Introduction
Breast cancer is a variable and complex disease, and
is a leading causes of mortality (1).
In the United States ~23,234 female fatalities are expected to
occur due to invasive breast cancer and ~39,620 novel cases of
invasive breast cancer are expected to be diagnosed in the next
decade (2). In the United States, it
is expected that ~12.5% of women will be diagnosed with breast
cancer in their lifetime (2). Breast
cancer cases in China accounted for 12.2% of global cases in 2008
(3). Data from the Human Genome
Project and the International HapMap Project have enabled
geneticists to study polygenic traits and diseases by genome-wide
association study (GWAS) (4). An
investigation of the genetic basis of common diseases is possible
by testing for variants that are significantly associated with
cases of disease over controls in a population. GWAS is widely used
in genetic epidemiology, resulting in >1,600 studies published
and reports of >11,000 single nucleotide polymorphisms (SNPs)
associated with hundreds of different diseases and traits (5). When interpreting GWAS results, it is
often difficult to identify the functional variant(s) underlying an
association (5). For example,
according to GWAS, 93% of SNPs associated with human phenotypes are
located outside of protein-coding regions (6). This emphasizes the requirement for a
method of annotation of the vast non-coding sequence in the human
genome.
Epigenetic maps serve an important role in
identifying specific functional variations that cause or contribute
to a number of diseases. Previous studies have reported that
epigenetic abnormalities may serve an important role in the initial
stages of a number of cancer types, including colorectal cancer and
prostate cancer (7,8). Methylation of one allele has been
indicated to affect gene activation mutations on the opposite
allele (9). The normal genome
commonly contains DNA methylation of 70–80% of all
5′-C-phosphate-G-3′ (CpG) dinucleotides (10). It has been hypothesized that this type
of DNA methylation prevents the inappropriate transcription of
repetitive sequences (7) and
maintains chromosomal stability (11). The remaining CpGs are grouped within
short DNA regions (0.2–1.0 kb in length) termed ‘CpG islands’
(CGIs). Human genes possess 40~50% of CGIs in or near the promoter
and/or the first exon, whereas a lack of methylation exists in
normal somatic cells (10).
Alterations in the methylation status of DNA are the most frequent
molecular changes observed in various types of cancer (12). Cancer studies focusing on genes with
hypermethylated promoter CGIs have revealed unique profiles of
hypermethylation that define each neoplasia (13–15).
Therefore, DNA methylation may serve as an effective biomarker for
cancer as it has been indicated to be associated with
tissue-specific gene silencing (13).
For instance, hypermethylation of the glutathione S-transferase
Pi-1 (GSTP1) gene promoter has been reported to occur in 80–90% of
patients with prostate cancer and is almost undetectable in
prostate tissues. Due to this high specificity, it is used as a
diagnostic biomarker (12,16,17).
Numerous studies have used methylation patterns as
diagnostic and prognostic biomarkers for breast cancer (18). Protocadherin (PCDH)-10 is a member of
the PCDH family and is located on the human chromosome 4q28.3.
Protein expression of PCDH has been predominantly identified in the
nervous system, serving an important role in signal transduction.
PCDH10 promotes cell-cell adhesion via Ca2+ in tissue
morphogenetic processes, and apoptosis by upregulating Fas cell
surface death receptor, Caspase 8, Jun proto-oncogene, AP-1
transcription factor subunit, Cyclin dependent kinase inhibitor 1A
and HIV-1 Tat interactive protein 2 (19–21).
Evidence suggests that members of the PCDH family can inhibit the
occurrence and progression of multiple carcinomas. Previous studies
have characterized PCDH10 expression downregulation by promoter
methylation, and its role as a tumor suppressor gene in the
alimentary system and the other carcinomas (22–24).
However, to the best of our knowledge, a limited number of studies,
have been conducted on PCDH10 expression in breast cancer (25,26). The
expression, methylation status, biological function and clinical
application of PCDH10 have yet to be determined.
The aim of the present study was to investigate a
novel methylation-based diagnostic tool for breast cancer and to
resolve the difficulties associated with the implementation of
methylation genes as biomarkers. It was demonstrated that PCDH10
could be an effective prognostic biomarker for patients with
various types of breast cancer, including ductal carcinoma in
situ (DCIS), invasive ductal carcinoma (IDC) and invasive
ductal carcinoma plus lymph-metastasis (IDC-L). The changes in
methylation status of sporadic breast cancer and hereditary breast
cancer (HpBC) were examined, along with overall survival (OS) rate
of patients and the association between PCDH10 methylation and
clinicopathological features.
Materials and methods
Patients and sample collection
A total of 392 samples of flash-frozen cancerous and
paired healthy breast tissues (≥5 cm distant from the tumor tissue)
were collected from patients with breast cancer, who underwent
mastectomy at the Harbin Medical University Cancer Hospital
(Heilongjiang, China) between May 2009 and October 2012. Serum
samples (1 ml) were obtained from 300 patients (47±18 years old)
with breast cancer, as well as from healthy subjects (45±12 years
old) at the Second Affiliated Hospital of Harbin Medical University
(Heilongjiang, China) between May 2009 and October 2012. The breast
cancer patients with other diseases were excluded from this study
according to clinical detection. The healthy subjects were from
patients who received physical examination and were identified as
healthy. The types of benign breast diseases included fibroadenoma,
desmoid tumors, benign phyllodes tumors, mastopathy, papilloma,
duct ectasia and hamartoma. The healthy serum sample (n=300) was
acquired from the Affiliated Tumor Prevention and Treatment
Institution of the Harbin Medical University (Heilongjiang, China)
between May 2009 and October 2012. All patients provided written
informed consent for tissue and serum collection, in consistence
with regulations of the institutional review board of the Harbin
Medical University (Heilongjiang, China). The present study was
completed in compliance with the Declaration of Helsinki and was
approved by the ethics committee of the Harbin Medical University
(Heilongjiang, China).
Immunohistochemistry and molecular
subtypes
Tissue sections (8 µm) were obtained from breast
tissues and stored at −25°C. These sections were stained with 10%
hematoxylin for 5 min and 0.5% eosin for 1 min at room temperature,
and then were examined by two independent pathologists from Harbin
Medical University Cancer Hospital (Harbin, China), who were blind
to the study, to ensure the integrity of the tumor sample (tumor
content >70%), and to verify that healthy tissue blocks
contained no tumor cells under light microscopy at ×100 and ×400
magnifications. Malignant samples were categorized into four groups
based on histopathology: i) DCIS; ii) IDC; iii) IDC-L; or iv) HpBC,
included patients with a first-degree relative with breast cancer,
patients with bilateral breast tumors, and <35-year-old patients
with early-onset breast cancer (27–31).
