Introduction
MicroRNAs (miRNAs and miRs) are a group of
endogenous small non-coding RNAs, which cause the degradation or
translational repression of its target mRNA by binding to it and
inhibiting the expression of its target gene (1). It has been demonstrated that abnormal
expression of miR-199b-5p is associated with various diseases and
certain processes, including tumorigenesis, tumor progression,
invasion and metastasis (2–7). In previous studies, miR-199b-5p has been
revealed to be closely associated with the prognosis of patients
with osteosarcoma and overall and disease-free survival rates were
significantly lower in patients exhibiting high levels of
miR-199b-5p expression compared with those exhibiting low levels.
It was also confirmed to serve a primary role in the regulation of
proliferation, migration and invasion of osteosarcoma cells in
vivo (2). Additionally, Fang
et al (3) demonstrated that
miR-199b-5p inhibits the proliferation, migration and clonogenicity
of HER2+ breast cancer cell lines by directly targeting HER2. It
was also demonstrated that downregulation of miR-199b-5p expression
was associated with poor prognosis in patients with breast cancer
(4). Triple-negative breast cancer
(TNBC) is aggressive and is associated with higher rates of
metastasis and poorer survival times than other breast cancer
subtypes (8). Furthermore, a lack of
effective therapy poses a significant challenge to physicians
(8). However, to the best of our
knowledge, the role of miR-199b-5p in TNBC has not been assessed.
miR-199b-5p can be used as a diagnostic marker and a novel
therapeutic target for various types of cancer, and has significant
clinical application value (2,5–7). Therefore, it is important to further
investigate the role of miR-199b-5p in TNBC. Discoidin domain
receptor tyrosine kinase 1 (DDR1) is a class of collagen receptor
that is overexpressed in a variety of epithelial malignancies,
including ovarian, breast, endometrial, esophageal and lung cancer
(9–19). A previous study demonstrated that a
DDR1Low/DDR2High protein profile is
associated with TNBC and may identify patients with invasive
carcinoma with a poor prognosis (20). High DDR1 expression may also serve a
role in breast cancer cell proliferation and/or survival (21,22).
To the best of our knowledge, the present study is
the first to demonstrate that miR-199b-5p inhibits the
proliferation, migration and invasion of TNBC cells. It was also
revealed that miR-199b-5p inhibited DDR1 gene expression by
directly targeting its 3′ untranslated region (UTR). These results
indicate that miR-199b-5p may be a novel alternate therapeutic
target for TNBC. In terms of clinical samples, the expression of
miR-199b-5p was significantly reduced in breast cancer tissues
compared with normal breast tissue.
Materials and methods
Patients and specimen collection
The present study was approved by the Ethics
Committee of Kunming Medical University (The Third Affiliated
Hospital of Kunming Medical University, Kunming, China) and
informed consent was obtained from all patients. Breast cancer
tissue and normal tissue (>5 cm distant from cancer tissue) were
collected from patients who were pathologically diagnosed with
breast cancer at the Tumor Hospital of Yunnan Province from
December 2015 to March 2016. None of the patients underwent any
therapy prior to specimen collection, including chemotherapy and
radiotherapy. Collected tissue samples were immediately stored in
RNAlater (Tiangen Biotech Co., Ltd., Beijing, China) at −80°C. A
total of 19 female patients with breast cancer were recruited into
the present study (age range, 39–68 years; mean average age, 53
years). The clinicopathological characteristics of each patient
were recorded, including age, tumor size, Tumor-Node-Metastasis
(TNM) stage, estrogen receptor (ER) status, progesterone receptor
(PR) status and lymph node involvement.
Cell lines and cell culture
The human triple negative breast cancer cell line,
HCC1937, and the mammary gland cell line, MCF-10A, were donated by
Professor Ceshi Chen from the Kunming Institute of Zoology
(Kunming, China), CAS (Chinese Academy of Sciences). was recovered
from cryopreservation in liquid nitrogen (−196°C) and used at an
early passage. 293 cells were purchased from the American Type
Culture Collection (Manassas, VA, USA). HCC1937 cells were
maintained in RPMI-1640 medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; HyClone; GE Healthcare Life Sciences), 10 U/ml penicillin and
0.1 mg/ml streptomycin (HyClone; GE Healthcare Life Sciences).
MCF-10A cells were maintained in Dulbecco's Modified Eagle's medium
(DMEM)-F12 medium (HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS, 10 U/ml penicillin and 0.1 mg/ml
streptomycin. 293 cells were maintained in Dulbecco's Modified
Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS, 10 U/ml penicillin and 0.1 mg/ml
streptomycin. HCC1937, MCF-10A and 293 cells were then incubated at
37°C in 5% CO2.
RNA isolation and reverse
transcription quantitative-polymerase chain reaction (RT-qPCR)
Tissue miRNA was extracted using the miRcute miRNA
isolation kit (Tiangen Biotech Co., Ltd.), according to the
manufacturer's protocol. cDNA was synthesized using the miRcute
Plus miRNA First-strand cDNA Synthesis kit (Tiangen Biotech Co.,
Ltd.). The thermocycling conditions were as follows: 60 min at 42°C
and 3 min at 95°C. miR-199b-5p expression was analyzed using the
miRcute Plus miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.)
with the 7500 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The thermocycling
conditions were as follows: An initial pre-denaturation step of 15
min at 95°C, followed by 40 cycles of denaturation at 94°C for 20
sec and annealing at 60°C for 34 sec. The primers sequences are as
follows miR-199b-5p, forward, 5′-CAGCCCAGTGTTTAGACTATC-3′ and
reverse, 5′-CAGTGCAGGGTCCGAGGT-3′, and U6, forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. Relative expression was calculated
using the 2−∆∆Cq method (23). The RT-qPCR assays were performed in
duplicate and the data were presented as the mean ± standard error
of the mean. Using SPSS 13.0 software (SPSS, Inc., Chicago, IL,
USA) for statistical analysis, the relative quantity (RQ) value was
defined as the ratio of miR-199b-5p expression in target samples to
that in control samples. An RQ <1 and ≥1 indicated low and high
expression of miR-199b-5p in the target samples, respectively.
