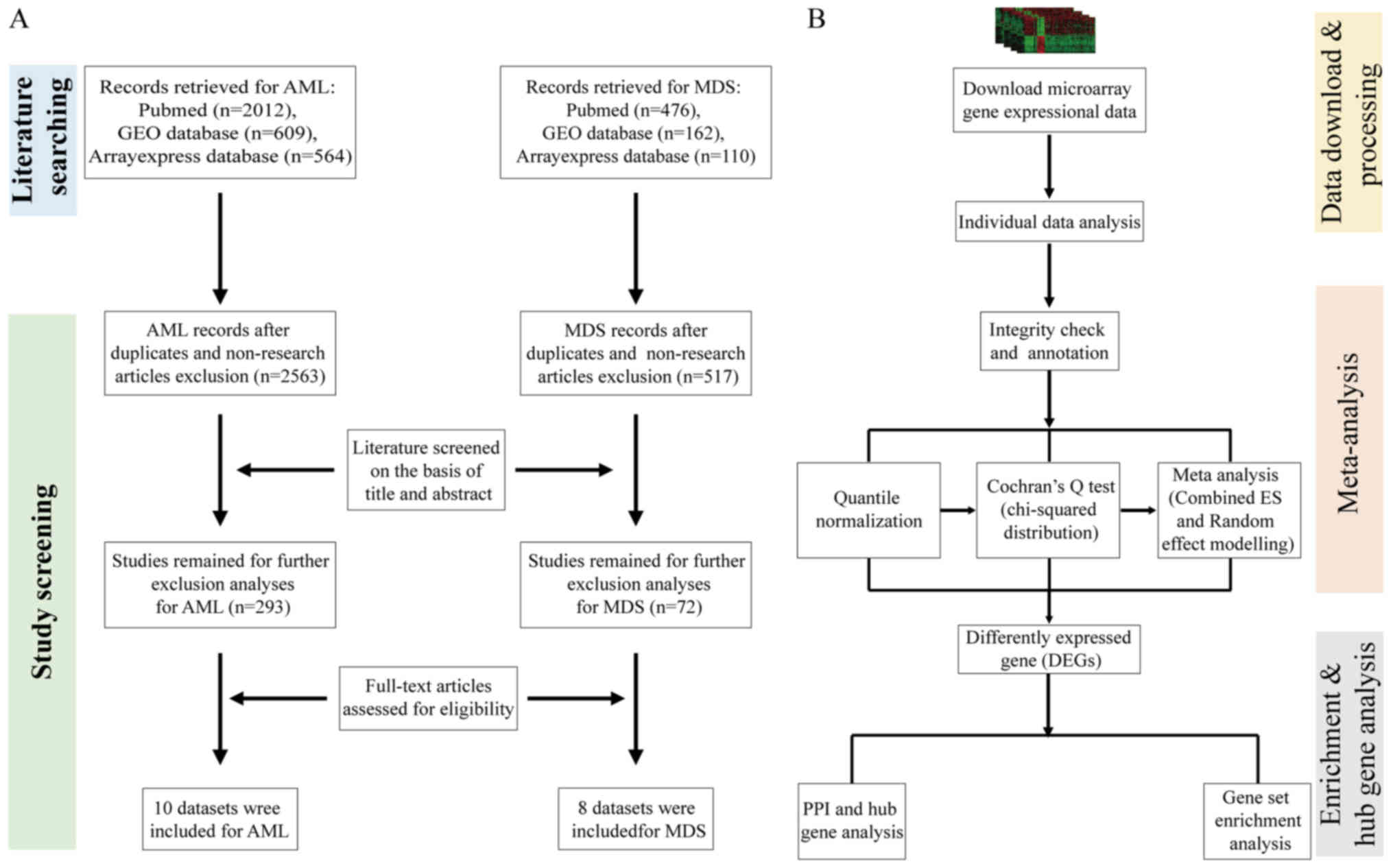
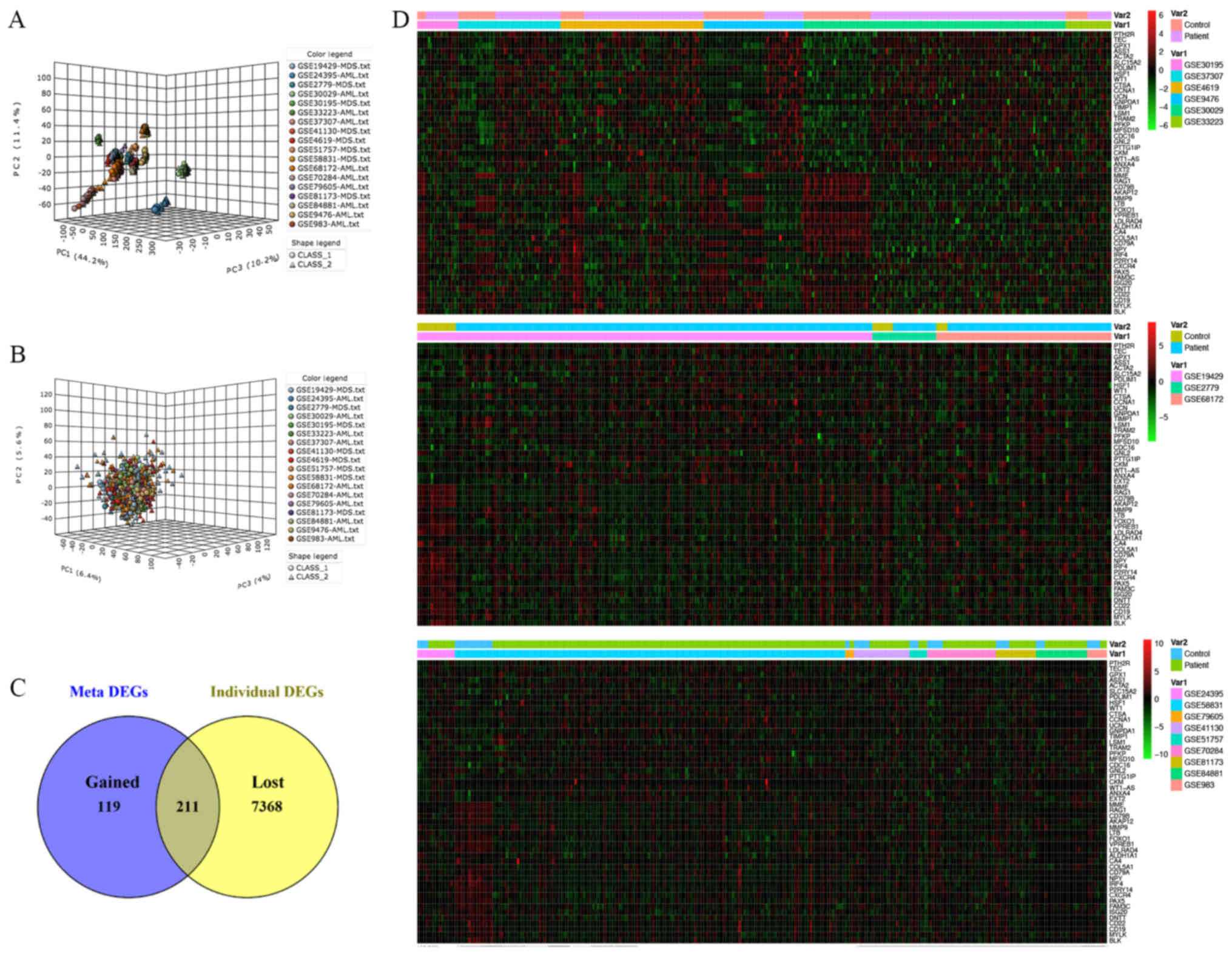
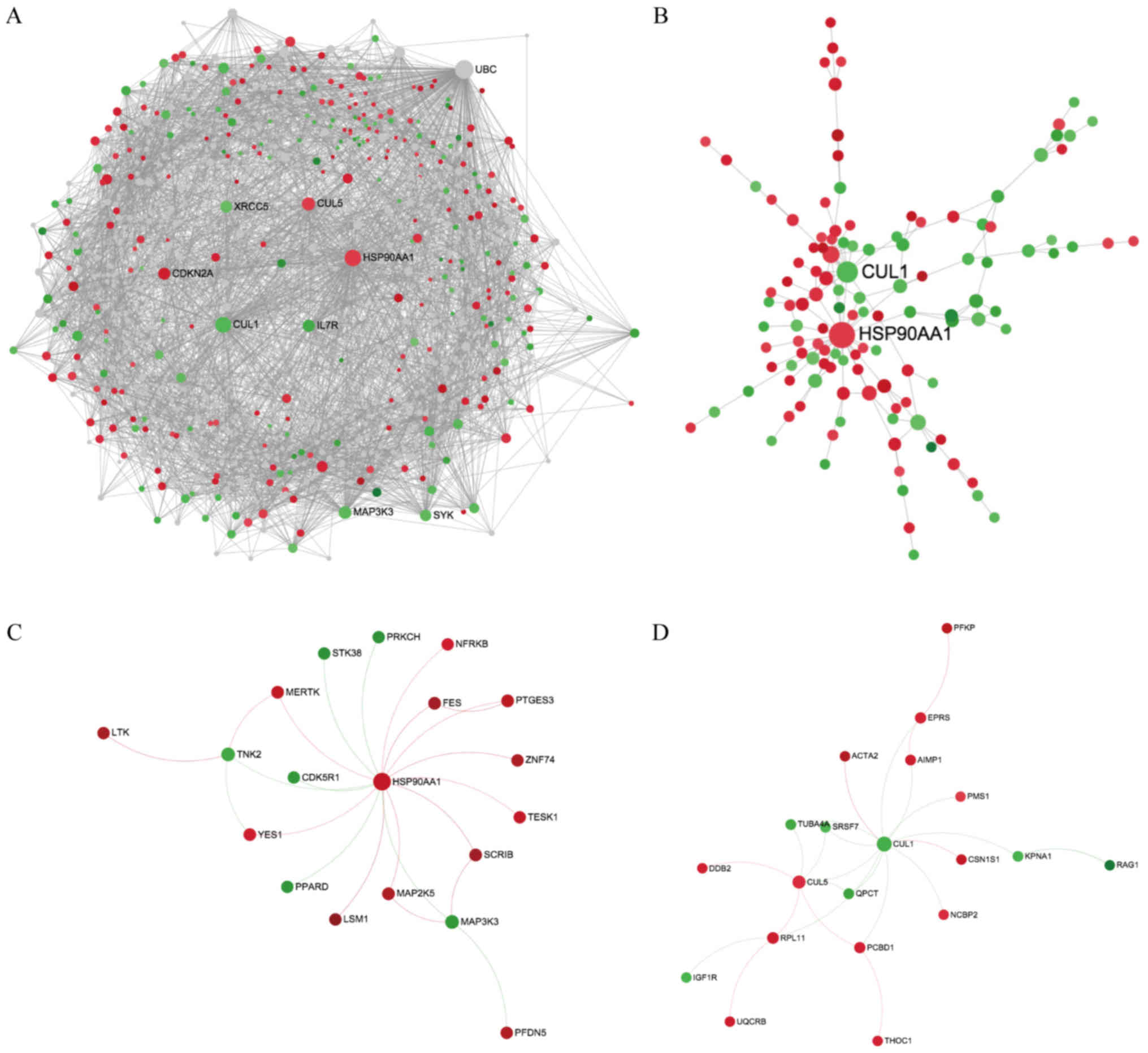
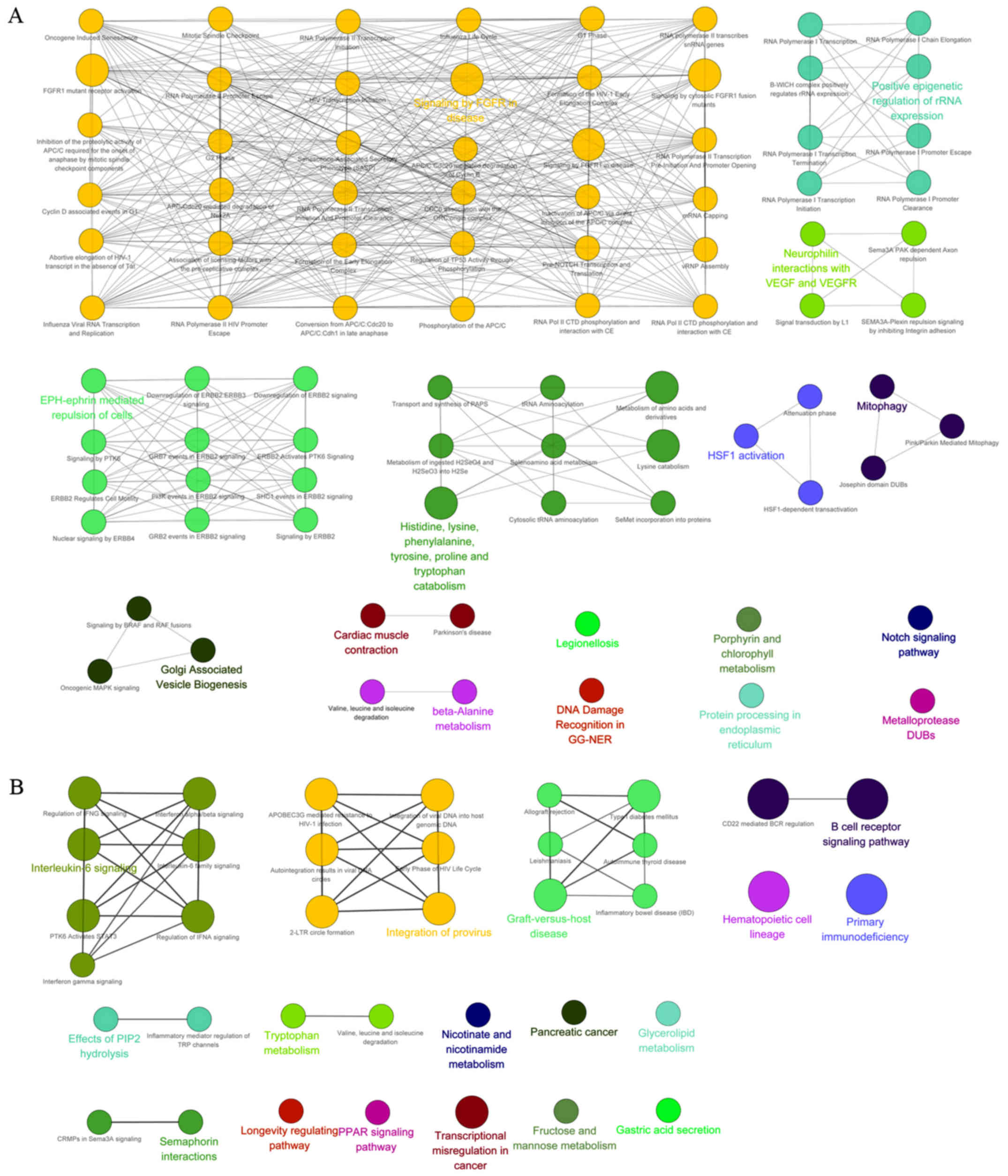
|
1
|
Morrison SJ and Scadden DT: The bone
marrow niche for haematopoietic stem cells. Nature. 505:327–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tenen DG, Hromas R, Licht JD and Zhang DE:
Transcription factors, normal myeloid development, and leukemia.
Blood. 90:489–519. 1997.PubMed/NCBI
|
|
3
|
Ozbalak M, Cetiner M, Bekoz H, Atesoglu
EB, Ar C, Salihoglu A, Tuzuner N and Ferhanoglu B: Azacitidine has
limited activity in ‘real life’ patients with MDS and AML: A single
centre experience. Hematol Oncol. 30:76–81. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greenberg PL, Young NS and Gattermann N:
Myelodysplastic syndromes. Hematology Am Soc Hematol Educ Program.
