Introduction
Hepatocellular carcinoma (HCC) develops in the liver
following long-term infection with the hepatitis B or C virus
(1). One of the biological
characteristics of HCC is that the cancer cells are similar to
immature hepatocytes (2).
Additionally, HCC cells are positive for cluster of differentiation
(CD)133, a cancer stem cell marker (3), and for biliary epithelial cell markers
(3). These results suggest that HCC
cells have the potential to differentiate into hepatocytes and
biliary epithelial cells (4).
Transcription factors serve an important role in
hepatocyte differentiation. For example, CCAAT/enhancer-binding
protein (C/EBP)α promotes the maturation of immature hepatocytes
(5), but it is downregulated in HCC
(6). Similarly, GATA binding protein
(GATA)4 and GATA6 are involved in hepatocyte differentiation.
Whilst GATA4 is associated with endoderm differentiation (7), GATA6 is required for liver bud formation
(8). In addition,
hematopoietically-expressed homeobox (HHEX) is essential for the
differentiation of human induced pluripotent stem (iPS) cells into
hepatocytes (9). These studies
indicate that GATA4, GATA6 and HHEX may be involved in the
carcinogenesis of HCC.
iPS cells are generated from adult cells upon the
introduction of reprogramming factors, including NANOG, sex
determining region Y box 2 (Sox2), Oct3/4 and Krüppel-like factor
(KLF)4 (10). The expression patterns
of these genes in HCC cells have yet to be elucidated. Therefore,
the present study investigated the expression patterns of these
transcription factors in order to identify potential novel
treatments for HCC. Human iPS cells were used as a model for
pluripotent stem cells to compare the expression patterns of these
genes.
Materials and methods
Cell culture
The 201B7 iPS cells (RIKEN BioResource Center,
Tsukuba, Japan) were cultured in ReproFF (ReproCELL, Yokohama,
Japan) in 6-well plates (Asahi Techno Glass Corporation, Funabashi,
Japan) coated with Matrigel™ (BD Biosciences, Franklin
Lakes, NJ, USA), in feeder-cell-free conditions at 37°C in an
incubator with 5% CO2. ReproFF was a complete medium and
ready to use. The cells were harvested with Accutase®
(Innovative Cell Technologies, Inc., San Diego, CA, USA) and seeded
onto 6-well plates or 96-well plates coated with Matrigel™ at a
density of 106 cells/well. The HLE, HLF, PLC/PRF/5,
Huh-7, Hep3B, Huh-6 and HepG2 HCC cell lines were also purchased
from RIKEN BioResource Center. The HCC cells were cultured in
Dulbecco's modified Eagle's medium (DMEM), supplemented with 10%
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at 37°C in an incubator with 5% CO2. The HCC
cells were trypsinized, harvested and plated onto 6-well plates or
96-well plates. The cells were observed under a light microscope
(CKX41N-31PHP; Olympus Corporation, Tokyo, Japan).
Transfection and cell proliferation
assay
The HLF cells were trypsinized, harvested and plated
onto 96-well flat-bottom plates (Asahi Techno Glass Corporation) at
a density of 1,000 cells/well, and then incubated for 24 h in DMEM
supplemented with 10% FBS at 37°C in an incubator with 5%
CO2. The cells were transfected with negative control
small interfering RNA (siRNA) of random sequences or with Oct3/4
siRNA at 20 or 200 nM using Lipofectamine® 2000 (all
from Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol, and cultured in DMEM supplemented with 10%
FBS at 37°C in 5% CO2. The sequence of the Oct3/4 siRNA
was 5′-CACCCUUUGUGUUCCCAAUUCCUUC-3′. The negative control siRNA was
Stealth RNAi™ siRNA Negative Control, MedGC (cat. no. 12935-300;
Thermo Fisher Scientific, Inc). Transfection with
Lipofectamine® 2000 and no nucleic acid material was
used as a mock transfection control. Furthermore, the siRNA-treated
cells were cultured for 72 h and an MTS colorimetric assay was
performed according to the manufacturer's protocol (CellTiter
96® Aqueous One Solution Cell Proliferation Assay;
Promega Corporation, Madison, WI, USA). MTS was ready to use
(11). Cells reduce MTS into
formazan, a colored product, which has a maximum absorbance at a
wavelength of 490 nm; this was measured using an iMark Microplate
Absorbance reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Scratch assay
HLF cells were plated onto 4-well chamber slides (BD
Biosciences) at a density of 5×104 cells/well. When the
cells were confluent, the cell sheets were scratched with a sterile
razor. The cells were immediately transfected with Oct3/4 siRNA or
the mock transfection control as aforementioned. After 2 days of
culture, the cells were stained with hematoxylin and eosin,
observed using an AX80 light microscope (magnification, ×100;
Olympus Corporation) and five distinct fields were imaged; the five
distinct fields were chosen randomly. The distance between the edge
of the cell sheets and the scratched line was measured. The
experiment was repeated three times.
