′-CAAAGCTCGCTCCGTCTGTAG-3′ |
| CA125 |
5′-TCATGAAGATCCTCACCGAG-3′ |
5′-GCATCCTCTTCAGTTACGTCC-3′ |
| TPA |
5′-ATGACTTCCTACAGCTATCG-3′ |
5′-AATGCTTTCTCCGCTCTG-3′ |
| ProGRB |
5′-CCCCTGGAAAGGGCTCAACAC-3′ |
5′-TCCAACCCAGGTCCTTCCTAAAGTC-3′ |
| Cyfra21-1 |
5′-GAAGGAAACCGAGCCTATTCAC-3′ |
5′-CCACAAGCATCAAACCACCA-3′ |
| NSE |
5′-TCATGAAGATCCTCACCGAG-3′ |
5′-CTTCATCTTCTCCCGCAGAG-3′ |
| β-actin |
5′-CGGAGTCAACGGATTTGGTC-3′ |
5′-AGCCTTCTCCATGGTCGTGA-3′ |
ELISA
A total of 10 ml blood samples were collected from
all patients and serum was obtained from using centrifugation at
4,500 × g for 10 min at 4°C. Serum levels of CEA (catalog no.
EHCEA; Thermo Fisher Scientific, Inc., Waltham, MA, USA), CA125
(EHMUC16; Thermo Fisher Scientific, Inc.), TPA (BMS258-2; Thermo
Fisher Scientific, Inc.), ProGRB (DY7847-05; Bio-Rad Laboratories,
Inc., Hercules, CA, USA), Cyfra21-1 (EHCYF211; Thermo Fisher
Scientific, Inc.) or NSE (DENL20; Bio-Rad Laboratories, Inc.) were
detected in subjects using ELISA kits according to the
manufacturer's protocols. Finally, the serum concentrations of CEA,
CA125, TPA, ProGRB, Cyfra21-1 and NSE were measured using an enzyme
microplate reader at 450 nm (Bio-Rad Laboratories, Inc.).
Immunohistochemistry staining
Cancer tissues from 328 patients who underwent
surgery for NSCLC were collected in the present study. The 328
tissue samples were fixed used 10% paraformaldehyde for 15 min at
37°C and stained with hematoxylin and eosin for 30 min at 37°C, and
final pathological diagnoses were confirmed. Immunohistochemical
analysis was used to identify expression levels of NSCLC tumor
markers CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE.
Paraffin-embedded tissue sections (4-µm thick) were prepared and
dewaxed in xylene for 12 h at 37°C, followed by rehydrating in a
gradient concentration of ethanol (100, 95 and 85%). The paraffin
sections were immersed in 3% H2O2 at room
temperature for 5 min at 37°C and antigen retrieval was performed
at 95°C for 30 min using antigen retrieval buffer [10 mM citrate
acid, 0.05% Tween-20 (pH 6.0)]. Following incubation with PBS
containing 10% bovine serum albumin (Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) for 2 h at 37°C, the tissue
sections were incubated at 4°C overnight with the following
anti-human antibodies: CEA (1:1,000; cat. no. ab133633; Abcam,
Cambridge, UK), CA125 (1:1,000; cat. no. ab10033; Abcam), TPA
(1:1,000; cat. no. ab157469; Abcam), ProGRB (1:1,000; cat. no.
ab158593; Abcam), Cyfra21-1 (1:1,000; cat. no. ab125831; Abcam) or
NSE (1:1,000; cat. no. ab79757; Abcam), followed by further
incubation with horseradish peroxidase-conjugated IgG monoclonal
antibody (cat. no. PV-6001, OriGene Technologies, Inc., Beijing,
China) at 4°C overnight with diaminobenzidine-peroxidase substrate
(Gene Tech, Biotechnology Co., Ltd., Shanghai, China. Slides were
finally counterstained with hematoxylin for 2 h at 37°C and mounted
with coverslips, and images were captured using a MicroChemi 4.2
instrument (Eastwin, Shenzen, China) at ×40, magnification.
Regression analysis
The serum levels of CEA, CA125, TPA, ProGRB,
Cyfra21-1 and NSE in the detective data (Y) were analyzed by
regression analysis in various tumor sizes in patients with NSCLC
using the least square convergence (20). Serum levels of CEA, CA125, TPA,
ProGRB, Cyfra21-1 and NSE were predicted according to tumor size.