136–161. 2002.PubMed/NCBI
|
|
5
|
Hope KJ, Jin L and Dick JE: Acute myeloid
leukemia originates from a hierarchy of leukemic stem cell classes
that differ in self-renewal capacity. Nat Immunol. 5:738–743. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vardiman JW, Harris NL and Brunning RD:
The World Health Organization (WHO) classification of the myeloid
neoplasms. Blood. 100:2292–2302. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adès L, Itzykson R and Fenaux P:
Myelodysplastic syndromes. Lancet. 383:2239–2252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Del Rey M, O'hagan K, Dellett M, Aibar S,
Colyer HA, Alonso ME, Díez-Campelo M, Armstrong RN, Sharpe DJ,
Gutiérrez NC, et al: Genome-wide profiling of methylation
identifies novel targets with aberrant hypermethylation and reduced
expression in low-risk myelodysplastic syndromes. Leukemia.
27:610–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gomez-Cabrero D, Abugessaisa I, Maier D,
Teschendorff A, Merkenschlager M, Gisel A, Ballestar E,
Bongcam-Rudloff E, Conesa A and Tegnér J: Data integration in the
era of omics: Current and future challenges. BMC Syst Biol. 8 Suppl
2:I12014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia J, Gill EE and Hancock RE:
NetworkAnalyst for statistical, visual and network-based
meta-analysis of gene expression data. Nat Protoc. 10:823–844.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haidich AB: Meta-analysis in medical
research. Hippokratia. 14 Suppl 1:S29–S37. 2010.
|
|
12
|
Johnson WE, Li C and Rabinovic A:
Adjusting batch effects in microarray expression data using
empirical Bayes methods. Biostatistics. 8:118–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pellagatti A, Cazzola M, Giagounidis A,
Perry J, Malcovati L, Della Porta MG, Jädersten M, Killick S, Verma
A, Norbury CJ, et al: Deregulated gene expression pathways in
myelodysplastic syndrome hematopoietic stem cells. Leukemia.
24:756–764. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sternberg A, Killick S, Littlewood T,
Hatton C, Peniket A, Seidl T, Soneji S, Leach J, Bowen D, Chapman
C, et al: Evidence for reduced B-cell progenitors in early
(low-risk) myelodysplastic syndrome. Blood. 106:2982–2991. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graubert TA, Shen D, Ding L, Okeyo-Owuor
T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et
al: Recurrent mutations in the U2AF1 splicing factor in
myelodysplastic syndromes. Nat Genet. 44:53–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pellagatti A, Cazzola M, Giagounidis AA,
Malcovati L, Porta MG, Killick S, Campbell LJ, Wang L, Langford CF,
Fidler C, et al: Gene expression profiles of CD34+ cells in
myelodysplastic syndromes: involvement of interferon-stimulated
genes and correlation to FAB subtype and karyotype. Blood.
108:337–345. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gerstung M, Pellagatti A, Malcovati L,
Giagounidis A, Porta MG, Jadersten M, Dolatshad H, Verma A, Cross
NC, Vyas P, et al: Combining gene mutation with gene expression
data improves outcome prediction in myelodysplastic syndromes. Nat
Commun. 6:59012015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kikushige Y, Shima T, Takayanagi S, Urata
S, Miyamoto T, Iwasaki H, Takenaka K, Teshima T, Tanaka T, Inagaki
Y and Akashi K: TIM-3 is a promising target to selectively kill
acute myeloid leukemia stem cells. Cell Stem Cell. 7:708–717. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Jonge HJ, Woolthuis CM, Vos AZ, Mulder
A, van den Berg E, Kluin PM, van der Weide K, de Bont ES, Huls G,
Vellenga E and Schuringa JJ: Gene expression profiling in the
leukemic stem cell-enriched CD34+ fraction identifies target genes
that predict prognosis in normal karyotype AML. Leukemia.