Transient transfection of miRNA
mimic
The miR-199b-5p mimic and negative control (NC)
miRNA were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). The miR-199b mimics sequence was
5′-CCCAGUGUUUAGACUAUCUGUUC-3′ and the NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. The miR-199b-5p mimic and NC (40 nM)
were transfected into cells using Lipofectamine RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.) in serum-free
conditions for 6 h prior to replacement with complete medium.
Western blotting and RT-qPCR were performed 48 h
post-transfection.
Luciferase reporter assay
To assess the mechanism by which miR-199b-5p
inhibits cell proliferation and migration, an miRNA target search
was performed using TargetScan (http://www.targetscan.org). The results demonstrated
that the 3′UTR of DDR1 contained a highly conserved putative
miR-199b-5p binding site. Luciferase reporter plasmids,
pRL-DDR1-3′UTR wild type (WT) and pRL-DDR1-3′UTR mutant (MUT)
plasmids were synthesized and verified by Shanghai Genomeditech
Co., Ltd (Shanghai, China). For the luciferase reporter assay, 293
cells were co-transfected with pRL-DDR1-3′UTR WT or pRL-DDR1-3′UTR
MUT, and miR-199b-5p mimics or NC using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The pRL-TK plasmid
(Promega Corporation, Madison, WI, USA) was used as control. The
successful overexpression vector was incubated with LB
(Luria-Bertani) liquid medium overnight at 37°C and the plasmid was
prepared using a high purity Plasmid Extraction kit (Qiagen GmbH,
Hilden, Germany), according to the manufacturer's instructions. The
pRL-TK 3′-UTR target plasmid (10 ng) was transiently transfected
into 293 cells, using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with a pRL-TK 3′-UTR negative control plasmid (10
ng). Results were obtained according to the protocol of the
reportable gene detection kit (Shanghai Genomeditech Co., Ltd.,
Shanghai, China) and luciferase activities were analyzed using a
Dual Luciferase Reporter assay system (Promega Corporation,
Madison, WI, USA) 48 h after transfection. Firefly luciferase
activity was used for normalization.
Western blot analysis
Total protein from cells was extracted using 1%
radioimmunoprecipitation lysis buffer (Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China) containing
phenylmethanesulfonylfluoride (Beijing Solarbio Science &
Technology Co., Ltd.). The miR-199b-5p mimic group was transfected
with miR-199b-5p mimics, the negative control group transfected
with negative control miR, the control group was untreated. The
protein concentration of total cell lysates was measured using a
BCA protein assay kit (Tiangen Biotech Co., Ltd.). Proteins (20 µg)
were separated by 10% SDS-PAGE and transferred to polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MD, USA). Membranes
were blocked at room temperature with 5% non-fat milk for 1 h and
incubated at 4°C overnight with the following antibodies: DDR1
(dilution, 1:500; cat. no. ENT1307; Elabscience, Wuhan, China) and
GAPDH (dilution, 1:3,000; cat. no. 21612; Signalway Antibody LLC,
College Park, MD, USA). Membranes were then washed with TBST three
times for 10 min, incubated with a goat anti-rabbit immunoglobulin
G secondary antibody (dilution, 1:3,000; cat. no. L3012; Signalway
Antibody LLC) for 2 h at room temperature and washed with
Tris-buffered saline with Tween 3 times for 10 min. The protein was
detected by chemiluminescence using an ECL detection reagent
(Beijing Solarbio Science & Technology Co., Ltd.). Finally, the
quantification of western blotting was performed using Image J
software (version 1.51; National Institutes of Health, Bethesda,
MD, USA).
Wound-healing assay
HCC1937 cells were transfected with the miR-199b-5p
mimic and negative control for 48 h. A marker pen and a ruler were
first used to draw 5 lines on each well of a 6-well culture dish.
Subsequently, 2×106 HCC1937 cells cultured for 12 h at
37°C in 5% CO2, and then a 200-µl pipette tip was used
to scratch two or more parallel wounds of ~200 mm in width,
perpendicular to the marker lines. Wounds were observed following
incubation at 37°C in 5% CO2 for 12 and 24 h under a
phase contrast microscope (magnification, ×100) to assess healing
rate.
Cell migration and invasion
assays
The miR-199b-5p mimic and negative control were
transfected into HCC1937 cells as aforementioned. Transwell
chambers without and with Matrigel (Corning Incorporated, Corning,
NY, USA) were used to assess cell migration and invasion,
respectively. A total of 5×104 cells in 200 µl RPMI-1640
medium without FBS were placed into the cell culture inserts (BD
Biosciences Japan, Co., Ltd, Tokyo, Japan). A total of 600 µl
RPMI-1640 medium containing 10% FBS was added to the bottom
chamber. Cells were then incubated for 24 h at 37°C in 5%
CO2, fixed with paraformaldehyde and stained with 0.1%
crystal violet at room temperature for 20 min. Cell migration and
invasion were analyzed by counting the cells on the underside of
the membrane using a light microscope (magnification, ×100) in 5
random fields of view.
Cell proliferation assay
The effect of miR-199b-5p on the proliferation of
HCC1937 cells was assessed using a CCK-8 assay. Cells in 200 µl of
DMEM were seeded into 96-well plates (2×103/well) and
incubated at 37°C in 5% CO2 for 48 h. Cell proliferation
was evaluated every 12 h for 1 day. A total of 10 µl CCK-8
(Biosharp, Hefei, China) was added to each well and incubated for
an additional 4 h at 37°C in 5% CO2. A CCK8
Enzyme-linked immune monitor (Thermo Fisher Scientific, Inc.) was
utilized to assess absorbance values at a wavelength of 450 nm.