The estrogen receptor (ER) mouse monoclonal antibody
was obtained from Ventana Medical Systems, Inc. (1:200 dilution;
cat. no. 760-2596; Tucson, AZ, USA) and progesterone receptor (PR)
mouse monoclonal antibody from Dako (1:200 dilution; cat. no.
M3569; Agilent Technologies, Inc., Santa Clara, CA, USA). The
sections were incubated with antibodies at 4°C overnight. The bound
antibodies were detected using peroxidase-conjugated goat
anti-mouse IgG (ready-to-use secondary antibody; cat. no. TA130004;
OriGene Technologies, Inc., Beijing, China) at 37°C for 2 h, and
the final staining was completed with DAB (OriGene Technologies,
Inc.) at room temperature for 3 min. Nuclear labeling revealed that
>1% of cells were ER- or PR-positive (32). Human epidermal growth factor receptor
2 (HER-2) immunohistochemistry was performed using the DAKO
Herceptest kit (Agilent Technologies, Inc.), according to the
manufacturer's protocol. Cases were scored according to the number
of positive cells as follows: 0 (negative, <5%); 1+ (weak
positive 6–25%); 2+ (positive, 26–50%); and 3+ (strong positive,
>50%). Fluorescence in situ hybridization analysis for
HER-2 amplification was performed for all samples with a score of
2+(equivocal) using the Path Vision kit (Abbott Pharmaceutical Co.,
Ltd., Lake Bluff, IL, USA), according to manufacturer's protocol.
Samples with a 3+ IHC score or a HER-2 fluorescence in situ
hybridization amplification ratio >2.2 were considered to be
HER-2-positive. Samples were divided into one of four categories,
according to the accepted and previously validated IHC surrogate
profiles of breast cancer (33).
Mouse monoclonal antibody P53 (1:100 dilution; cat. no. 760-2542)
and Ki-67 (1:100 dilution; cat. no. M7240) antibodies were obtained
for nuclear labeling from Ventana Medical Systems, Inc. The
incubation process and secondary antibody was identical to that
aforementioned. For P53, a labeling score of >30% was regarded
as aberrant overexpression, which has been previously associated
with p53 mutation (34). A cut-off
point of 13% Ki67 expression was used to categorize high- or
low-proliferation tumors (35).
Luminal tumors were categorized as immunoreactive for ER and/or PR,
negative for HER-2 expression or exhibiting low proliferation. ER+
and/or PR+ tissues that were also HER-2+ and/or exhibiting high
proliferation were considered to be luminal B tumors. HER-2
subtypes were defined as ER-, PR-, and HER-2+. Based on published
criteria, all basal-like cases were considered to have a
triple-negative phenotype (ER-/PR-/HER-2-).
DNA extraction
Genomic DNA was extracted from the fresh-frozen
primary breast tumor tissues and the matched healthy breast
tissues. Samples were pre-treated with 20 mg/ml proteinase K
(Promega Corporation, Madison, WI, USA) at 55°C overnight. DNA was
extracted using the AxyPrep™ Multisource Genomic DNA
Miniprep kit (Axygen; Corning Incorporated, Corning, NY, USA),
according to manufacturer's protocol, and ~5 ml of peripheral blood
was collected prior to the physical examination or surgery. All
samples were analyzed in the laboratory within 4 h. Circulating
free DNA was obtained from 1 ml of serum using the QIAamp UltraSens
Virus kit (cat. no. 53706; Qiagen GmbH, Hilden, Germany).
Bisulfite conversion and MethyLight
assay by quantitative polymerase chain reaction (qPCR)
The bisulfite conversion of genomic DNA and
MethyLight assay was performed using the EZ DNA Methylation kit
(Zymo Research Corp., Irvine, CA, USA), according to the
manufacture's protocol. For each bisulfite sequencing PCR (BSP)
reaction, the PCR mixture included 1.5 mM MgCl2, 200 µM
dNTP, 1 µM of forward and reverse primers (sequences in Table I), 2.5 units of Platinum Taq
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
1× Platinum Taq buffer to achieve a final reaction volume of 50 µl.
The detailed method was described in a previous study (36). The PCR products were extracted using a
1% agarose gel, ligated into the pGEM-T vector (Promega
Corporation). The proportion of PCR products and the conversion of
Escherichia coli (strain DH5α) was conducted according to
standardized procedures. Blue-white screening was used to select a
minimum of 10 positive bacterial clones, from which plasmid DNA was
isolated using a QIAprep Spin Miniprep kit (Qiagen, Inc., Valencia,
CA, USA), according the to the manufacture's protocol. Clones were
screened by digesting 1 µg of plasmid DNA with BstZI
(Promega Corporation) and 1% agarose gel electrophoresis was
performed to confirm the insert and plasmid size as reported
previously (37). Positive clones
were sequenced by Invitrogen (Thermo Fisher Scientific, Inc.).
 | Table I.The sequences of probes and primers
used polymerase chain reaction and methylation analysis. |
Table I.
The sequences of probes and primers
used polymerase chain reaction and methylation analysis.
| Primer | Sequence |
|---|
|
Methylation-specific primers |
|
|
Forward |
5′-TCGTTAAATAGATACGTTACGC-3′ |
|
Reverse |
5′-TAAAAACTAAAAACTTTCCGCG-3′ |
| TaqMan
MGB probe |
5′-TGGTTAAGGGTTCGGTGGT-3′ |
| Globin reference
primers |
|
|
Forward |
5′-AGGTAGAAAAGGAGAATGAAGATAAA-3′ |
|
Reverse |
5′-CTTTCCACTCTTTTCTCATTCTCTC-3′ |
| TaqMan MGB
probe |
5′-AGGAGGATAAGGAAGAGGGGAAATAGG-3′ |
The probable promoter CpG island methylated sites
were selected to design probes for the PCDH10 gene, in accordance
with the results of BSP sequencing. CpG methylation of the PCDH10
gene was detected using methylation-specific primers (Table I) and the TaqMan® MGB-based
probe fluorescence real-time qPCR kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
TaqMan® MGB qPCR was performed with primers specific to
the bisulfite-converted methylated sequences of particular loci, as
well as with globin reference primers and TaqMan® MGB
probes (Table I). TaqMan MGB probes
increased assay specificity and facilitated flexible analysis due
to their small size. The thermocycling conditions were as follows:
Denaturation at 96°C for 5 min; 35 cycles of amplification at 95°C
for 10 sec/cycle; and then 60°C for 30 sec. The quantification of
the methylation rate (%) at a promoter was calculated using the
2−ΔΔCq method (38). All
samples were assayed in duplicate, to confirm the accuracy of the
2−ΔΔCq method and the amplification efficiency. Globin
was analyzed using serial dilutions of DNA with a 100-fold range
and gene-specific primers for each gene and globin (Table I). The ΔCq value [Cq (target gene)-Cq
(reference gene)] was obtained for each DNA dilution and log DNA
dilution vs. ΔCq was plotted.