25:1825–1833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bacher U, Schnittger S, Macijewski K,
Grossmann V, Kohlmann A, Alpermann T, Kowarsch A, Nadarajah N, Kern
W, Haferlach C and Haferlach T: Multilineage dysplasia does not
influence prognosis in CEBPA-mutated AML, supporting the WHO
proposal to classify these patients as a unique entity. Blood.
119:4719–4722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schneider V, Zhang L, Rojewski M, Fekete
N, Schrezenmeier H, Erle A, Bullinger L, Hofmann S, Götz M, Döhner
K, et al: Leukemic progenitor cells are susceptible to targeting by
stimulated cytotoxic T cells against immunogenic
leukemia-associated antigens. Int J Cancer. 137:2083–2092. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Virtaneva K, Wright FA, Tanner SM, Yuan B,
Lemon WJ, Caligiuri MA, Bloomfield CD, De La Chapelle A and Krahe
R: Expression profiling reveals fundamental biological differences
in acute myeloid leukemia with isolated trisomy 8 and normal
cytogenetics. Proc Natl Acad Sci USA. 98:1124–1129. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Von Der Heide EK, Neumann M, Vosberg S,
James AR, Schroeder MP, Ortiz-Tanchez J, Isaakidis K, Schlee C,
Luther M, Jöhrens K, et al: Molecular alterations in bone marrow
mesenchymal stromal cells derived from acute myeloid leukemia
patients. Leukemia. 31:1069–1078. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stirewalt DL, Meshinchi S, Kopecky KJ, Fan
W, Pogosova-Agadjanyan EL, Engel JH, Cronk MR, Dorcy KS, McQuary
AR, Hockenbery D, et al: Identification of genes with abnormal
expression changes in acute myeloid leukemia. Genes Chromosomes
Cancer. 47:8–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stegmaier K, Ross KN, Colavito SA,
O'malley S, Stockwell BR and Golub TR: Gene expression-based
high-throughput screening(GE-HTS) and application to leukemia
differentiation. Nat Genet. 36:257–263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vidal M, Cusick ME and Barabási AL:
Interactome networks and human disease. Cell. 144:986–998. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schramm SJ, Li SS, Jayaswal V, Fung DC,
Campain AE, Pang CN, Scolyer RA, Yang YH, Mann GJ and Wilkins MR:
Disturbed protein-protein interaction networks in metastatic
melanoma are associated with worse prognosis and increased
functional mutation burden. Pigment Cell Melanoma Res. 26:708–722.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Zhang X and Fan S: Meta-analysis
of salt-related gene expression profiles identifies common
signatures of salt stress responses in Arabidopsis. Plant Syst
Evol. 303:757–774. 2017. View Article : Google Scholar
|
|
29
|
Greenberg P, Cox C, Lebeau MM, Fenaux P,
Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, et al:
International scoring system for evaluating prognosis in
myelodysplastic syndromes. Blood. 89:2079–2088. 1997.PubMed/NCBI
|
|
30
|
Bonardi F, Fusetti F, Deelen P, Van
Gosliga D, Vellenga E and Schuringa JJ: A proteomics and
transcriptomics approach to identify leukemic stem cell (LSC)
markers. Mol Cell Proteomics. 12:626–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sato K, Mano H, Ariyama T, Inazawa J,
Yazaki Y and Hirai H: Molecular cloning and analysis of the human
Tec protein-tyrosine kinase. Leukemia. 8:1663–1672. 1994.PubMed/NCBI
|
|
32
|
Goncalves AC, Alves R, Baldeiras I,
Cortesão E, Carda JP, Branco CC, Oliveiros B, Loureiro L, Pereira
A, Costa Nascimento JM, et al: Genetic variants involved in
oxidative stress, base excision repair, DNA methylation, and folate
metabolism pathways influence myeloid neoplasias susceptibility and
prognosis. Mol Carcinog. 56:130–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pei S, Minhajuddin M, Callahan KP, Balys
M, Ashton JM, Neering SJ, Lagadinou ED, Corbett C, Ye H, Liesveld
JL, et al: Targeting aberrant glutathione metabolism to eradicate
human acute myelogenous leukemia cells. J Biol Chem.