Statistical analyses
Statistical analysis was performed using SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA). Data are presented as the
mean ± standard error of the mean and were analyzed using a
Student's t-test (between two groups) or one-way analysis of
variance followed by Tukey's test (multiple groups). The expression
of miR-199b-5p and clinicopathological parameters were analyzed
using a χ2 test. P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using GraphPad Prism 7.0a Macintosh Version (GraphPad
Software Inc., La Jolla, CA, USA).
Results
miR-199b-5p promotes cell migration
and invasion
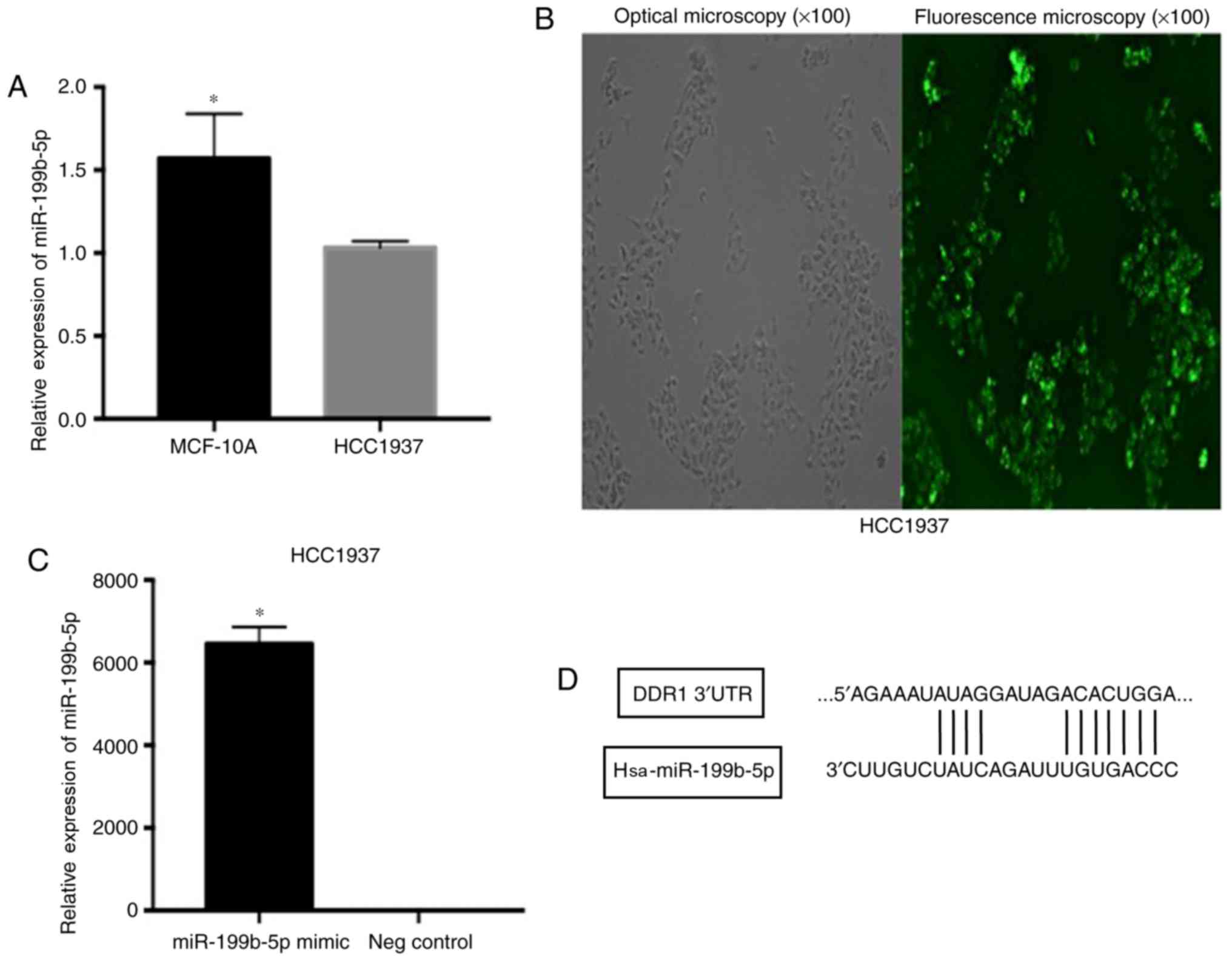
The TNBC cell line, HCC1937, exhibited the lower
expression of miR-199b-5p compared with the MCF-10A cell line
(Fig. 1A). To determine the effect of
miR-199b-5p on HCC1937 cell migration and invasion, cells were
treated with a miR-199b-5p mimic and negative control miR.