The predicted curve that results in the lowest sum of squares is
the best fit using Spearman's rank correlation analysis. If the fit
is robust, then the parameters of the observed curve may be
inferred from those of the predicted curve.
Statistical analysis
All data are presented as the mean ± standard error.
All data were analyzed using SPSS Statistics (version 19.0; IBM
Corp., Armonk, NY, USA). Student's t-test were used to make
comparisons between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Serum levels of tumor markers in
patients with lung cancer
In the present study, the serum levels of tumor
markers were analyzed in 328 patients with NSCLC and age-matched
healthy volunteers. The results demonstrated that serum levels of
CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE were increased in
patients with NSCLC compared with those in the healthy volunteers
(Table III). As presented in
Table IV, serum tumor marker
expression levels were associated with a poor TNM stage of NSCLC.
These results indicated that tumor markers of CEA, CA125, TPA,
ProGRB, Cyfra21-1 and NSE are associated with the progression of
NSCLC.
 | Table III.Serum levels of tumor markers in
patients with NSCLC. |
Table III.
Serum levels of tumor markers in
patients with NSCLC.
| Marker | Healthy patients
(ng/ml) | NSCLC
patients(ng/ml) |
|---|
| CEA | 2.6±1.2 | 53.4±12.5 |
| CA125 | 3.0±2.4 | 42.5±13.5 |
| TPA | 3.8±1.6 | 35.8±8.4 |
| ProGRB | 4.7±1.4 | 41.4±9.5 |
| Cyfra21-1 | 5.0±1.5 | 46.3±9.6 |
| NSE | 6.2±2.3 | 31.5±7.6 |
 | Table IV.Association between tumor marker
expression levels and Tumor-Node-Metastasis stage. |
Table IV.
Association between tumor marker
expression levels and Tumor-Node-Metastasis stage.
| Marker | T stage | N stage | M stage |
|---|
| CEA | 31.2±10.4 | 53.4±12.5 | 65.8±15.7 |
| CA125 | 22.4±10.8 | 42.6±14.6 | 56.6±16.2 |
| TPA | 41.2±13.7 | 55.6±16.8 | 83.5±26.8 |
| ProGRB | 26.8±10.8 | 42.7±11.4 | 58.6±15.7 |
| Cyfra21-1 | 32.6±16.6 | 58.4±17.0 | 78.5±14.9 |
| NSE | 23.6±8.5 | 41.0±9.8 | 56.6±13.5 |
Marker gene expression in NSCLC
tissues
The expression of CEA, CA125, TPA, ProGRB, Cyfra21-1
and NSE marker genes was examined in NSCLC tissues. It was
indicated that gene expression levels of CEA, CA125, TPA, ProGRB,
Cyfra21-1 and NSE were upregulated in NSCLC tissues compared with
those in the corresponding adjacent non-tumor tissues, as
determined using the quantitative polymerase chain reaction
(Fig. 1). Immunohistochemistry
demonstrated that expression levels of CEA, CA125, TPA, ProGRB,
Cyfra21-1 and NSE were notably upregulated in NSCLC tissues
compared with those in the corresponding adjacent non-tumor tissues
(Fig. 2). These results indicated
that NSCLC tissues exhibited increased expression levels of tumor
markers compared with adjacent non-tumor tissues.