MethyLight is a high-throughput assay for DNA
methylation based on real time qPCR. MethyLight requires only
minute amounts of modest quality DNA, making it clinically
applicable and compatible for use with small biopsies and
paraffin-embedded tissues (39,40).
Researchers have used percentage of methylated reference (PMR) to
evaluate positive methylation. However, the cut-off value of PMR
using MethyLight has been reported to vary among studies (41–44). This
may be a result of not including matched normal tissue, but only
Sss I-treated human peripheral white-blood-cell DNA from the same
patient or from healthy subjects as a control. This method of
comparison cannot accurately reflect the number of positive
methylation cases, as methylation modification is influenced by
many factor, including lifestyle, environmental exposures,
ethnicity, age and tissue heterogeneity (40,45). The
present study used breast cancer tissue and matched healthy breast
tissue and methylation percentage of a particular area was assessed
using the 2−ΔΔCq method. A value ≥1.5 (allelic gene
methylation) was considered to be positive. MethyLight was also
used to determine the methylation frequency of serum DNA (46,47).
Statistical analysis
The levels of methylation and mRNA expression were
analyzed using Fisher's exact test and Kruskal-Wallis test.
Receiver operating characteristic (ROC) curve analysis,
Mann-Whitney U test, Kaplan-Meier survival curves and Cox
proportional hazards regression model were used to assess
prognostic associations. All tests were performed using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Epigenetic identification of the
PCDH10 CGI as a hypermethylated sequence in breast cancer
Using bioinformatics analysis to identify DNA
fragments (n=31) of the PCDH family, the proximal promoter and exon
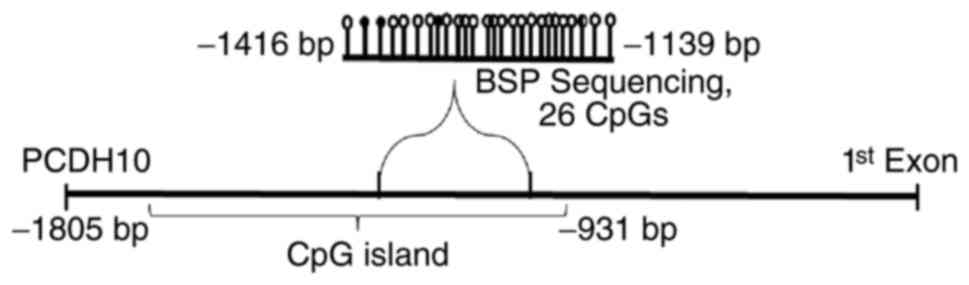
1 of the PCDH10 gene were located in 4q28.3 (48). The PCDH10 gene promoter region
comprises two classic CpG islands: CGI-1, −2133 to −854 bp from the
transcription start site (TSS), and CGI-2, +367 to +1972 bp from
the TSS (Accession no. NM_032961.1). PCDH10 gene promoters in tumor
tissue were prescreened and matched with healthy tissues from 6
patients with breast cancer to detect the presence of aberrant
methylation targets and to design TaqMan® MGB detection
probes, in order to investigate methylation status. Following
comparison of the methylation frequency between breast cancer
tissues and matched healthy breast tissues, it was determined that
PCDH10 gene methylation was significantly increased in breast
cancer. The CpG site analysis demonstrated that the PCDH10 promoter
is a typical CGI (Fig. 1) (49).
Prevalence of PCDH10 methylation and
mRNA expression in breast cancer tissues