Observation with fluorescence microscope (Fig. 1B) and the results of RT-qPCR assay
(Fig. 1C) demonstrated that the
relative expression of miR-199b-5p was significantly elevated in
the mimic group compared with the negative control. A wound-healing
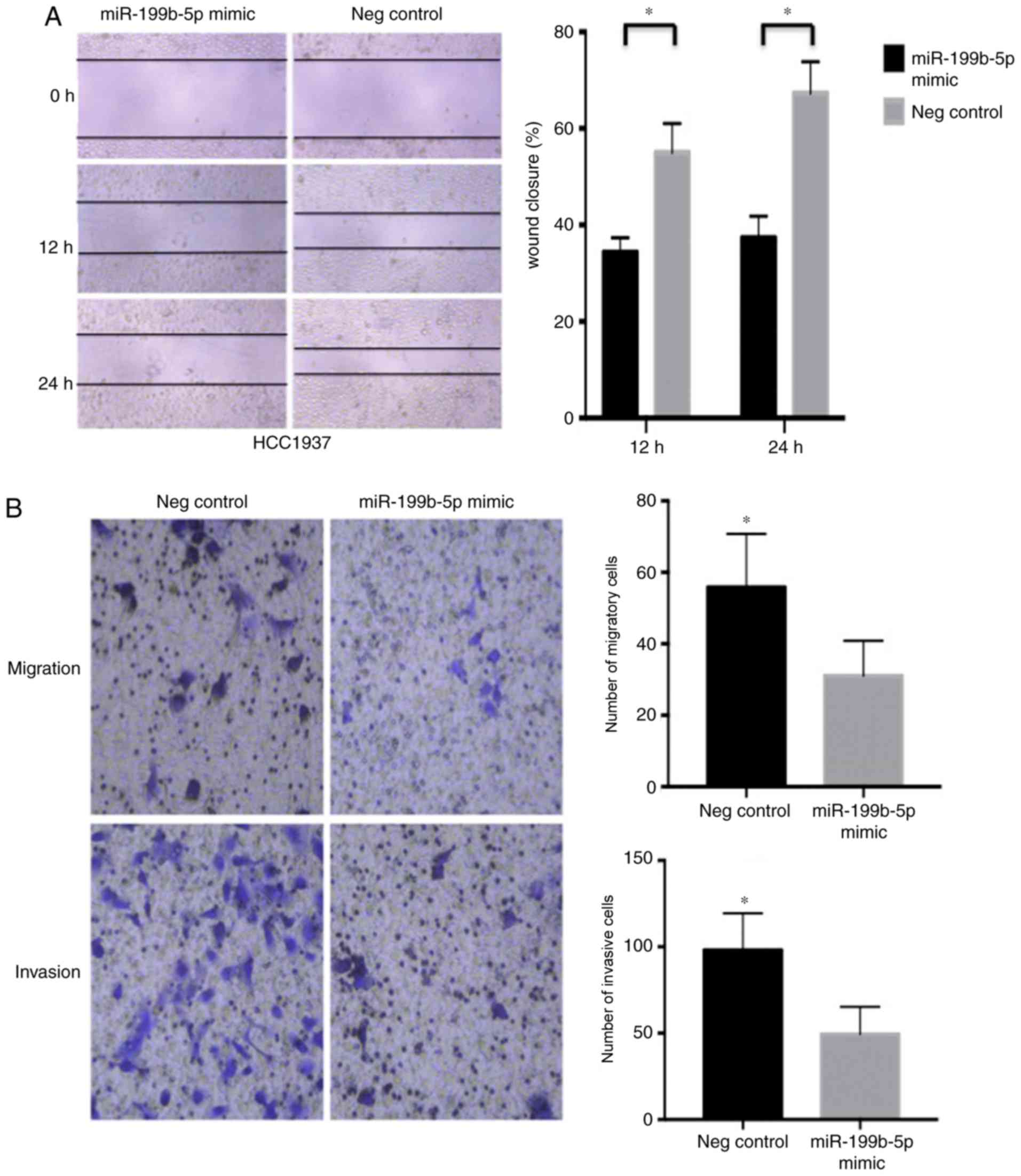
assay demonstrated that the miR-199b-5p mimic significantly
decreased the migration rate of HCC1937 cells (Fig. 2A) compared with the negative control.
Furthermore, the Transwell migration assay revealed that
miR-199b-5p mimics significantly decreased the number of migrated
cells compared with the negative control group. The effect of the
miR-199b-5p mimic on HCC1937 cell invasion was assessed using a
Matrigel invasion assay, which demonstrated similar results to the
migration assay (Fig. 2B).
miR-199b-5p inhibits HCC1937 cell
proliferation in vitro
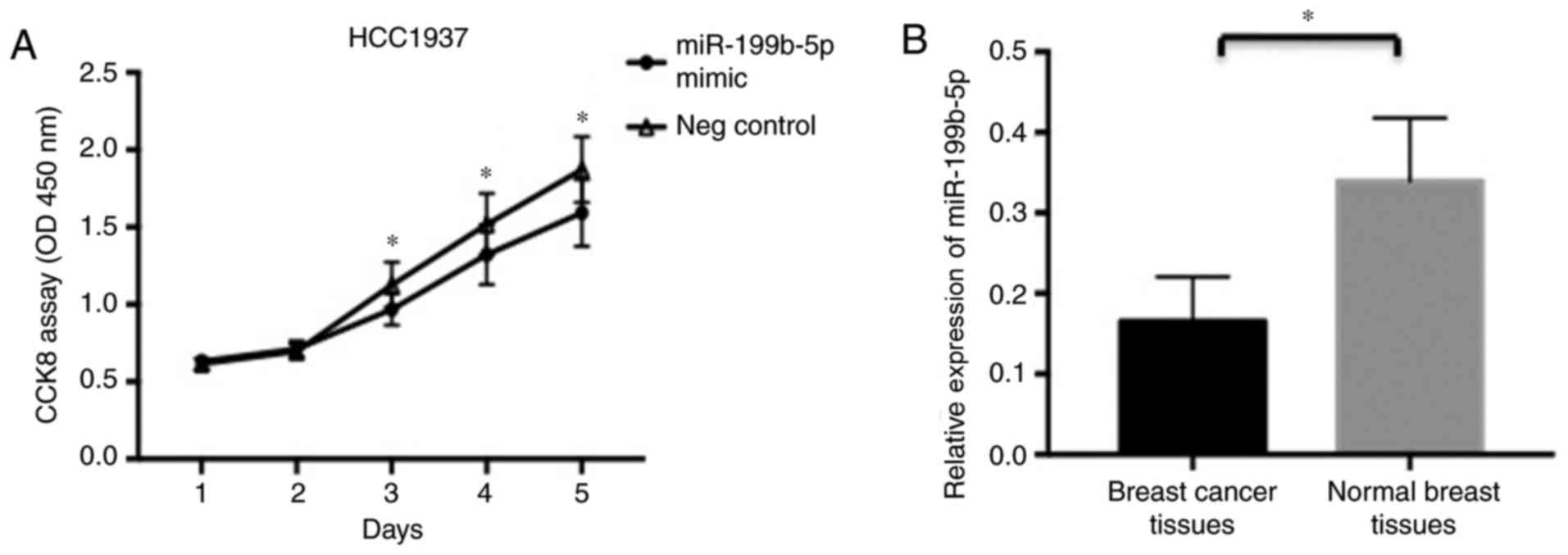
To assess the role of miR-199b-5p in the
proliferation of HCC1937 cells, a gain-of-function approach was
utilized. HCC1937 cells were transfected with a miR-199b-5p mimic
and negative control miR. Furthermore, cell proliferation was
evaluated using a CCK-8 assay at different time points. It was
revealed that the upregulation of miR-199b-5p significantly
inhibited HCC1937 cell proliferation at days 3–5 in comparison with
the control group (Fig. 3A).