Reverse transcription polymerase chain
reaction (RT-PCR) and RT-quantitative PCR (RT-qPCR)
Total RNA (5 µg) was isolated using
Isogen® (Nippon Gene Co., Ltd., Tokyo, Japan) from cells
cultured in 6-well plates, was utilized for first-strand
complementary (c)DNA synthesis, using SuperScript® III
First Strand Synthesis and oligo(dT) and following the
manufacturer's protocol (Thermo Fisher Scientific, Inc.). RNA from
human fetal and adult liver was purchased from Clontech
Laboratories, Inc. (Mountain View, CA, USA). PCR was performed
using Taq DNA polymerase (Thermo Fisher Scientific, Inc.) and
products were separated using gel electrophoresis in 2% agarose in
1X TAE (40 mM Tris-acetate/1 mM EDTA). PCR primer (Thermo Fisher
Scientific, Inc.) sequences, annealing temperatures, reaction cycle
numbers and amplicon lengths for RT-PCR are presented in Table I. RT-qPCR was performed using the Fast
SYBR-Green Master Mix (Thermo Fisher Scientific, Inc.) and analyzed
with the MiniOpticon™ Detection System (Bio-Rad Laboratories,
Inc.). RT-qPCR was performed for 40 cycles of two steps consisting
of 5 sec denaturation at 95°C and 5 sec annealing-extension at
60°C. PCR primers (Thermo Fisher Scientific, Inc.), annealing
temperatures, reaction cycle numbers and amplicon lengths for
RT-qPCR are listed in Table II.
GAPDH and ribosomal protein L19 (RPL19) were used as internal
controls for RT-PCR and RT-qPCR, respectively. RPL19 was used as an
endogenous control to monitor the quantity of mRNA as a
constitutively expressed housekeeping gene (12). The gene expression levels were
analyzed using the automated MiniOpticon™ system based on the ΔΔ
cycle threshold (ΔΔCq) method (13).
The relative expression was calculated as the expression level of a
specific gene divided by the expression level of RPL19. The
experiments were performed three times, and triplicates were used
in each experiment.
 | Table I.Primer sequences for reverse
transcription polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription polymerase chain reaction.
| Primer name | Sequence | Description | Product size, bp | Annealing
temperature, °C | No. of cycles | GenBank accession
number |
|---|
| OMC21 |
5′-ACCTGACCTGCCGTCTAGAA-3′ | GAPDH, forward | 350 | 63 | 40 | BC025925 |
| OMC22 |
5′-TCCACCACCCTGTTGCTGTA-3′ | GAPDH, reverse |
|
|
|
|
| OMC271 |
5′-CTTGCTGCAGAAGTGGGTGGAGGAA-3′ | Oct3/4, forward | 187 | 60 | 40 | NM_002701 |
| OMC272 |
5′-CTGCAGTGTGGGTTTCGGGCA-3′ | Oct3/4, reverse |
|
|
|
|
| OMC273 |
5′-AAAAAAGGAAGACAAGGTCCCG-3′ | NANOG, forward | 299 | 56 | 40 | NM_024865 |
| OMC274 |
5′-ATCCCTGGTGGTAGGAAGAGTAAAG-3′ | NANOG, reverse |
|
|
|
|
| OMC277 |
5′-TTCTCCAACGACCAGACCATCG-3′ | HHEX, forward | 348 | 56 | 40 | NM_002729 |
| OMC278 |
5′-TTTTATCGCCCTCAATGTCCAC-3′ | HHEX, reverse |
|
|
|
|
| OMC279 |
5′-TTCATCACGGCGGCTTGGATTGTC-3′ | GATA6, forward | 299 | 56 | 40 | NM_005257 |
| OMC280 |
5′-GTGTTGTGGGGGAAGTATTTTTGC-3′ | GATA6, reverse |
|
|
|
|
| OMC281 |
5′-GAAAACGGAAGCCCAAGAACC-3′ | GATA4, forward | 218 | 56 | 40 | NM_002052 |
| OMC282 |
5′-AGACATCGCACTGACTGAGAACG-3′ | GATA4, reverse |
|
|
|
|
| OMC283 |
5′-TGCGGCAAAACCTACACAAAG-3′ | KLF4, forward | 379 | 57 | 40 | NM_004235 |
| OMC284 |
5′-CACTCACAAGATGACTCAGTTGGG-3′ | KLF4, reverse |
|
|
|
|
| OMC285 |
5′-AGAAAAACGAGGGAAATGGGAG-3′ | Sox2, forward | 161 | 53 | 40 | NM_003106 |
| OMC286 |
5′-TTGCGTGAGTGTGGATGGGATTGG-3′ | Sox2, reverse | 187 | 65 | 40 |
|
 | Table II.Primer sequences and conditions for
reverse transcription-quantitative polymerase chain reaction. |
Table II.