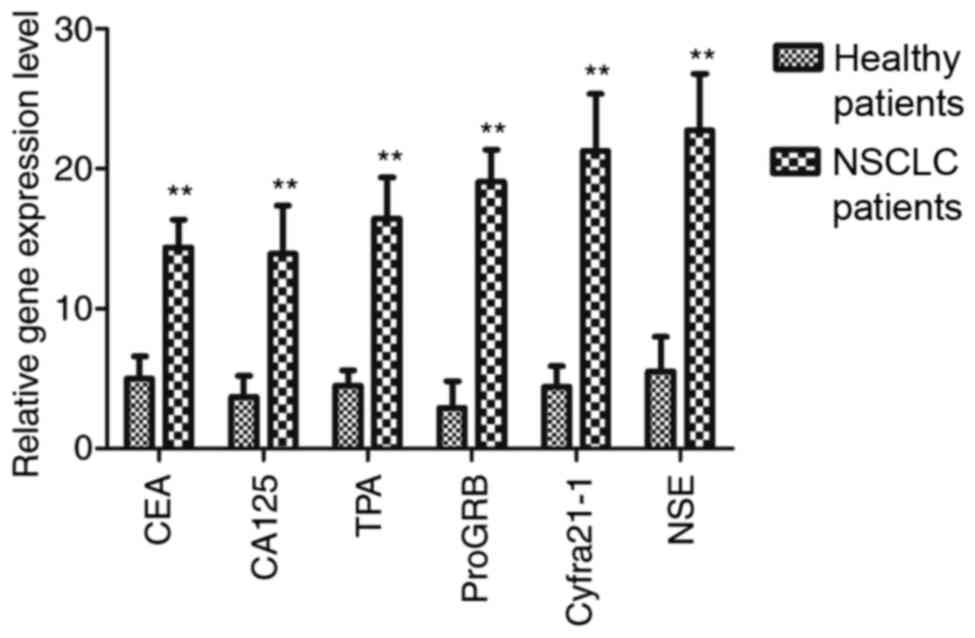 | Figure 1.Gene expression levels of CEA, CA125,
TPA, ProGRB, Cyfra21-1 and NSE in NSCLC tissues and adjacent
non-tumor tissues, as determined using the quantitative polymerase
chain reaction. **P<0.01 vs. healthy patients. CEA,
carcinoembryonic antigen; CA125, cancer antigen 125; TPA, tissue
polypeptide antigen; ProGRB, pro-gastrin-releasing peptide;
Cyfra21-1, cytokeratin fragment 21-1; NSE, neuron-specific enolase;
NSCLC, non-small cell lung cancer. |
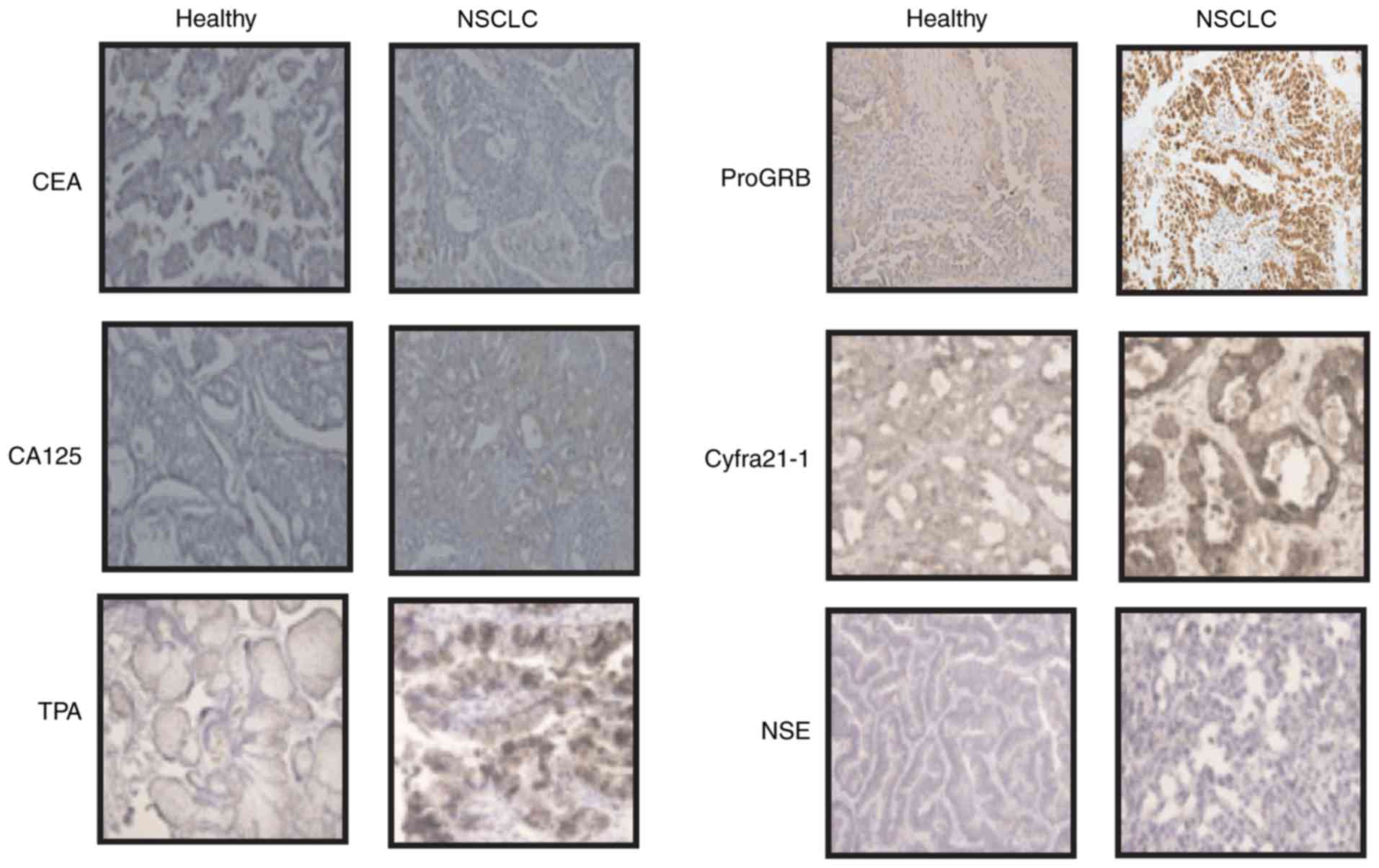 | Figure 2.Expression levels of CEA, CA125, TPA,
ProGRB, Cyfra21-1 and NSE in NSCLC tissues compared with those in
corresponding adjacent non-tumor tissues, as determined by
immunohistochemistry. Magnification, ×40. CEA, carcinoembryonic