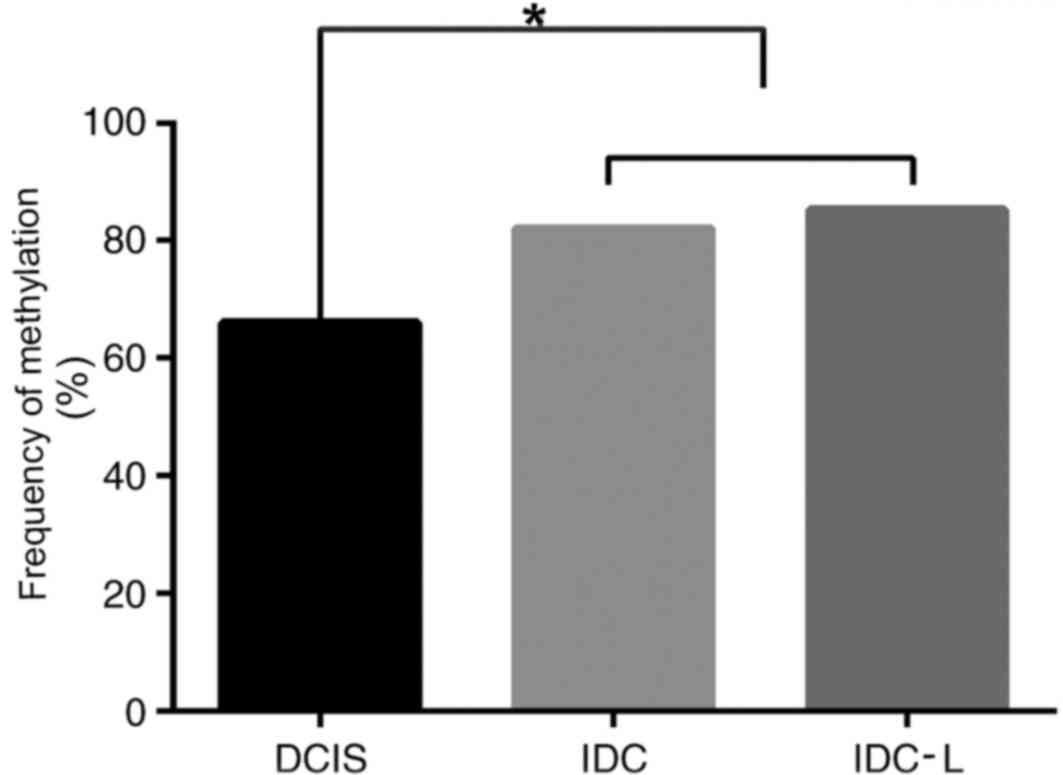
PCDH10 displayed widespread methylation of the
aberrant promoter CpG island. The frequency of PCDH10 methylation
significantly increased with disease progression from in
situ to invasive cancer. However, no significant difference in
methylation was observed between IDC and IDC-L (P<0.05; Fig. 2). The results of methylation frequency
analysis for PCDH10 are indicated in Table II. There was no significant
difference between the frequency of PCDH10 gene methylation in
sporadic breast cancer tissues and HpBC tissues (P>0.05). PCDH10
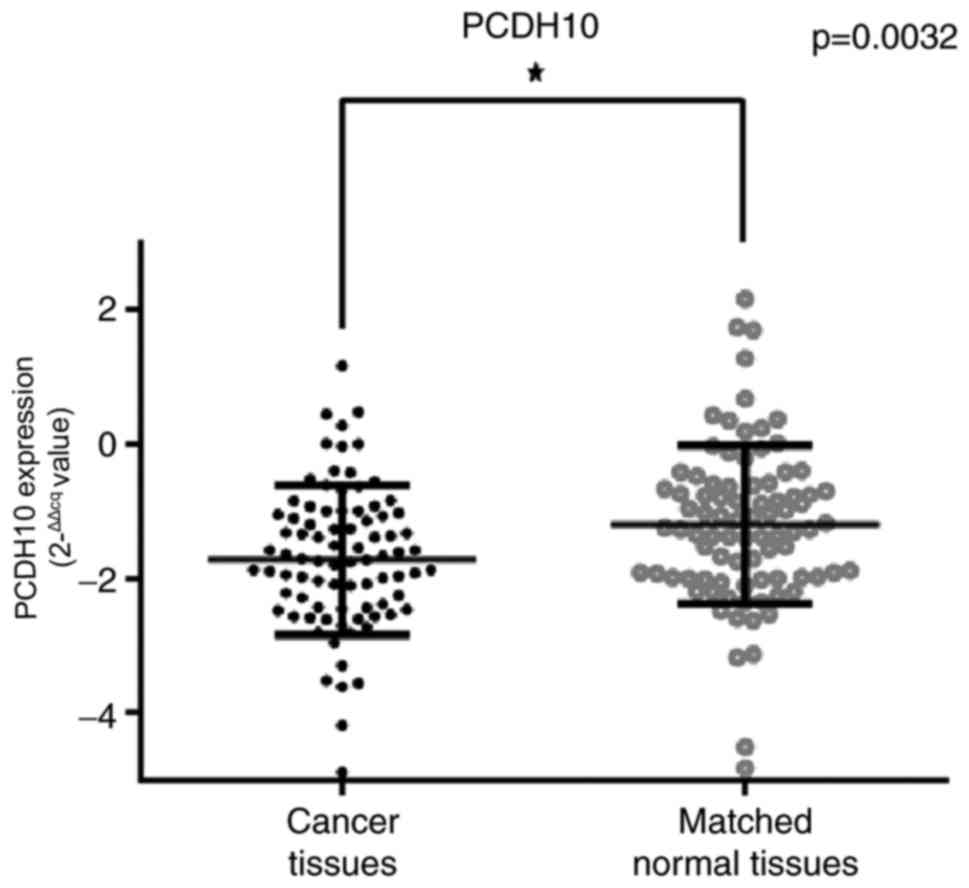
mRNA expression in breast cancer tissues and paired healthy breast
tissues was calculated using the 2−ΔΔCq method (38). PCDH10 mRNA expression was indicated to
be significantly decreased in breast cancer tissues, compared with
matched healthy breast tissues, according to qPCR results
(P=0.0032; Fig. 3).
 | Table II.Frequency of PCDH10 methylation in
patients with sporadic and hereditary breast cancer. |
Table II.
Frequency of PCDH10 methylation in
patients with sporadic and hereditary breast cancer.
|
| Breast cancer
tissue methylation frequency (%) |
|---|
|
|
|
|---|
|
| Sporadic
(n=296) |
|
|---|
|
|
|
|
|---|
| Methylated
gene | DCIS | IDC | IDC-L | HpBC (n=96) |
|---|
| PCDH10 | 66 | 82 | 85.32 | 72.37 |
Association of PCDH10 methylation with
clinicopathological characteristics and breast cancer
prognosis
The association between PCDH10 methylation and
various clinicopathological features was also examined (Table III). PCDH10 methylation was
significantly associated with tumor size (P=0.004). In the tumor
size ≥2 cm group, the rate of PCDH10 methylation (76.1%) was
significantly increased compared with unmethylated PCDH10 (56.6%)
(P=0.004). However, no association was observed between PCDH10
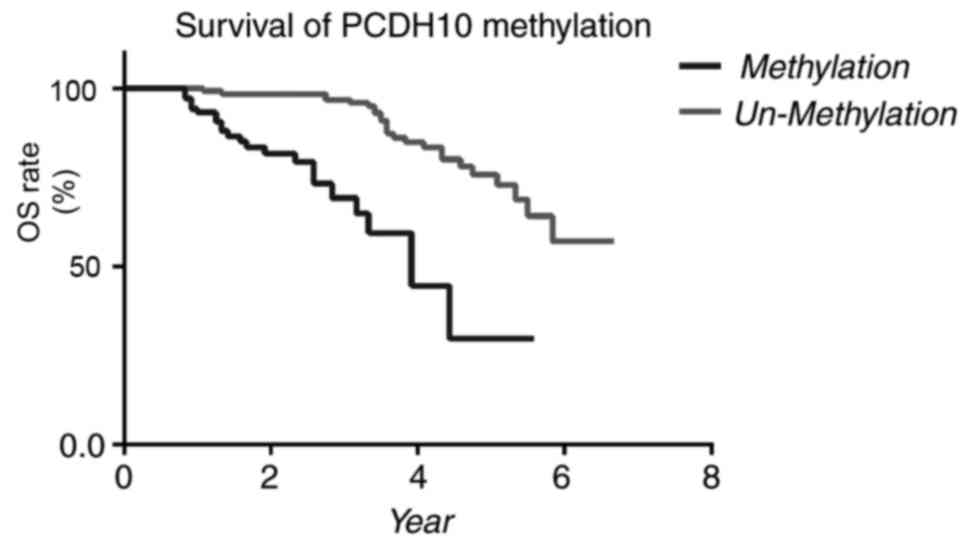
methylation and other clinicopathological factors (Fig. 4). Patients exhibiting methylated
PCDH10 had significantly lower OS times (P<0.0001, log-rank
test) compared with patients exhibiting unmethylated PCDH10. In the
Cox proportional hazards regression model, univariate and
multivariate survival analyses were applied to assess the
association between PCDH10 methylation and clinicopathological
features. Univariate analyses of OS rate demonstrated that tumor
size (P<0.001), lymph node metastasis (LNM) (P<0.001), ER
(P=0.023) and PCDH10 methylation (P=0.006) were associated with OS
rate, while other factors were not associated with OS rate.
Multivariate analysis revealed that tumor size (P<0.001), LNM
(P<0.001) and PCDH10 methylation (P=0.012) were independent
predictors of prognosis (Table
IV).
 | Table III.Association between PCDH10
methylation and different clinicopathological parameters. |
Table III.