Primer sequences and conditions for
reverse transcription-quantitative polymerase chain reaction.
| Primer name | Sequence | Description | Product size, bp | Annealing
temperature, °C | No. of cycles | GenBank accession
number |
|---|
| OMC311 |
5′-CCGTTTTTGGCTCTGTTTTG-3′ | NANOG, forward | 187 | 60 | 40 | NM_024865 |
| OMC312 |
5′-TCATCGAAACACTCGGTGAA-3′ | NANOG, reverse |
|
|
|
|
| OMC321 |
5′-CGAATGCCAGAGAAGGTCAC-3′ | RPL19, forward | 157 | 60 | 40 | BC095445 |
| OMC322 |
5′-CCATGAGAATCCGCTTGTTT-3′ | RPL19, reverse |
|
|
|
|
Statistical analysis
One-way analysis of variance was applied for
statistical analysis. JMP 10.0.2 software (SAS Institute Inc., Cary
NC, USA) was used for statistical analysis. P<0.05 was
considered to indicate a statistically significant result.
Results
RT-PCR indicates the expression levels
of transcription factors in HCC cells
To reveal the expression patterns of transcription
factors in HCC cells, RT-PCR was performed (Fig. 1). Total RNA from fetal and adult liver
cells was analyzed to search for genes expressed in HCC cells but
not in fetal and adult liver. These genes may be involved in
proliferation of HCC. Whilst NANOG and Sox2 were not expressed in
any of the HCC cell lines, Oct3/4 was expressed in HLE, HLF and
Hep3B cells (Lanes 5, 6 and 9, respectively), and GATA4, GATA6 was