antigen; CA125, cancer antigen 125; TPA, tissue polypeptide
antigen; ProGRB, pro-gastrin-releasing peptide; Cyfra21-1,
cytokeratin fragment 21-1; NSE, neuron-specific enolase; NSCLC,
non-small cell lung cancer. |
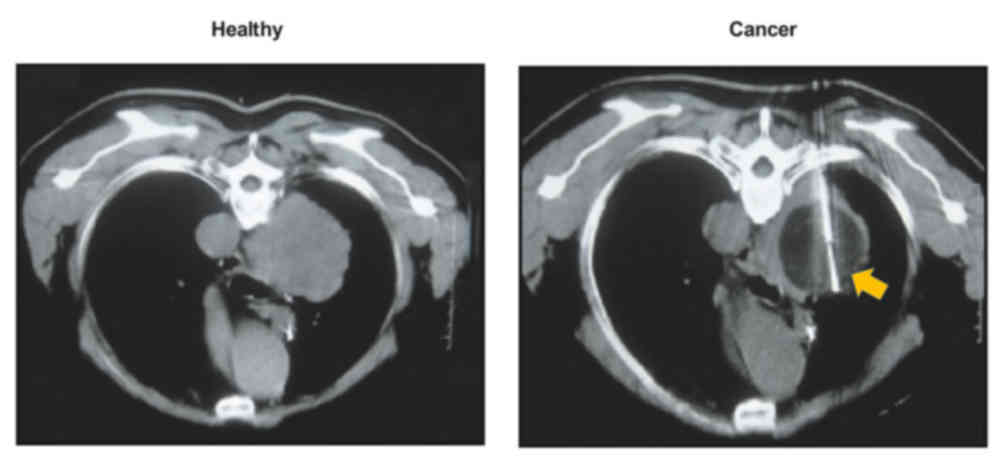
Tumor size detection by CT
The tumor size was determined by CT in the present
study. As shown in Fig. 3, CT may be
used to determine NSCLC tumor size. Among the patients, CT combined
with gene expression identified 328 cases, including 204
adenocarcinoma cases, 75 large cell carcinoma cases and 49 squamous
cell carcinoma cases determined by immunohistochemistry (Table V). These results indicated that CT
combined with marker gene detection may be used to diagnose human
NSCLC.
 | Table V.Tumor size and tumor type. |
Table V.
Tumor size and tumor type.