Association between PCDH10
methylation and different clinicopathological parameters.
|
|
| Unmethylated
PCDH10 | Methylated
PCDH10 |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Variables | Total, n | n | % | n | % | χ2 | P-value |
|---|
| Age (years) |
|
|
|
|
| 0.798 | 0.372 |
|
<45 | 128 | 60 | 36.1 | 68 | 30.1 |
|
|
|
≥45 | 264 | 106 | 63.9 | 158 | 69.9 |
|
|
| Grade |
|
|
|
|
| 0.168 | 0.682 |
|
I+II | 360 | 154 | 92.8 | 206 | 91.2 |
|
|
|
III | 32 | 12 | 7.2 | 20 | 8.8 |
|
|
| Tumor size
(cm) |
|
|
|
|
| 8.325 | 0.004a |
|
<2 | 126 | 72 | 43.4 | 54 | 23.9 |
|
|
| ≥2 | 266 | 94 | 56.6 | 172 | 76.1 |
|
|
| LNM |
|
|
|
|
| 0.040 | 0.841 |
|
Negative | 214 | 92 | 55.4 | 122 | 54.0 |
|
|
|
Positive | 178 | 74 | 44.6 | 104 | 46.0 |
|
|
| ER |
|
|
|
|
| 0.221 | 0.638 |
|
Negative | 106 | 42 | 25.3 | 64 | 28.3 |
|
|
|
Positive | 286 | 124 | 74.7 | 162 | 71.7 |
|
|
| PR |
|
|
|
|
| 0.950 | 0.330 |
|
Negative | 138 | 52 | 31.3 | 86 | 38.1 |
|
|
|
Positive | 254 | 114 | 68.7 | 140 | 61.9 |
|
|
| HER-2 |
|
|
|
|
| 2.276 | 0.131 |
|
Negative | 280 | 128 | 77.1 | 152 | 67.3 |
|
|
|
Positive | 112 | 38 | 22.9 | 74 | 32.7 |
|
|
| P53 |
|
|
|
|
| 0.285 | 0.594 |
|
Negative | 324 | 140 | 84.3 | 184 | 81.4 |
|
|
|
Positive | 68 | 26 | 15.7 | 42 | 18.6 |
|
|
| Ki-67 |
|
|
|
|
| 0.791 | 0.374 |
|
<20% | 198 | 90 | 54.2 | 108 | 47.8 |
|
|
|
≥20% | 194 | 76 | 45.8 | 118 | 52.2 |
|
|
| Molecular
subtype |
|
|
|
|
| 3.399 | 0.334 |
| Luminal
A | 226 | 102 | 61.4 | 124 | 54.9 |
|
|
| Luminal
B | 64 | 24 | 14.5 | 40 | 17.7 |
|
|
|
HER-2 | 50 | 14 | 8.4 | 36 | 15.9 |
|
|
|
TNBC | 52 | 26 | 15.7 | 26 | 11.5 |
|
|
 | Table IV.Cox proportional hazards assessment
of prognostic factors. |
Table IV.
Cox proportional hazards assessment
of prognostic factors.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≥45 vs. <45
years) | 0.879 | (0.544–1.416) | 0.591 |
|
|
|
| Grade (III vs.
II+I) | 0.732 | (0.282–1.780) | 0.482 |
|
|
|
| Tumor size (≥2 vs.
<2 cm) | 3.712 | (2.039–7.569) |
<0.00a | 3.115 | (1.597–6.274) |
<0.00a |
| LNM (positive vs.
negative) | 2.531 | (1.903–4.181) |
<0.001a | 2.337 | (1.939–3.842) |
<0.001a |
| ER (positive vs.
negative) | 0.682 | (0.464–0.929) | 0.026a |
|
|
|
| PR (positive vs.
negative) | 0.744 | (0.539–1.197) | 0.127 |
|
|
|
| Her2 (positive vs.
negative) | 1.547 | (0.983–2.119) | 0.115 |
|
|
|
| p53 (positive vs.
negative) | 1.411 | (0.886–2.124) | 0.298 |
|
|
|
| Ki-67 (positive vs.
negative) | 1.269 | (0.846–1.692) | 0.534 |
|
|
|
| PCDH10 (methylated
vs. unmethylated) | 1.780 | (1.322–3.117) | 0.006a | 1.798 | (1.231–3.071) | 0.011a |
The results of the present study demonstrated that
the frequency of gene methylation was significantly increased in
breast cancer tissues compared with control. Methylation was
investigated as a possible diagnostic predictor of breast cancer.
Identical specific probes were used to assess PCDH10 methylation in
matched serum samples using MethyLight. Samples included 300
age-matched healthy controls and 300 age-matched patients with
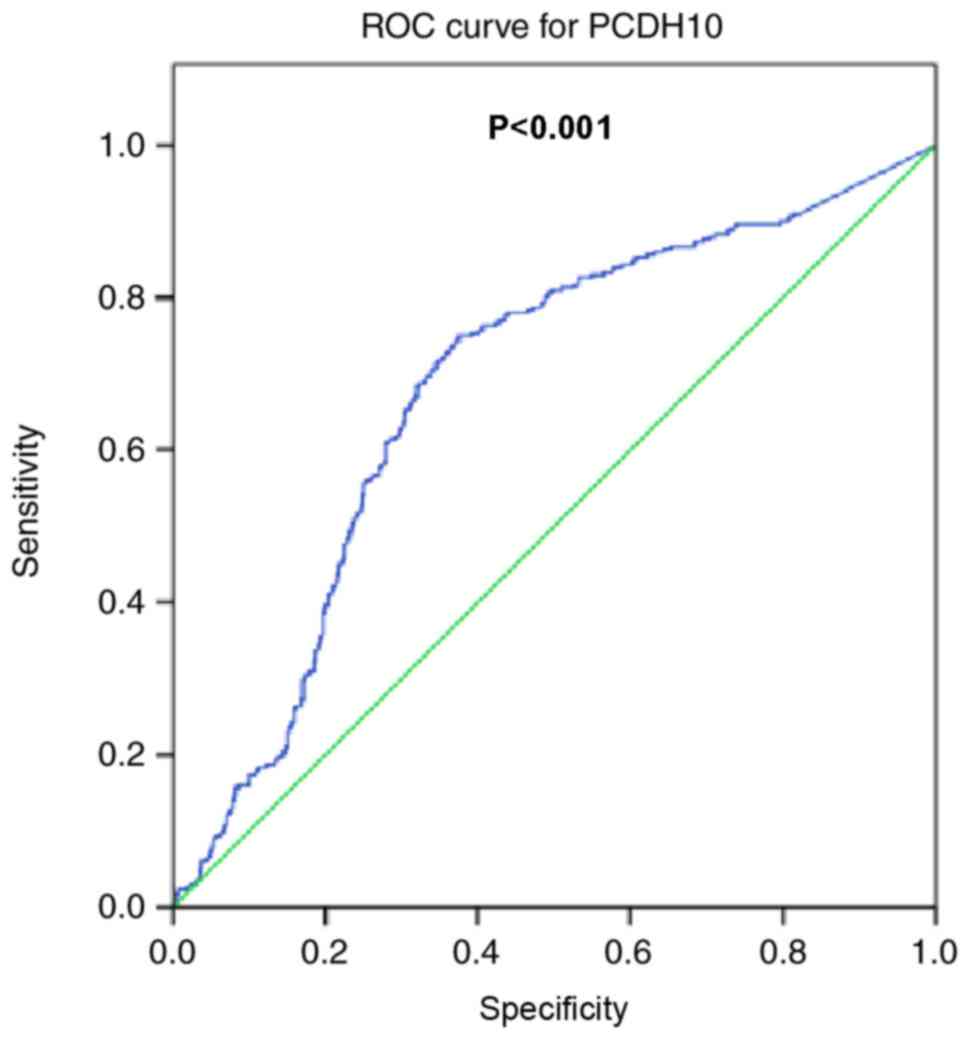
benign breast diseases. ROC analysis of PCDH10 indicated a
sensitivity of 75%, a specificity of 62.5%, and an area under the
curve of 0.682 (95% confidence interval; 0.645–0.719; P<0.001;
Fig. 5).