expressed in HLF, Huh-7, Hep3B, Huh-6 and HepG2 cells. HHEX was
expressed in HLE, HLF, PLC/PRF/5, Hep3B, Huh-7 and HepG2 cells.
KLF4 was expressed in fetal and adult liver, ant not in 201B7
cells. Oct3/4 was expressed in HLE and HLF. Furthermore, NANOG,
Sox2 and Oct3/4 were highly expressed in 201B7 cells while not in
fetal and adult liver cells (Lane 2). Nanog and Sox2 were not
expressed in HCC cells analyzed in the present study; therefore
Nanog and Sox2 may not be associated with HCC carcinogenesis.
Oct3/4 was expressed in HLE, HLF and Hep3B cells. Therefore, Oct3/4
may have a role in the proliferation of the HLE, HLF and Hep3B cell
lines.
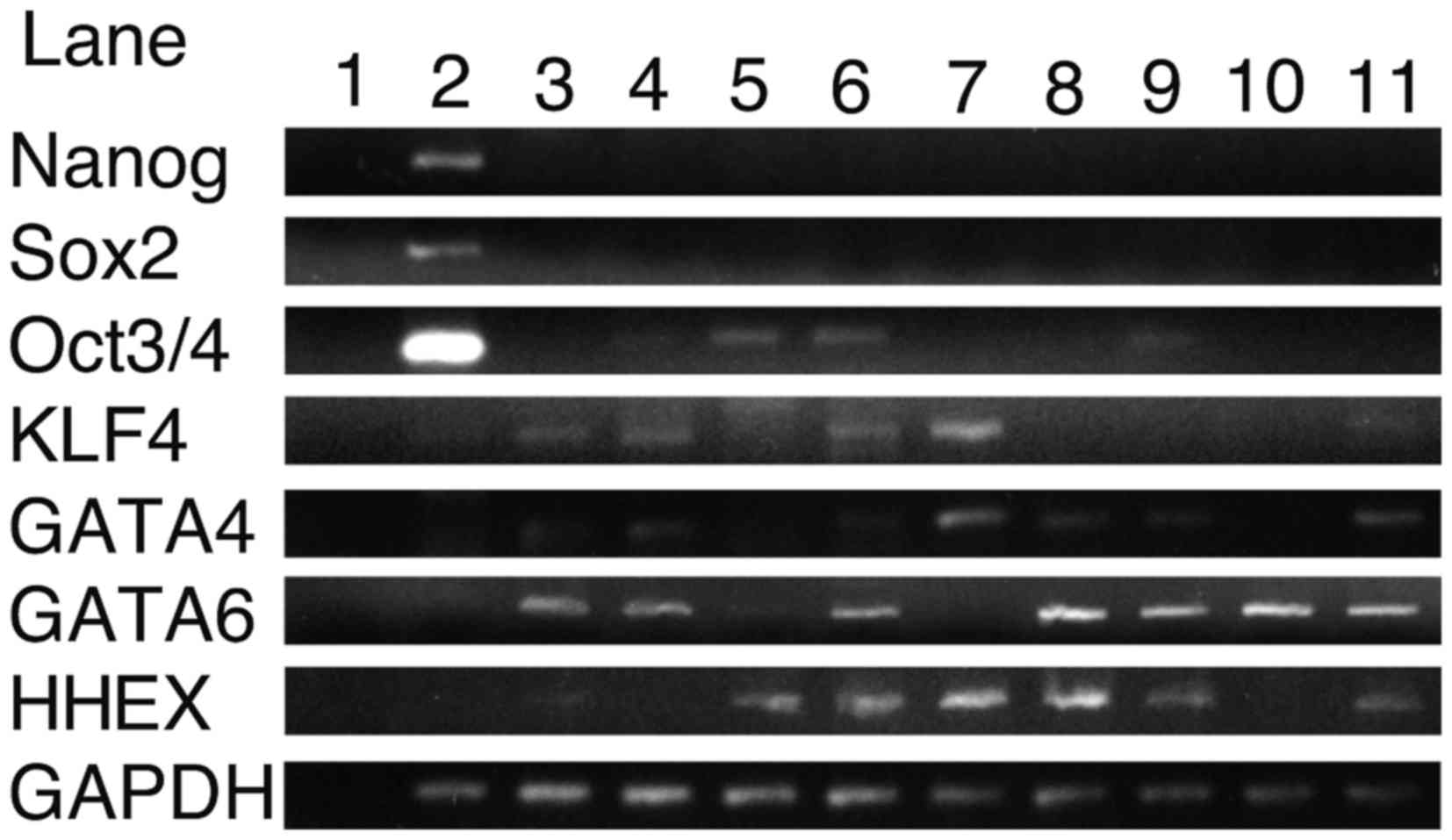 | Figure 1.Expression levels of NANOG, Sox2,
Oct3/4, KLF4, GATA4, GATA6 and HHEX in control and HCC cell lines
as measured by reverse transcription polymerase chain reaction.
Lane 1, H2O; 2, 201B7; 3, fetal liver; 4, adult liver;
5, HLE; 6, HLF; 7, PLC/PRF/5; 8, Huh-7; 9, Hep3B; 10, Huh-6 and 11,
HepG2 cells. HCC, hepatocellular carcinoma; Sox2, sex determining
region Y box 2; KLF4, Krüppel-like factor 4; GATA, GATA binding
protein; HHEX, hematopoietically expressed homeobox. |
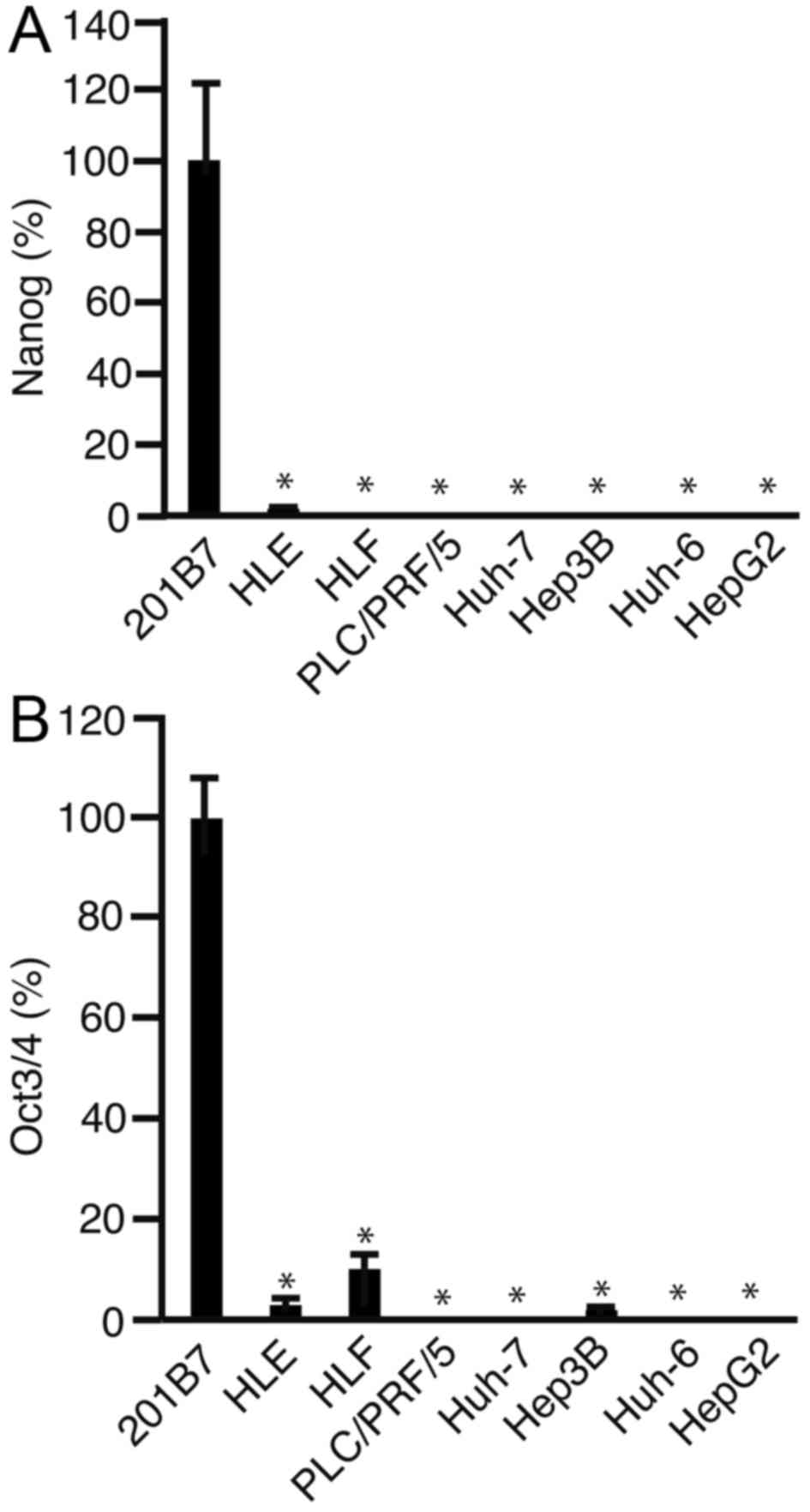
RT-qPCR indicates the expression
levels of NANOG and Oct3/4 in HCC cells
RT-qPCR was performed to analyze the expression