| Tumor type | n | Tumor size, mm |
|---|
| Adenocarcinoma | 204 | 3.2–8.4 |
| Large cell
carcinoma | 75 | 4.0–12.5 |
| Squamous cell
carcinoma | 49 | 3.1–15.0 |
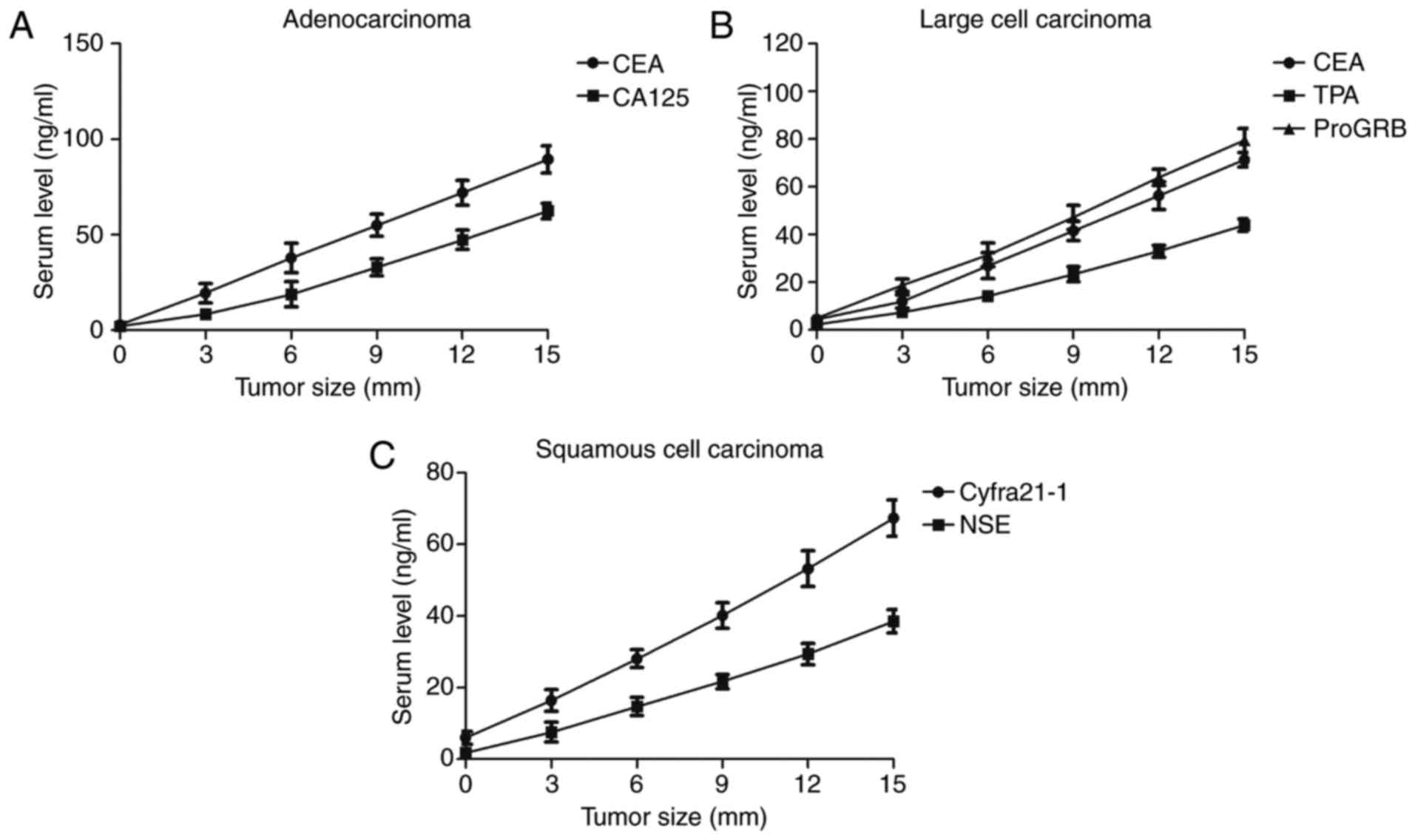
Analysis of the correlation between
serum levels of marker gene proteins and tumor size
Spearman's rank correlation analysis was used to
determine the correlation between serum levels of marker gene
proteins and tumor size calculated using CT in a total of 328
patients with NSCLC. Spearman's rank correlation analysis
demonstrated that CEA and CA125 serum levels were correlated with
CT-diagnosed adenocarcinoma tumor size (Fig. 4A). Furthermore, large cell carcinoma
tumor size was correlated with serum levels of CEA, TPA and ProGRB
(Fig. 4B). Results indicated that
Cyfra21-1 and NSE were associated with the squamous cell carcinoma
cases diagnosed by CT (Fig. 4C).
These results indicated that serum levels of marker gene proteins
was associated with NSCLC size determined by CT, which is
beneficial for human NSCLC diagnosis.