The expression of miR-199b-5p in
breast cancer tissue is significantly reduced compared with normal
breast tissue
A total of 19 samples were obtained from patients
with breast cancer who did not receive any treatment prior to
surgery. Breast cancer tissues and normal adjacent breast tissue
(>5 cm from the lesion) were excised. Compared with normal
breast tissues, the expression of miR-199b-5p in breast cancer
tissues was significantly decreased (Fig.
3B). Furthermore, there was no significant association between
miR-199b-5p expression and any assessed characteristics, including
age, tumor size and TNM stage were observed (Table I).
 | Table I.Comparative analysis of patient
clinicopathological characteristics and miR-199b-5p in breast
cancer tissue samples. |
Table I.
Comparative analysis of patient
clinicopathological characteristics and miR-199b-5p in breast
cancer tissue samples.
|
|
| miR-199b-5p |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | No. | High | Low | χ2 | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
<50 | 10 | 2 | 8 | 0.014 | 0.906 |
| ≥50 | 9 | 2 | 7 |
|
|
| Tumor size |
|
|
|
|
|
| <3.0
cm | 10 | 3 | 7 | 1.017 | 0.313 |
| ≥3.0
cm | 9 | 1 | 9 |
|
|
| ER |
|
|
|
|
|
|
<10% | 7 | 1 | 6 | 0.305 | 0.518 |
|
≥10% | 12 | 3 | 9 |
|
|
| PR |
|
|
|
|
|
|
<10% | 8 | 2 | 6 | 0.130 | 0.719 |
|
≥10% | 11 | 2 | 9 |
|
|
| Ki67 |
|
|
|
|
|
|
<10% | 7 | 3 | 4 | 3.170 | 0.075 |
|
≥10% | 12 | 1 | 11 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 7 | 1 | 6 | 0.305 | 0.518 |
|
Negative | 12 | 3 | 9 |
|
|
| TNM stage |
|
|
|
|
|
| I | 5 | 0 | 5 | 1.953 | 0.377 |
| II | 8 | 2 | 6 |
|
|
|
III | 6 | 2 | 4 |
|
|
The expression of miR-199b-5p is
negatively associated with DDR1 in breast cancer tissues
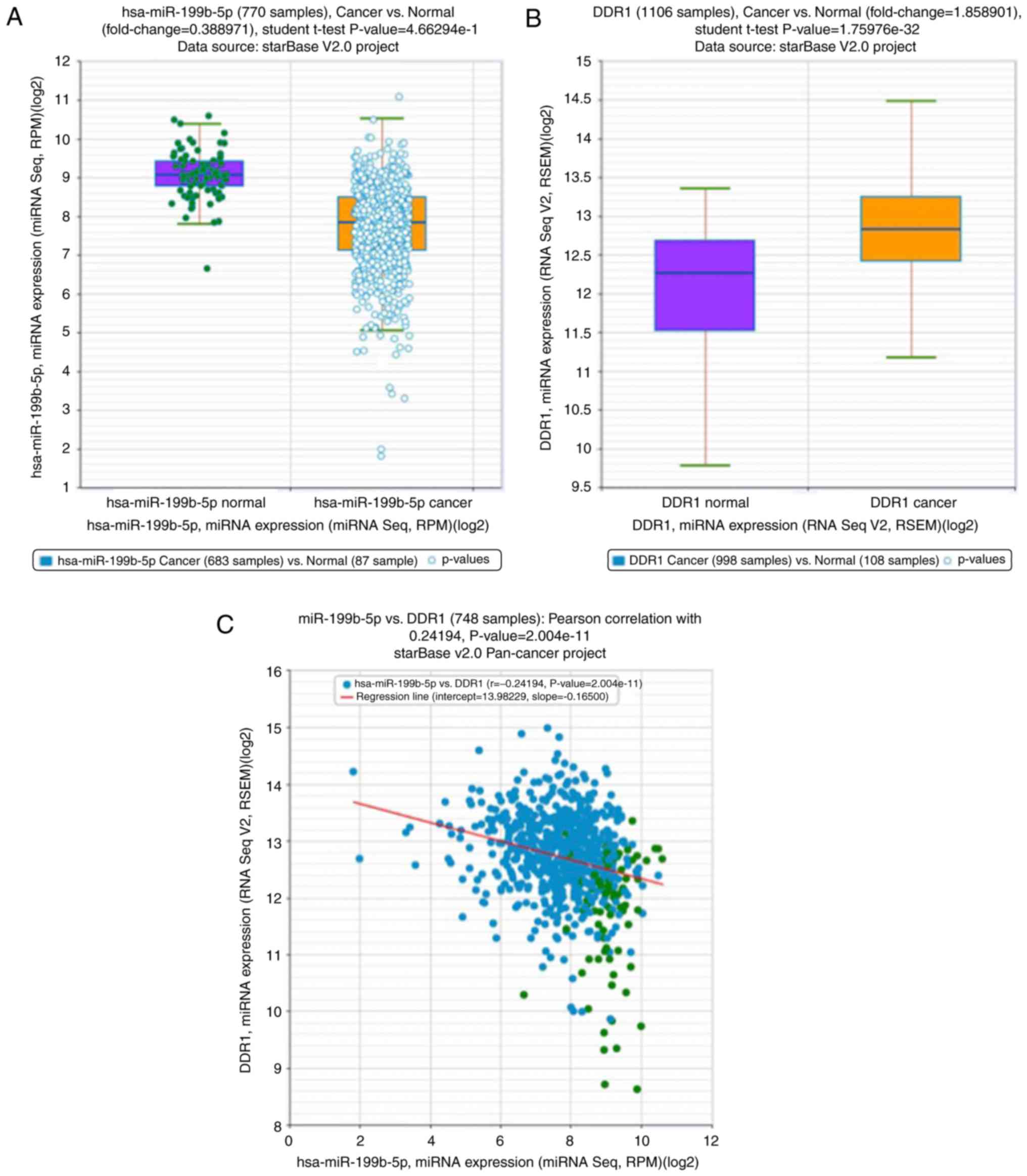
The expression of miR-199b-5p and DDR1 in breast
cancer and the association between them were analyzed using public
databases (http://starbase.sysu.edu.cn). Compared with normal
breast tissue, the expression of miR-199b-5p in breast cancer
tissues was significantly reduced (Fig.