Discussion
Detection of hypermethylation is an important tool
in the diagnosis of cancer and in prognostic and therapeutic
guidance (23,50,51).
Cadherins, which serve a crucial role in cellular adhesion, are
involved in tumorigenesis and progression (52). There are three primary types of
cadherin: Classical cadherins, desmosomal cadherins and PCDHs.
E-cadherin has been reported to act as a tumor suppressor and to
participate in carcinogenesis, particularly in breast and gastric
cancer (53). PCDHs constitute one of
the largest subgroups of the cadherin superfamily. Previous studies
have indicated that various types of cancer are associated with
abnormal promoter methylation of cadherin family genes (54). It has been reported that the
expression of PCDH is dependent on epigenetic modifications
(55). In addition, promoter
methylation has been indicated to trigger inactivation of
PCDH-17/20 in various types of cancer. Therefore, aberrant promoter
methylation may be a competent diagnostic and prognostic biomarker
for cancer (55–57).
It has been reported that PCDH10 suppresses
proliferation, metastasis and invasion of cancer cells (58–60).
Promoter methylation and transcriptional silencing of PCDH1 has
been reported in various types of cancer, including myeloma,
nasopharyngeal, esophageal, colorectal, cervical, lung and
hepatocellular carcinoma cell lines (26,61–64). As
the present study demonstrated, silencing of PCDH10 methylation
also occurs in breast cancer cells.
MethyLight was used in the present study to identify
hypermethylation-silenced genes in tumor tissues. These genes may
be candidate tumor suppressor genes (TSGs). PCDH10 is frequently
silenced by methylation in a tumor-specific manner (26). The results of the present study
suggest that PCDH10 serves a critical role in cancer suppression,
and that PCDH10 silencing is associated with tumor growth and
progression. In addition, PCDH10 participates in nervous system
development (26), and functions as a
TSG in various types of cancer. Numerous studies have reported that
the frequency of methylation significantly increases with cancer
progression, and that methylation may be a potential prognostic
predictor in breast cancer (47,65).
Breast cancer tissues and paired healthy tissues
were examined and PCDH10 methylation was identified in ~72% of
breast cancer tissues. It was also determined that the rate of low
PCDH10 expression rate was ~70%. DNA methylation can silence TSGs
and, therefore, we hypothesized that decreased PCDH10 expression
may be attributed to its methylation. Using a large sample size, it
was concluded that PCDH10 methylation occurs in the majority of
breast cancer cases. The present study demonstrated that PCDH10
methylation is associated with tumor size (P=0.004). In the tumor
size ≥2 cm group, methylated PCDH10 tissues were more prevalent
than unmethylated PCDH10 tissues. This indicates that DNA
methylation may serve an important role in breast cancer. The
present study, to the best of our knowledge, is the first to
investigate the prognostic value of PCDH10 gene promoter
methylation in patients with breast cancer. Patients with PCDH10
methylation exhibited notably reduced OS rates compared with
patients exhibiting unmethylated PCDH10 (P=0.005, log-rank test).
Therefore, PCDH10 methylation is indicated to be independently
associated with poor prognosis in patients with invasive breast
cancer. The present study is the first, to the best of our
knowledge, to illustrate the association between DNA methylation,
clinicopathological characteristics and survival in Chinese
patients with breast cancer.
Furthermore, cell-free DNA detected in serum and
extracted from cancer cells has been used as a non-invasive
biomarker to facilitate diagnosis and prognostic guidance for
various types of cancer, including gastric cancer (66,67).
Another study also detected PCDH10 methylation in serum, providing
further evidence of clinical relevance in other types of cancer,
including prostate cancer (68).
Methylation detection has the potential to improve the existing
understanding of the development of cancer, including tumor
differentiation, stage and distant metastasis.
In conclusion, the results of the present study
suggest that frequency of PCDH10 promoter methylation is increased
in human breast cancer tissues, compared with adjacent non-tumor
tissues. PCDH10 was concluded to be an important TSG, which
restricted the progression of breast cancer. In addition, the
methylation status of PCDH10 may be a useful diagnostic and
prognostic biomarker for breast cancer. However, it has been
reported that environmental and lifestyle factors can alter the
status of DNA methylation (69,70). The
present study did not exclude the effects of smoking and alcohol
intake on methylation, and therefore, further investigation is
required to consider this factor.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The data used and analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
QZ designed the study and revised the paper. WL and
JW performed the studies. GS, XY and DL performed the statistical
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This research was completed in compliance with the
Helsinki Declaration. The data collection and analysis were
conducted without disclosing patients' identities. All patients
provided written informed consent for tissue and serum collection,
in consistence with regulations of the institutional review board
of the Harbin Medical University (Heilongjiang, China).
Patient consent for publication
All patients provided written informed consent for
publication, in consistence with regulations of the institutional
review board of the Harbin Medical University (Heilongjiang,
China).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Seneviratne S, Lawrenson R, Scott N, Kim
B, Shirley R and Campbell I: Breast cancer biology and ethnic
disparities in breast cancer mortality in new zealand: A cohort
study. PLoS One. 10:e01235232015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frazer KA, Murray SS, Schork NJ and Topol
EJ: Human genetic variation and its contribution to complex traits.