Efficacy of comprehensive analysis of
marker gene detection and CT for NSCLC diagnosis
Finally, the diagnostic efficacy of marker gene
detection combined with CT was investigated in a total of 328
patients with NSCLC. It was demonstrated that comprehensive
analysis of marker gene detection and CT was able to diagnose 320
patients with NSCLC, whereas CT alone diagnosed 256 patients with
NSCLC and marker gene detection diagnosed 270 patients with NSCLC
(Table VI). It was determined that
only 8 patients with NSCLC could not be diagnosed using marker gene
detection combined with CT. These results indicated that
comprehensive analysis of marker gene detection and CT is an
efficient method for the diagnosis of patients with NSCLC.
 | Table VI.Diagnostic efficacy of marker gene
detection and CT in 328 cases. |
Table VI.
Diagnostic efficacy of marker gene
detection and CT in 328 cases.
| Diagnosis
method | n |
|---|
| Marker gene
detection | 270 |
| CT | 256 |
| Marker gene
detection combined with CT | 320 |
Discussion
Lung cancer is a respiratory disease that is a major
cause of cancer-associated mortality resulting from air
contamination caused by industrial pollution globally (21). It has been indicated that the early
diagnosis of patients with NSCLC is beneficial for the treatment of
cancer (22). Serum tumor markers
have been demonstrated to possess the ability to predict the
efficacy of pemetrexed-based chemotherapy (23). In the present study, the efficacy of
marker gene detection and CT for human lung cancer diagnosis was
comprehensively analyzed. It was demonstrated that serum levels of
CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE were significantly
upregulated in patients with NSCLC. Results of the present study
also indicated that marker gene expression levels were correlated
with tumor size determined by CT for patients with NSCLC, according
to Spearman's rank correlation analysis.
Currently, there are numerous cancer diagnostic
methods, including ultrasound, X-ray screening, CT, magnetic
resonance imaging and immunological detection (24–26). CT
(fluorodeoxyglucose PET/CT) is frequently performed in hilar and
mediastinal lymph node staging of NSCLC (27). The present study demonstrated that CT
could approximately measure the NSCLC tumor size. A previous study
indicated that PET/CT has become feasible for fast imaging and may
be used for cancer staging in patients with a malignant condition
(28). In addition, three-dimensional
CT measurement may be useful for diagnosing nodal metastases
(29). Notably, the present study
determined that single CT diagnosis is insufficient in the
diagnosis of patients with NSCLC.
The clinical value of a cancer diagnosis with tumor
marker determination has been identified in lung cancer (30,31). The
present study demonstrated that tumor marker expression combined
with CT is an efficient method of diagnosing patients with NSCLC. A
previous study demonstrated the clinical value of CEA and CA125 to
evaluate the relapse, metastasis and prognosis of patients with
resectable NSCLC (32). The present
study demonstrated that serum levels of CEA and CA125 levels were
associated with CT-diagnosed adenocarcinoma tumor size. In
addition, Wang et al (33)
indicated that the serum concentrations of Cyfra21-1, NSE and CEA
may be diagnostic indicators of meningeal carcinomatosis of lung
cancer. Results of the present study indicated that Cyfra21-1 and
NSE were associated with the squamous cell carcinoma cases. A
previous meta-analysis indicated that the serum level of ProGRB is
a promising biomarker for SCLC diagnosis (34). In the present study, it was indicated
that the large cell carcinoma tumor size was associated with serum
levels of CEA TPA and ProGRB. These data have identified that the
tumor markers CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE are useful
in determining tumor size.
In conclusion, the present study investigated the
effects of tumor marker genes combined with CT in diagnosing
patients with NSCLC. The results demonstrated that tumor markers
CEA, CA125, TPA, ProGRB, Cyfra21-1 and NSE were significantly
upregulated in NSCLC tissues. Furthermore, it was determined that
tumor marker serum levels were associated with tumor size.
Importantly, tumor markers combined with CT may contribute to
diagnosing the progression and treatment of NSCLC cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
MC conducted major experiments in this study. XS,
GL, KC and ZL conducted a small number of experiments. CS and DX
analyzed the experimental data in the present study. LL designed
all experiments in the present study.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the China Japan Union Hospital (Changchun, China;
approval no. 20130701XA).
Patient consent for publication
All patients provided written informed consent prior
to the examinations.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Paleiron N, Bylicki O, André M, Rivière E,
Grassin F, Robinet G and Chouaïd C: Targeted therapy for localized
non-small-cell lung cancer: A review. Onco Targets Ther.