4A). These results are congruent with those that were obtained
from the present study. Furthermore, compared with normal breast
tissue, the expression of DDR1 in breast cancer tissues was
significantly upregulated (Fig. 4B).
The expression of miR-199b-5p was also demonstrated to be
negatively correlated to DDR1 (Fig.
4C).
DDR1 is a candidate target of
miR-199b-5p
To assess the mechanism by which miR-199b-5p
inhibits cell proliferation and migration, an miRNA target search
was performed using TargetScan (http://www.targetscan.org). The results demonstrated
that the 3′UTR of DDR1 contained a highly conserved putative
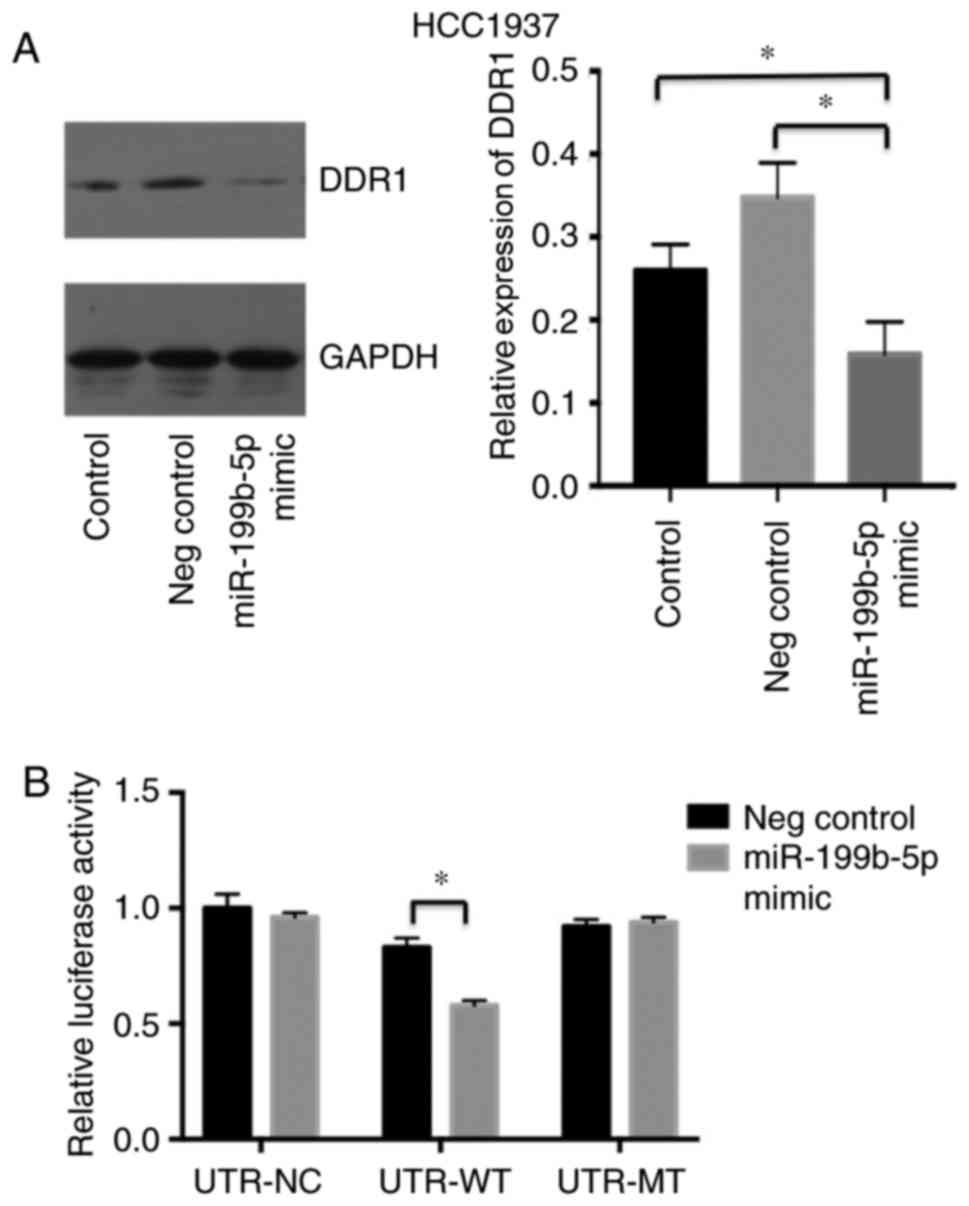
miR-199b-5p binding site. Western blotting also revealed that the
expression of DDR1 was significantly decreased in the miR-199b-5p
mimic group compared with the negative control group and the
control group (Fig. 5A). In addition,
pRL-DDR1 3′UTR-wild type (WT) and DDR1 pRL-3′UTR-mutant (MUT)
plasmids were generated to assess the interaction between
miR-199b-5p and its putative target sequences. The results of the
luciferase reporter assay demonstrated that the luciferase activity
of DDR1 3′UTR-luc-WT was decreased upon transfection with the
miR-199b-5p mimic, compared with those transfected with negative
control (Fig. 5B). These results
indicated that miR-199b-5p directly inhibited DDR1 expression by
binding to the 3′UTR of DDR1.