Nat Rev Genet. 10:241–251. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rivera CM and Ren B: Mapping human
epigenomes. Cell. 155:39–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer-a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feinberg AP, Ohlsson R and Henikoff S: The
epigenetic progenitor origin of human cancer. Nat Rev Genet.
7:21–33. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esteller M, Fraga MF, Guo M,
Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J,
Vaurs-Barrière C, Bignon YJ, Ramus S, et al: DNA methylation
patterns in hereditary human cancers mimic sporadic tumorigenesis.
Hum Mol Genet. 10:3001–3007. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Craig JM and Bickmore WA: The distribution
of CpG islands in mammalian chromosomes. Nat Genet. 7:376–382.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eden A, Gaudet F, Waghmare A and Jaenisch
R: Chromosomal instability and tumors promoted by DNA
hypomethylation. Science. 300:4552003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fernandez AF, Assenov Y, Martin-Subero JI,
Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC,
Galm O, et al: A DNA methylation fingerprint of 1628 human samples.
Genome Res. 22:407–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moarii M, Boeva V, Vert JP and Reyal F:
Changes in correlation between promoter methylation and gene
expression in cancer. BMC Genomics. 16:8732015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu YY, Sun CX, Liu YK, Li Y, Wang L and
Zhang W: Genome-wide screen of ovary-specific DNA methylation in
polycystic ovary syndrome. Fertil Steril. 104(145–153): e62015.
|
|
16
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Neste L, Herman JG, Otto G, Bigley JW,
Epstein JI and Van Criekinge W: The epigenetic promise for prostate
cancer diagnosis. Prostate. 72:1248–1261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lo PK and Sukumar S: Epigenomics and
breast cancer. Pharmacogenomics. 9:1879–1902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan M, Su Y, Kang W, Gao R, Li X and
Zhang G: Aberrant expression and functions of protocadherins in
human malignant tumors. Tumour Biol. 37:12969–12981. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Otani K, Li X, Arakawa T, Chan FK and Yu
J: Epigenetic-mediated tumor suppressor genes as diagnostic or
prognostic biomarkers in gastric cancer. Expert Rev Mol Diagn.
13:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN,
Geng H, Tian LW, Wong YP, Tong JH, Ying JM, et al: Methylation of
protocadherin 10, a novel tumor suppressor, is associated with poor
prognosis in patients with gastric cancer. Gastroenterology.
136(640–651): e12009.
|
|
22
|
Lv J, Zhu P, Yang Z, Li M, Zhang X, Cheng
J, Chen X and Lu F: PCDH20 functions as a tumour-suppressor gene
through antagonizing the Wnt/β-catenin signalling pathway in
hepatocellular carcinoma. J Viral Hepat. 22:201–211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang C, Peng Y, Yang F, Qin R, Liu W and
Zhang C: PCDH8 is frequently inactivated by promoter
hypermethylation in liver cancer: Diagnostic and clinical
significance. J Cancer. 7:446–452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee NK, Lee JH, Kim WK, Yun S, Youn YH,
Park CH, Choi YY, Kim H and Lee SK: Promoter methylation of PCDH10
by HOTAIR regulates the progression of gastrointestinal stromal
tumors. Oncotarget. 7:75307–75318. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyamoto K, Fukutomi T, Akashi-Tanaka S,
Hasegawa T, Asahara T, Sugimura T and Ushijima T: Identification of
20 genes aberrantly methylated in human breast cancers. Int J
Cancer. 116:407–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying J, Li H, Seng TJ, Langford C,
Srivastava G, Tsao SW, Putti T, Murray P, Chan AT and Tao Q:
Functional epigenetics identifies a protocadherin PCDH10 as a
candidate tumor suppressor for nasopharyngeal, esophageal and
multiple other carcinomas with frequent methylation. Oncogene.
25:1070–1080. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Foulkes WD: Inherited susceptibility to
common cancers. N Engl J Med. 359:2143–2153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pharoah PD, Day NE, Duffy S, Easton DF and
Ponder BA: Family history and the risk of breast cancer: A
systematic review and meta-analysis. Int J Cancer. 71:800–809.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Porter P: ‘Westernizing’ women's risks?
Breast cancer in lower-income countries. N Engl J Med. 358:213–216.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lynch HT, Silva E, Snyder C and Lynch JF:
Hereditary breast cancer: Part I. Diagnosing hereditary breast
cancer syndromes. Breast J. 14:3–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Majdak-Paredes EJ and Fatah F: Hereditary
breast cancer syndromes and clinical implications. J Plast Reconstr
Aesthet Surg. 62:181–189. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ogura H, Akiyama F, Kasumi F, Kazui T and
Sakamoto G: Evaluation of HER-2 status in breast carcinoma by
fluorescence in situ hybridization and immunohistochemistry. Breast
Cancer. 10:234–240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Soong R, Robbins PD, Dix BR, Grieu F, Lim
B, Knowles S, Williams KE, Turbett GR, House AK and Iacopetta BJ:
Concordance between p53 protein overexpression and gene mutation in
a large series of common human carcinomas. Hum Pathol.
27:1050–1055. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hugh J, Hanson J, Cheang MC, Nielsen TO,
Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, et
al: Breast cancer subtypes and response to docetaxel in
node-positive breast cancer: Use of an immunohistochemical
definition in the BCIRG 001 trial. J Clin Oncol. 27:1168–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu Y, Yan W, Liu X, Jia Y, Cao B, Yu Y, Lv
Y, Brock MV, Herman JG, Licchesi J, et al: DACT2 is frequently
methylated in human gastric cancer and methylation of DACT2
activated Wnt signaling. Am J Cancer Res. 4:710–724.
2014.PubMed/NCBI
|
|
37
|
Koontz L: Agarose gel electrophoresis.
Methods Enzymol. 529:35–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: MethyLight: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kagan J, Srivastava S, Barker PE, Belinsky
SA and Cairns P: Towards clinical application of methylated DNA
sequences as cancer biomarkers: A Joint NCI's EDRN and NIST
workshop on standards, methods, assays, reagents and tools. Cancer
Res. 67:4545–4549. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ogino S, Kawasaki T, Brahmandam M, Cantor
M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ,
Laird PW, Loda M and Fuchs CS: Precision and performance
characteristics of bisulfite conversion and real-time PCR
(MethyLight) for quantitative DNA methylation analysis. J Mol
Diagn. 8:209–217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Widschwendter M, Siegmund KD, Müller HM,
Fiegl H, Marth C, Müller-Holzner E, Jones PA and Laird PW:
Association of breast cancer DNA methylation profiles with hormone
receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Widschwendter M, Apostolidou S, Jones AA,
Fourkala EO, Arora R, Pearce CL, Frasco MA, Ayhan A, Zikan M,
Cibula D, et al: HOXA methylation in normal endometrium from
premenopausal women is associated with the presence of ovarian
cancer: A proof of principle study. Int J Cancer. 125:2214–2218.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho YH, Shen J, Gammon MD, Zhang YJ, Wang
Q, Gonzalez K, Xu X, Bradshaw PT, Teitelbaum SL, Garbowski G, et
al: Prognostic significance of gene-specific promoter
hypermethylation in breast cancer patients. Breast Cancer Res
Treat. 131:197–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang W and Srivastava S: Strategic
approach to validating methylated genes as biomarkers for breast
cancer. Cancer Prev Res (Phila). 3:16–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Clark SJ and Melki J: DNA methylation and
gene silencing in cancer: Which is the guilty party? Oncogene.