levels of Oct3/4. The expression levels of NANOG (Fig. 2A) and Oct3/4 (Fig. 2B) were significantly lower in all the
HCC cell lines, as compared with in the 201B7 iPS cells
(P<0.05). The expression levels of Oct3/4 were significantly
lower in HLE, HLF and Hep3B cells, compared with those in 201B7
cells (P<0.05). In addition, the expression level of Oct3/4 was
significantly higher in HLF cells, compared with in HLE (P<0.05)
and Hep3B cells (P<0.05). These results suggest that Oct3/4 has
a role in carcinogenesis in HLF cells. Therefore, HLF cells were
used for further investigation.
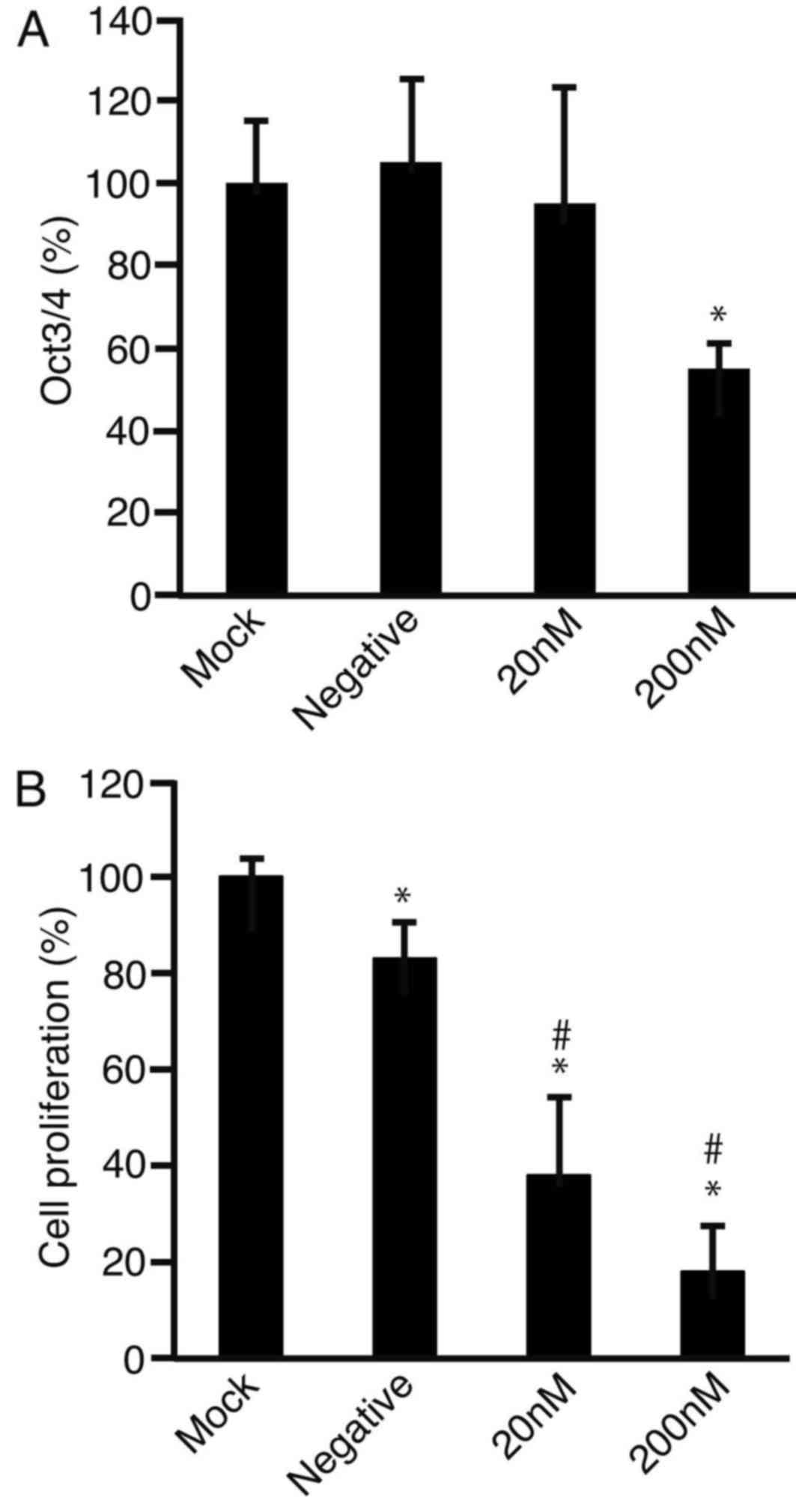
Oct3/4 siRNA transfection reduces the
expression levels Oct3/4 and cell proliferation
HLF cells were transfected with Oct3/4 siRNA and
after 2 days of culture, RNA was isolated and subjected to RT-qPCR
to analyze the expression levels of Oct3/4 (Fig. 3A). It was observed that the expression
levels of Oct3/4 were significantly downregulated following
treatment with 200 nM targeted-siRNA (P<0.05). The MTS assay
demonstrated that cell proliferation was suppressed with Oct3/4
siRNA at 20 and 200 nM compared with the mock control (both
P<0.01; Fig. 3B). Negative control
siRNA also decreased cell proliferation compared with mock
(P<0.05); however, Oct3/4 siRNA significantly decreased cell