Taken together, the data of the present study
indicate that miR-199b-5p serves as a potent oncogenic miRNA in
HCC1937 cells by affecting cell proliferation, migration and
invasion, possibly by targeting DDR1.
Discussion
MiRNA is a non-coding RNA comprised of 19–24
nucleotides in eukaryotes, whose primary function is to regulate
the expression of target genes (24).
According to a previous study, 60–70% of human and other mammalian
genes are regulated by miRNAs (25).
In several types of cancer, including hepatocellular carcinoma,
renal cell carcinoma, osteosarcoma and acute myeloid leukemia,
miR-199b-5p has been revealed to be a biomarker (2,5–7). A further study demonstrated that the
downregulation of miR-199b-5p is associated with poor prognosis in
patients with breast cancer (4). The
study also revealed that miR-199b-5p inhibited HER2+ breast cancer
cell proliferation, migration and clonogenicity by directly
targeting HER2 (3). However, the role
of miR-199b-5p in TNBC has not yet been assessed. The present study
demonstrated that different levels of miR-199b-5p were exhibited in
different cells. The TNBC cell line HCC1937 exhibited the lower
expression of miR-199b-5p compared with the MCF-10A cell line. To
assess the effect of miR-199b-5p on the physiological processes of
TNBC cells, CCK-8, wound-healing and Transwell assays were
performed. The results of these assays indicated that the
overexpression of miR-199b-5p inhibited breast cancer cell
proliferation, migration and invasion.
The expression of miR-199b-5p in breast cancer and
normal breast tissue was detected using RT-qPCR and the
clinicopathological characteristics of miR-199b-5p in tissues were
recorded. The results demonstrated that miR-199b-5p expression in
breast cancer tissue was significantly reduced compared with normal
breast tissue. This is congruent with the result obtained from
public databases. Furthermore, no statistically significant
differences were according to age, tumor size or TNM stage.
However, this may be a result of the relatively small sample size.
Nevertheless, the loss of expression of miR-199b-5p in breast
cancer may contribute to its carcinogenesis and progression.
DDR1 is a class of collagen receptor, expressed on
various human leukocytes, including neutrophils, monocytes,
lymphocytes and podocytes (9). DDR1
is also overexpressed in a variety of epithelial malignancies,
including ovarian, breast, endometrial, esophageal and lung cancer
(10–15). Of note, a previous study demonstrated
that a DDR1Low/DDR2High protein profile is
associated with TNBC and may be used to identify invasive
carcinomas with a poor prognosis (20). High DDR1 expression may also serve a
role in breast cancer cell proliferation and/or survival (21,22). A
previous study has also indicated that the inhibition of DDR1
expression significantly enhances breast cancer cell
chemosensitivity to genotoxic drugs (16). Furthermore, Valencia et al
(15) demonstrated that the
inhibition of DDR1 decreased cell survival, homing and colonization
in lung cancer bone metastasis. Additionally, DDR1 overexpression
promoted the migration and invasion of hepatocellular carcinoma and
glioma cells (17,18). Shen et al (19) also revealed that the decreased
expression of miR-199a-5p in hepatocellular carcinoma contributed
to increases in cell invasion via the functional deregulation of
DDR1 activity. In the current study, the expression of DDR1 in
patients with breast cancer were retrieved from a public database.
Compared with normal breast tissue, the expression of DDR1 in
breast cancer tissues was significantly upregulated. Thus, DDR1 may
contribute to disease aggressiveness in a subset of invasive
carcinomas.
In conclusion, the present study demonstrated that
miR-199b-5p directly modulates DDR1 expression by binding to the
3′UTR of DDR1. The overexpression of miR-199b-5p in HCC1937 cells
also significantly reduced the expression of DDR1. Furthermore, the
luciferase activity of DDR1 3′UTR-luc-WT was decreased upon
transfection with miR-199b-5p mimics compared with control groups.
However, no marked changes in luciferase activity were observed in
the DDR1 3′UTR-luc-MUT group. The association of DDR1 and
miR-199b-5p in breast cancer tissue was analyzed using public
databases. The results demonstrated that the expression of
miR-199b-5p is negatively associated with that of DDR1. The present
study hypothesized that miR-199b-5p may inhibit the proliferation,
migration and invasion of TNBC cells by inhibiting the expression
of DDR1. Thus, DDR1 signaling may provide a novel target for the
therapeutic intervention of TNBC in the future. However, few
studies have assessed the regulation of miR-199b-5p, which should
be further explored. Furthermore, the gene targeted by miR-199b-5p
in TNBC, perhaps DDR2, requires determination in future studies.
Due limitations of the present study, further analysis of the
association between miR-199b-5p expression, clinicopathological
characteristics and the prognosis of patients with breast cancer is
required, using a larger sample size . The results of the present
study indicated that miR-199b-5p may serve a primary role in cancer
suppression, potentially acting as a prognostic biomarker for
breast cancer and potentially providing a novel approach for the
management of TNBC.
Acknowledgements
The authors would like to thank Professor Ceshi Chen
from the Kunming Institute of Zoology (Kunming, China) for the
donation of Human breast cancer (HCC1937) and mammary gland
(MCF-10A) cell lines.
Funding
This research was supported by the Initial
Scientific Research Fund of Doctors from the Tumor Hospital of
Yunnan Province (grant no. 201509) and Internal Research Projects
of Yunnan Provincial Medicine and Health Units (grant no.