21:5380–5387. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Muggerud AA, Rønneberg JA, Wärnberg F,
Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I,
Børresen-Dale AL, Kristensen VN, et al: Frequent aberrant DNA
methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma
in situ and early invasive breast cancer. Breast Cancer Res.
12:R32010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wolverton T and Lalande M: Identification
and characterization of three members of a novel subclass of
protocadherins. Genomics. 76:66–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qiu GH, Tan LK, Loh KS, Lim CY, Srivastava
G, Tsai ST, Tsao SW and Tao Q: The candidate tumor suppressor gene
BLU, located at the commonly deleted region 3p21.3, is an
E2F-regulated, stress-responsive gene and inactivated by both
epigenetic and genetic mechanisms in nasopharyngeal carcinoma.
Oncogene. 23:4793–4806. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jeschke J, Van Neste L, Glöckner SC, Dhir
M, Calmon MF, Deregowski V, Van Criekinge W, Vlassenbroeck I, Koch
A, Chan TA, et al: Biomarkers for detection and prognosis of breast
cancer identified by a functional hypermethylome screen.
Epigenetics. 7:701–709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Morris MR, Ricketts C, Gentle D,
Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F and
Maher ER: Identification of candidate tumour suppressor genes
frequently methylated in renal cell carcinoma. Oncogene.
29:2104–2117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Su YJ, Chang YW, Lin WH, Liang CL and Lee
JL: An aberrant nuclear localization of E-cadherin is a potent
inhibitor of Wnt/beta-catenin-elicited promotion of the cancer stem
cell phenotype. Oncogenesis. 4:e1572015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Patra SK and Bettuzzi S: Epigenetic
DNA-methylation regulation of genes coding for lipid
raft-associated components: A role for raft proteins in cell
transformation and cancer progression (review). Oncol Rep.
17:1279–1290. 2007.PubMed/NCBI
|
|
55
|
Imoto I, Izumi H, Yokoi S, Hosoda H,
Shibata T, Hosoda F, Ohki M, Hirohashi S and Inazawa J: Frequent
silencing of the candidate tumor suppressor PCDH20 by epigenetic
mechanism in non-small-cell lung cancers. Cancer Res. 66:4617–4626.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Haruki S, Imoto I, Kozaki K, Matsui T,
Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T and Inazawa
J: Frequent silencing of protocadherin 17, a candidate tumour
suppressor for esophageal squamous cell carcinoma. Carcinogenesis.
31:1027–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Uyen TN, Sakashita K, Al-Kzayer LF,
Nakazawa Y, Kurata T and Koike K: Aberrant methylation of
protocadherin 17 and its prognostic value in pediatric acute
lymphoblastic leukemia. Pediatr Blood Cancer. 64:2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qiu C, Bu X and Jiang Z: Protocadherin-10
acts as a tumor suppressor gene, and is frequently downregulated by
promoter methylation in pancreatic cancer cells. Oncol Rep.
36:383–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shi D, Murty VV and Gu W: PCDH10, a novel
p53 transcriptional target in regulating cell migration. Cell
Cycle. 14:857–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang Y, Jiang Y, Jiang M, Zhang J, Yang B,
She Y, Wang W, Deng Y and Ye Y: Protocadherin 10 inhibits cell
proliferation and induces apoptosis via regulation of DEP domain
containing 1 in endometrial endometrioid carcinoma. Exp Mol Pathol.
100:344–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang KH, Liu HW, Lin SR, Ding DC and Chu
TY: Field methylation silencing of the protocadherin 10 gene in
cervical carcinogenesis as a potential specific diagnostic test
from cervical scrapings. Cancer Sci. 100:2175–2180. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tang X, Yin X, Xiang T, Li H, Li F, Chen L
and Ren G: Protocadherin 10 is frequently downregulated by promoter
methylation and functions as a tumor suppressor gene in non-small
cell lung cancer. Cancer Biomark. 12:11–19. 2013. View Article : Google Scholar
|
|
63
|
Li Y, Yang ZS, Song JJ, Liu Q and Chen JB:
Protocadherin-10 is involved in angiogenesis and methylation
correlated with multiple myeloma. Int J Mol Med. 29:704–710. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhong X, Zhu Y, Mao J, Zhang J and Zheng
S: Frequent epigenetic silencing of PCDH10 by methylation in human
colorectal cancer. J Cancer Res Clin Oncol. 139:485–490. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Novak P, Jensen TJ, Garbe JC, Stampfer MR
and Futscher BW: Stepwise DNA methylation changes are linked to
escape from defined proliferation barriers and mammary epithelial
cell immortalization. Cancer Res. 69:5251–5258. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pimson C, Ekalaksananan T, Pientong C,
Promthet S, Putthanachote N, Suwanrungruang K and Wiangnon S:
Aberrant methylation of PCDH10 and RASSF1A genes in blood samples
for non-invasive diagnosis and prognostic assessment of gastric
cancer. Peer J. 4:e21122016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hou YC, Deng JY, Zhang RP, Xie XM, Cui JL,
Wu WP, Hao XS and Liang H: Evaluating the clinical feasibility: The
direct bisulfite genomic sequencing for examination of methylated
status of protocadherin10 (PCDH10) promoter to predict the
prognosis of gastric cancer. Cancer Biomark. 15:567–573. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Deng QK, Lei YG, Lin YL, Ma JG and Li WP:
Prognostic value of Protocadherin10 (PCDH10) methylation in serum
of prostate cancer patients. Med Sci Monit. 22:516–521. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Donkin I and Barres R: Sperm epigenetics
and influence of environmental factors. Mol Metab. Feb
27–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ingerslev LR, Donkin I, Fabre O, Versteyhe
S, Mechta M, Pattamaprapanont P, Mortensen B, Krarup NT and Barrès
R: Endurance training remodels sperm-borne small RNA expression and
methylation at neurological gene hotspots. Clin Epigenetics.
10:122018. View Article : Google Scholar : PubMed/NCBI
|