proliferation compared with the negative control siRNA at 20 and
200 nM (both P<0.01).
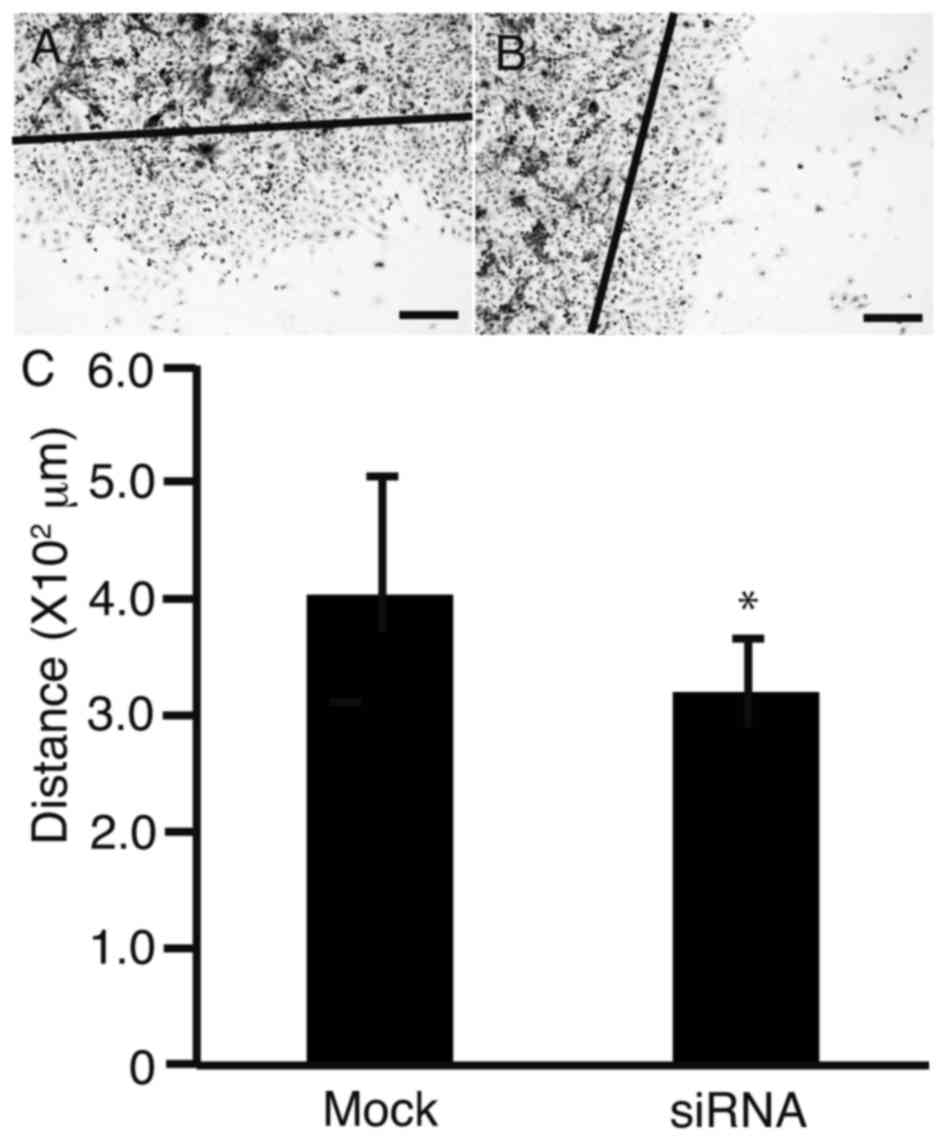
Oct3/4 siRNA reduces the ability of
HLF cells to migrate
To clarify the effect of Oct3/4 siRNA on cell
motility, a scratch assay was performed. HLF cells were scratched
with a sterile razor and transfected with Oct3/4 siRNA. After 2
days of culture, the transfected cells were imaged (Fig. 4A). The distance between the edge of
the cell sheet and the scratched line was measured (Fig. 4B). It was observed that the distance
significantly decreased in the layer of cells transfected with
Oct3/4 siRNA, as compared with the mock transfection control
(P<0.05).
Discussion
HCC cells are hypo-methylated in the promoter region
of NANOG (14). Cancer stem cells are
rare and a population may be enriched for cancer stem cells using
serum-free media (15). In the
present study, NANOG and Sox2 were not expressed in any of the HCC
cell lines. Therefore, it is hypothesized that the promoter region
of NANOG may be methylated. However, it remains to be elucidated as
to why Sox2 is not expressed in HCC cells.
Oct3/4 is involved in pluripotency along with NANOG,
Sox2 and KLF4 (16). OCT3/4 is
expressed in HCC cells (17), as well
as being highly expressed in the cancer stem cells of HCC (18). Oct3/4 is upregulated by growth
factors, including the insulin-like growth factor-1 via protein
kinase B (19,20). These previous studies suggest that
Oct3/4 may affect the stemness of HCC cells, and also their
phenotype. Higher expression levels of Oct3/4 are predictive of
poor prognosis in patients with HCC (21). This previous study suggests that
Oct3/4 has a role in carcinogenesis or cancer progression (21). In the present study, Oct3/4 siRNA
significantly suppressed the proliferation and motility of HLF
cells. These results indicate that Oct3/4 may be a potential
therapeutic target for the treatment of HCC. This hypothesis was
supported by Murakami et al (22), who demonstrated that Oct3/4 was a
potentially effective target for differentiation therapy.
One limitation of the present study is that stemness
markers were not analyzed in HLF cells following transfection with
Oct3/4 siRNA. Therefore, the differentiation state of the cells was
not determined. An investigation into the differentiation state of
HCC cells transfected with Oct3/4 siRNA may facilitate further
understanding. In conclusion, Oct3/4 is expressed in HLE, HLF and
Hep3B cells, and Oct3/4 siRNA suppresses the proliferation and
motility of HLF cells.
Acknowledgements
This study was supported by a Grant-in-Aid for
Scientific Research from the Japan Society for the Promotion of
Science (grant no. 15K09032).