9:4099–4104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li SJ, Huang J, Zhou XD, Zhang WB, Lai YT
and Che GW: Clinicopathological and prognostic significance of
Oct-4 expression in patients with non-small cell lung cancer: A
systematic review and meta-analysis. J Thorac Dis. 8:1587–1600.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aokage K, Yoshida J, Hishida T, Tsuboi M,
Saji H, Okada M, Suzuki K, Watanabe S and Asamura H: Limited
resection for early-stage non-small cell lung cancer as
function-preserving radical surgery: A review. Jpn J Clin Oncol.
47:7–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brody H: Lung cancer. Nature. 513:S12014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moro-Sibilot D, Smit E, de Castro Carpeño
J, Lesniewski-Kmak K, Aerts JG, Villatoro R, Kraaij K, Nacerddine
K, Dyachkova Y, Smith KT, et al: Non-small cell lung cancer
patients with brain metastases treated with first-line
platinum-doublet chemotherapy: Analysis from the European FRAME
study. Lung Cancer. 90:427–432. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barnett SA, Downey RJ, Zheng J, Plourde G,
Shen R, Chaft J, Akhurst T, Park BJ and Rusch VW: Utility of
routine PET imaging to predict response and survival after
induction therapy for non-small cell lung cancer. Ann Thorac Surg.
101:1052–1059. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loganadane G, Martinetti F, Mercier O,
Krhili S, Riet FG, Mbagui R, To H, Le Péchoux C and Levy A:
Stereotactic ablative radiotherapy for early stage non-small cell
lung cancer: A critical literature review of predictive factors of
relapse. Cancer Treat Rev. 50:240–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JO, Davis F, Butts C and Winget M:
Waiting time intervals for non-small cell lung cancer diagnosis and
treatment in alberta: Quantification of intervals and
identification of risk factors associated with delays. Clin Oncol
(R Coll Radiol). 28:750–759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng D and Chen H: Advances in liquid
biopsy and its clinical application in the diagnosis and treatment
of non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 19:394–398.
2016.(In Chinese). PubMed/NCBI
|
|
10
|
Benitez-Majano S, Fowler H, Maringe C, Di
Girolamo C and Rachet B: Deriving stage at diagnosis from multiple
population-based sources: Colorectal and lung cancer in England. Br
J Cancer. 115:391–400. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sheu CC, Chang MY, Chang HC, Tsai JR, Lin
SR, Chang SJ, Hwang JJ, Huang MS and Chong IW: Combined detection
of CEA, CK-19 and c-met mRNAs in peripheral blood: A highly
sensitive panel for potential molecular diagnosis of non-small cell
lung cancer. Oncology. 70:203–211. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chanvorachote P, Luanpitpong S, Chunhacha
P, Promden W and Sriuranpong V: Expression of CA125 and cisplatin
susceptibility of pleural effusion-derived human lung cancer cells
from a Thai patient. Oncol Lett. 4:252–256. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Passowicz-Muszyńska E, Gisterek I,
Marciniak M, Kornafel J, Kolodziej J and Jankowska R: Tumor markers
TPA and Cyfra 21.1 in patients with non-small cell lung cancer
after surgery and chemotherapy. Pol Merkur Lekarski. 13:294–297.
2002.(In Polish). PubMed/NCBI
|
|
14
|
Tang JH, Zhang XL, Zhang ZH, Wang R, Zhang
HM, Zhang ZL, Wang JH and Ren WD: Diagnostic value of tumor marker
pro-gastrin-releasing peptide in patients with small cell lung
cancer: A systematic review. Chin Med J (Engl). 124:1563–1568.
2011.PubMed/NCBI
|
|
15
|
Lazarev SM, Massard ZH, Reshetov AV,
Nikolaev GV, Volgin GN, Osipov EV, Lomteva EIu, Nokhrin AV and
Kakysheva OE: Role of biological tumor markers CEA, Cyfra-21, NSE,
TU M2-PK in diagnosis and treatment of lung cancer. Vestn Khir Im I
I Grek. 169:39–43. 2010.(In Russian). PubMed/NCBI
|
|
16
|
Raz DJ, Wu GX, Consunji M, Nelson RA, Kim
H, Sun CL, Sun V and Kim JY: The effect of primary care physician
knowledge of lung cancer screening guidelines on perceptions and
utilization of low-dose computed tomography. Clin Lung Cancer.