288:33542–33558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miraki-Moud F, Ghazaly E, Ariza-Mcnaughton
L, Hodby KA, Clear A, Anjos-Afonso F, Liapis K, Grantham M, Sohrabi
F, Cavenagh J, et al: Arginine deprivation using pegylated arginine
deiminase has activity against primary acute myeloid leukemia cells
in vivo. Blood. 125:4060–4068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li G, Song Y, Zhang Y, Wang H and Xie J:
miR-34b targets HSF1 to suppress cell survival in acute myeloid
leukemia. Oncol Res. 24:109–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miwa H, Beran M and Saunders GF:
Expression of the Wilms' tumor gene (WT1) in human leukemias.
Leukemia. 6:405–409. 1992.PubMed/NCBI
|
|
37
|
Feldhahn N, Arutyunyan A, Stoddart S,
Zhang B, Schmidhuber S, Yi SJ, Kim YM, Groffen J and Heisterkamp N:
Environment-mediated drug resistance in Bcr/Abl-positive acute
lymphoblastic leukemia. Oncoimmunology. 1:618–629. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Boultwood J, Pellagatti A, Watkins F,
Campbell LJ, Esoof N, Cross NC, Eagleton H, Littlewood TJ, Fidler C
and Wainscoat JS: Low expression of the putative tumour suppressor
gene gravin in chronic myeloid leukaemia, myelodysplastic syndromes
and acute myeloid leukaemia. Br J Haematol. 126:508–511. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Armengol G, Canellas A, Alvarez Y, Bastida
P, Toledo JS, Mdel Pérez-Iribarne M, Camós M, Tuset E, Estella J,
Coll MD, et al: Genetic changes including gene copy number
alterations and their relation to prognosis in childhood acute
myeloid leukemia. Leuk Lymphoma. 51:114–124. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seedhouse CH, Mills KI, Ahluwalia S,
Grundy M, Shang S, Burnett AK, Russell NH and Pallis M: Distinct
poor prognostic subgroups of acute myeloid leukaemia, FLT3-ITD and
P-glycoprotein-positive, have contrasting levels of FOXO1. Leuk
Res. 38:131–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gasparetto M, Pei S, Minhajuddin M, Khan
N, Pollyea DA, Myers JR, Ashton JM, Becker MW, Vasiliou V,
Humphries KR, et al: Targeted therapy for a subset of acute myeloid
leukemias that lack expression of aldehyde dehydrogenase 1A1.
Haematologica. 102:1054–1065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Barabási AL, Gulbahce N and Loscalzo J:
Network medicine: A network-based approach to human disease. Nat
Rev Genet. 12:56–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Taipale M, Jarosz DF and Lindquist S:
HSP90 at the hub of protein homeostasis: Emerging mechanistic
insights. Nat Rev Mol Cell Biol. 11:515–528. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Flandrin-Gresta P, Solly F, Aanei CM,
Cornillon J, Tavernier E, Nadal N, Morteux F, Guyotat D, Wattel E
and Campos L: Heat Shock Protein 90 is overexpressed in high-risk
myelodysplastic syndromes and associated with higher expression and
activation of focal adhesion kinase. Oncotarget. 3:1158–1168. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tian WL, He F, Fu X, Lin JT, Tang P, Huang
YM, Guo R and Sun L: High expression of heat shock protein 90 alpha
and its significance in human acute leukemia cells. Gene.
542:122–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Flandrin P, Guyotat D, Duval A, Cornillon
J, Tavernier E, Nadal N and Campos L: Significance of heat-shock
protein (HSP) 90 expression in acute myeloid leukemia cells. Cell
Stress Chaperones. 13:357–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin S, Li J, Zhou W, Qian W, Wang B and
Chen Z: BIIB021, an Hsp90 inhibitor, effectively kills a
myelodysplastic syndrome cell line via the activation of caspases
and inhibition of PI3K/Akt and NF-κB pathway proteins. Exp Ther
Med. 7:1539–1544. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lazenby M, Hills R, Burnett AK and
Zabkiewicz J: The HSP90 inhibitor ganetespib: A potential effective
agent for Acute Myeloid Leukemia in combination with cytarabine.