2016NS078).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL conceived and designed the present study; AW
performed all in vitro experiments and the luciferase
reporter assay, and drafted the manuscript; YC performed PCR and
western blotting; YFL performed the statistical analysis; and YL
critically revised the manuscript for important intellectual
content and was involved in analysing and interpreting of data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Kunming Medical University (Kunming, China). Written
informed consent was obtained from all patients.
Patient consent for publication
All patients provided consent for publication of
this manuscript.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng H, Zhang Z, Dai X, Chen Y, Ye J and
Jin Z: Increased expression of microRNA-199b-5p associates with
poor prognosis through promoting cell proliferation, invasion and
migration abilities of human osteosarcoma. Pathol Oncol Res.
22:253–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang C, Zhao Y and Guo B: MiR-199b-5p
targets HER2 in breast cancer cells. J Cell Biochem. 114:1457–1463.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fang C, Wang FB, Li Y and Zeng XT:
Down-regulation of miR-199b-5p is correlated with poor prognosis
for breast cancer patients. Biomed Pharmacother. 84:1189–1193.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Favreau AJ, McGlauflin RE, Duarte CW and
Sathyanarayana P: miR-199b, a novel tumor suppressor miRNA in acute
myeloid leukemia with prognostic implications. Exp Hematol Oncol.
5:42016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Song B, Song W, Liu J, Sun A, Wu
D, Yu H, Lian J, Chen L and Han J: Underexpressed microRNA-199b-5p
targets hypoxia-inducible factor-1α in hepatocellular carcinoma and
predicts prognosis of hepatocellular carcinoma patients. J
Gastroenterol Hepatol. 26:1630–1637. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Navrátil J, Fabian P, Palácová M,
Petráková K, Vyzula R and Svoboda M: Triple negative breast cancer.
Klin Onkol. 28:405–415. 2015.(In Czech). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yoshimura T, Matsuyama W and Kamohara H:
Discoidin domain receptor 1: A new class of receptor regulating
leukocyte-collagen interaction. Immunol Res. 31:219–230. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Turashvili G, Bouchal J, Baumforth K, Wei
W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J,
Srovnal J, et al: Novel markers for differentiation of lobular and
ductal invasive breast carcinomas by laser microdissection and
microarray analysis. BMC Cancer. 7:552007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Neuhaus B, Bühren S, Böck B, Alves F,
Vogel WF and Kiefer F: Migration inhibition of mammary epithelial
cells by Sky is blocked in the presence of DDR1 receptors. Cell Mol
Life Sci. 68:3757–3770. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Quan J, Yahata T, Adachi S, Yoshihara K
and Tanaka K: Identification of receptor tyrosine kinase, discoidin
domain receptor 1 (DDRl), as a potential biomarker for serous
ovarian cancer. Int J Mol Sci. 12:971–982. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Colas E, Perez C, Cabrera S, Pedrola N,
Monge M, Castellvi J, Eyzaguirre F, Gregorio J, Ruiz A, Llaurado M,
et al: Molecular markers of endometrial carcinoma detected in
uterine aspirates. Int J Cancer. 129:2435–2444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nemoto T, Ohashi K, Akashi T, Johnson JD
and Hirokawa K: Overexpression of protein tyrosine kinases in human
esophageal cancer. Pathobiology. 65:195–203. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valencia K, Ormazábal C, Zandueta C,
Luis-Ravelo D, Antón I, Pajares MJ, Agorreta J, Montuenga LM,
Martínez-Canarias S, Leitinger B and Lecanda F: Inhibition of
collagen receptor discoidin domain receptor-1 (DDR1) reduces cell
survival, homing, and colonization in lung cancer bone metastasis.
Clin Cancer Res. 18:969–980. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Das S, Ongusaha PP, Yang YS, Park JM,
Aaronson SA and Lee SW: Discoidin domain receptor 1 receptor
tyrosine kinase induces cyclooxygenase-2 and promotes
chemoresistance through nuclear factor-kappaB pathway activation.
Cancer Res. 66:8123–8130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park HS, Kim KR, Lee HJ, Choi HN, Kim DK,
Kim BT and Moon WS: Overexpression of discoidin domain receptor 1
increases the migration and invasion of hepatocellular carcinoma
cells in association with matrix metalloproteinase. Oncol Rep.
18:1435–1441. 2007.PubMed/NCBI
|
|
18
|
Ram R, Lorente G, Nikolich K, Urfer R,
Foehr E and Nagavarapu U: Discoidin domain receptor-1a (DDR1a)
promotes glioma cell invasion and adhesion in association with
matrix metalloproteinase-2. J Neurooncol. 76:239–248. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G and
Beckebaum S: Role of microRNA-199a-5p and discoidin domain receptor
1 in human hepatocellular carcinoma invasion. Mol Cancer.
9:2272010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Toy KA, Valiathan RR, Núñez F, Kidwell KM,
Gonzalez ME, Fridman R and Kleer CG: Tyrosine kinase discoidin
domain receptors DDR1 and DDR2 are coordinately deregulated in
triple-negative breast cancer. Breast Cancer Res Treat. 150:9–18.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duncan JS, Whittle MC, Nakamura K, Abell
AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV,
Darr DB, et al: Dynamic reprogramming of the kinome in response to
targeted MEK inhibition in triple-negative breast cancer. Cell.
149:307–321. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marcotte R, Brown KR, Suarez F, Sayad A,
Karamboulas K, Krzyzanowski PM, Sircoulomb F, Medrano M, Fedyshyn
Y, Koh JLY, et al: Essential gene profiles in breast, pancreatic,
and ovarian cancer cells. Cancer Discov. 2:172–189. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chalmin F, Ladoire S, Mignot G, Vincent J,
Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau
D, et al: Membrane-associated Hsp72 from tumor-derived exosomes
mediates STAT3-dependent immunosuppressive function of mouse and
human myeloid-derived suppressor cells. J Clin Invest. 120:457–471.
2010.PubMed/NCBI
|
|
25
|
Ebert MS and Sharp PA: MicroRNA sponges:
Progresss and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|