19:51–57. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawakami W, Takemura A, Yokoyama K,
Nakajima K, Yokoyama S and Koshida K: The use of positron emission
tomography/computed tomography imaging in radiation therapy: A
phantom study for setting internal target volume of biological
target volume. Radiat Oncol. 10:12015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hayes AF and Rockwood NJ: Regression-based
statistical mediation and moderation analysis in clinical research:
Observations, recommendations, and implementation. Behav Res Ther.
98:39–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fenton-Ambrose L and Kazerooni EA:
Preventative care: Lung-cancer screens now worth the cost. Nature.
514:352014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herth F, Kirsch CM and Stoelben E:
Diagnosis of non-small-cell lung carcinoma (NSCLC). Onkologie.
29(Suppl 2): S3–S6. 2006.(In German). View Article : Google Scholar
|
|
23
|
Fiala O, Pesek M, Finek J, Svaton M,
Sorejs O, Bortlicek Z, Kucera R and Topolcan O: Prognostic
significance of serum tumor markers in patients with advanced-stage
NSCLC treated with pemetrexed-based chemotherapy. Anticancer Res.
36:461–466. 2016.PubMed/NCBI
|
|
24
|
Barrio-Muñoz M1, Abad-Gairín C,
Amengual-Guedán JM and Prats-López J: Diagnosis of prostate cancer
by analyzing oxidative stress in human seminal plasma: Developing
unsophisticated tools for noninvasive prostate cancer diagnosis.
Eur J Cancer Prev. 25:518–523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang L, Wang J, Li J, Zhang H, Guo S, Yan
M, Zhu Z, Lan B, Ding Y, Xu M, et al: Identification of serum
biomarkers for gastric cancer diagnosis using a human proteome
microarray. Mol Cell Proteomics. 15:614–623. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu Y, Guo M, Zhang L, Xu T, Wang L and Xu
G: Biomarker triplet NAMPT/VEGF/HER2 as a de novo detection panel
for the diagnosis and prognosis of human breast cancer. Oncol Rep.
35:454–462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li S, Zheng Q, Ma Y, Wang Y, Feng Y, Zhao
B and Yang Y: Implications of false negative and false positive
diagnosis in lymph node staging of NSCLC by means of
¹8F-FDG PET/CT. PLoS One. 8:e785522013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HS, Lee KS, Ohno Y, van Beek EJ and
Biederer J: PET/CT versus MRI for diagnosis, staging, and follow-up
of lung cancer. J Magn Reson Imaging. 42:247–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takahashi Y, Takashima S, Hakucho T,
Miyake C, Morimoto D, Jiang BH, Numasaki H, Tomita Y, Nakanishi K
and Higashiyama M: Diagnosis of regional node metastases in lung
cancer with computer-aided 3D measurement of the volume and
CT-attenuation values of lymph nodes. Acad Radiol. 20:740–745.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang YJ, Cheng DY, Fang X and Li XX: The
clinical diagnosis value of fibro-optic bronchoscope examination
combined with tumor marker determination to lung cancer. Sichuan Da
Xue Xue Bao Yi Xue Ban. 38:312–315. 2007.(In Chinese). PubMed/NCBI
|
|
31
|
Shi GL, Hu XL, Yue SD and Song CX: The
value of serum tumor marker in the diagnosis of lung cancer.
Zhonghua Zhong Liu Za Zhi. 27:299–301. 2005.PubMed/NCBI
|
|
32
|
Pollán M, Varela G, Torres A, de la Torre
M, Ludeña MD, Ortega MD, Pac J, Freixenet J, Gómez G, Sebastián F,
et al: Clinical value of p53, c-erbB-2, CEA and CA125 regarding
relapse, metastasis and death in resectable non-small cell lung
cancer. Int J Cancer. 107:781–790. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang P, Piao Y, Zhang X, Li W and Hao X:
The concentration of CYFRA 21-1, NSE and CEA in cerebro-spinal
fluid can be useful indicators for diagnosis of meningeal
carcinomatosis of lung cancer. Cancer Biomark. 13:123–130. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H and Qian J: Serum
pro-gastrin-releasing peptide in diagnosis of small cell lung
cancer: A meta-analysis. J Cancer Res Ther. 12 Suppl:C260–C263.
2016. View Article : Google Scholar : PubMed/NCBI
|