References
|
1
|
Cameron AM: Screening for viral hepatitis
and hepatocellular cancer. Surg Clin North Am. 95:1013–1021. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tomizawa M, Kondo F and Kondo Y: Growth
patterns and interstitial invasion of small hepatocellular
carcinoma. Pathol Int. 45:352–358. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsujikawa H, Masugi Y, Yamazaki K, Itano
O, Kitagawa Y and Sakamoto M: Immunohistochemical molecular
analysis indicates hepatocellular carcinoma subgroups that reflect
tumor aggressiveness. Hum Pathol. 50:24–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tomizawa M, Garfield S, Factor V and
Xanthopoulos KG: Hepatocytes deficient in CCAAT/enhancer binding
protein alpha (C/EBP alpha) exhibit both hepatocyte and biliary
epithelial cell character. Biochem Biophys Res Commun. 249:1–5.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamasaki H, Sada A, Iwata T, Niwa T,
Tomizawa M, Xanthopoulos KG, Koike T and Shiojiri N: Suppression of
C/EBP alpha expression in periportal hepatoblasts may stimulate
biliary cell differentiation through increased Hnf6 and Hnf1b
expression. Development. 133:4233–4243. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomizawa M, Wang YQ, Ebara M, Saisho H,
Watanabe K, Nakagawara A and Tagawa M: Decreased expression of the
CCAAT/enhancer binding protein alpha gene involved in hepatocyte
proliferation in human hepatocellular carcinomas. Int J Mol Med.
9:597–600. 2002.PubMed/NCBI
|
|
7
|
Zaret KS and Carroll JS: Pioneer
transcription factors: Establishing competence for gene expression.
Genes Dev. 25:2227–2241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guye P, Ebrahimkhani MR, Kipniss N,
Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG and Weiss R:
Genetically engineering self-organization of human pluripotent stem
cells into a liver bud-like tissue using Gata6. Nat Commun.
7:102432016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takayama K, Inamura M, Kawabata K,
Sugawara M, Kikuchi K, Higuchi M, Nagamoto Y, Watanabe H, Tashiro
K, Sakurai F, et al: Generation of metabolically functioning
hepatocytes from human pluripotent stem cells by FOXA2 and HNF1α
transduction. J Hepatol. 57:628–636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomizawa M, Shinozaki F, Motoyoshi Y,
Sugiyama T, Yamamoto S and Ishige N: Niclosamide suppresses
migration of hepatocellular carcinoma cells and downregulates
matrix metalloproteinase-9 expression. Oncol Lett. 10:3515–3518.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davies B and Fried M: The L19 ribosomal
protein gene (RPL19): Gene organization, chromosomal mapping, and
novel promoter region. Genomics. 25:372–380. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tam S, Clavijo A, Engelhard EK and
Thurmond MC: Fluorescence-based multiplex real-time RT-PCR arrays
for the detection and serotype determination of foot-and-mouth
disease virus. J Virol Methods. 161:183–191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang XQ, Ng RK, Ming X, Zhang W, Chen L,
Chu AC, Pang R, Lo CM, Tsao SW, Liu X, et al: Epigenetic regulation
of pluripotent genes mediates stem cell features in human
hepatocellular carcinoma and cancer cell lines. PLoS One.
8:e724352013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Setoguchi T, Taga T and Kondo T: Cancer
stem cells persist in many cancer cell lines. Cell Cycle.
3:414–415. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakai-Futatsugi Y and Niwa H:
Transcription factor network in embryonic stem cells: Heterogeneity
under the stringency. Biol Pharm Bull. 36:166–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu G, Wilson G, Zhou G, Hebbard L, George
J and Qiao L: Oct4 is a reliable marker of liver tumor propagating
cells in hepatocellular carcinoma. Discov Med. 20:219–229.
2015.PubMed/NCBI
|
|
18
|
Zhu P, Wang Y, He L, Huang G, Du Y, Zhang
G, Yan X, Xia P, Ye B, Wang S, et al: ZIC2-dependent OCT4
activation drives self-renewal of human liver cancer stem cells. J
Clin Invest. 125:3795–3808. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang TS, Wu YC, Chi CC, Su WC, Chang PJ,
Lee KF, Tung TH, Wang J, Liu JJ, Tung SY, et al: Activation of
IL6/IGFIR confers poor prognosis of HBV-related hepatocellular
carcinoma through induction of OCT4/NANOG expression. Clin Cancer
Res. 21:201–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Kong C, Chen X, Wang Z, Wan Z, Jia
L, Liu Q, Wang Y, Li W, Cui J, et al: Repetitive exposure to
low-dose X-irradiation attenuates testicular apoptosis in type 2
diabetic rats, likely via Akt-mediated Nrf2 activation. Mol Cell
Endocrinol. 422:203–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao RC, Zhou J, Chen KF, Gong J, Liu J,
He JY, Guan P, Li B and Qin Y: The prognostic value of combination
of CD90 and OCT4 for hepatocellular carcinoma after curative
resection. Neoplasma. 63:288–298. 2016.PubMed/NCBI
|
|
22
|
Murakami S, Ninomiya W, Sakamoto E,
Shibata T, Akiyama H and Tashiro F: SRY and OCT4 are required for
the acquisition of cancer stem cell-like properties and are
potential differentiation therapy targets. Stem Cells.
33:2652–2663. 2015. View Article : Google Scholar : PubMed/NCBI
|