Leuk Res. 39:617–624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zheng N, Schulman BA, Song L, Miller JJ,
Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al:
Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase
complex. Nature. 416:703–709. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: Cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ishino R, Minami K, Tanaka S, Nagai M,
Matsui K, Hasegawa N, Roeder RG, Asano S and Ito M: FGF7 supports
hematopoietic stem and progenitor cells and niche-dependent
myeloblastoma cells via autocrine action on bone marrow stromal
cells in vitro. Biochem Biophys Res Commun. 440:125–131. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang J and Li Y: Therapeutic uses of
FGFs. Semin Cell Dev Biol. 53:144–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hart KC, Robertson SC and Donoghue DJ:
Identification of tyrosine residues in constitutively activated
fibroblast growth factor receptor 3 involved in mitogenesis, Stat
activation, and phosphatidylinositol 3-kinase activation. Mol Biol
Cell. 12:931–942. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Webster MK and Donoghue DJ: Constitutive
activation of fibroblast growth factor receptor 3 by the
transmembrane domain point mutation found in achondroplasia. EMBO
J. 15:520–527. 1996.PubMed/NCBI
|
|
55
|
Kang S, Elf S, Dong S, Hitosugi T, Lythgoe
K, Guo A, Ruan H, Lonial S, Khoury HJ, Williams IR, et al:
Fibroblast growth factor receptor 3 associates with and tyrosine
phosphorylates p90 RSK2, leading to RSK2 activation that mediates
hematopoietic transformation. Mol Cell Biol. 29:2105–2117. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Airoldi I, Di Carlo E, Banelli B, Moserle
L, Cocco C, Pezzolo A, Sorrentino C, Rossi E, Romani M, Amadori A
and Pistoia V: The IL-12Rbeta2 gene functions as a tumor suppressor
in human B cell malignancies. J Clin Invest. 113:1651–1659. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jo SH, Schatz JH, Acquaviva J, Singh H and
Ren R: Cooperation between deficiencies of IRF-4 and IRF-8 promotes
both myeloid and lymphoid tumorigenesis. Blood. 116:2759–2767.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mangiavacchi A, Sorci M, Masciarelli S,
Larivera S, Legnini I, Iosue I, Bozzoni I, Fazi F and Fatica A: The
miR-223 host non-coding transcript linc-223 induces IRF4 expression
in acute myeloid leukemia by acting as a competing endogenous RNA.
Oncotarget. 7:60155–60168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Otto N, Manukjan G, Göhring G, Hofmann W,
Scherer R, Luna JC, Lehmann U, Ganser A, Welte K, Schlegelberger B
and Steinemann D: ICSBP promoter methylation in myelodysplastic
syndromes and acute myeloid leukaemia. Leukemia. 25:1202–1207.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pogosova-Agadjanyan EL, Kopecky KJ,
Ostronoff F, Appelbaum FR, Godwin J, Lee H, List AF, May JJ, Oehler
VG, Petersdorf S, et al: The prognostic significance of IRF8
transcripts in adult patients with acute myeloid leukemia. PLoS
One. 8:e708122013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xu L, Gu ZH, Li Y, Zhang JL, Chang CK, Pan
CM, Shi JY, Shen Y, Chen B, Wang YY, et al: Genomic landscape of
CD34+ hematopoietic cells in myelodysplastic syndrome and gene
mutation profiles as prognostic markers. Proc Natl Acad Sci USA.
111:8589–8594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cancer Genome Atlas Research Network, .
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A,
Hoadley K, Triche TJ Jr, Laird PW, et al: Genomic and epigenomic
landscapes of adult de novo acute myeloid leukemia. N Engl J Med.
368:2059–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|