Introduction
Gene expression is a differential process under
precise control, and the alteration of gene expression via diverse
biochemical mechanisms may influence the fate of the cells with
negative consequences, leading to the emergence of serious diseases
(1,2).
As a mechanism of gene regulation, alternative splicing acts at the
post-transcriptional level by sequential addition of pre-mRNA
transcripts, which results in the generation of various mature
mRNAs (3–5). This mechanism serves an essential role
in generating proteome diversity in malignancies through the
production of splice variants involved in key oncogenic pathways
and resistance to chemotherapeutic drugs (6). Thus, alternative splicing has been
considered as a novel class of biomarker for the diagnosis of
various types of cancer, providing informative, specific and
accurate analysis for the study of cancer (7,8).
Musashi-1 (Msi1) is an RNA-binding protein that
negatively regulates genes at the post-transcriptional level by
competing with eukaryotic translation initiation factor 4 G (eIF4G)
(9). In mammals, Msi1 has been
described as a marker of adult stem cells and progenitor cells in
the central nervous system by regulating Notch signaling (10). A plethora of research demonstrates
that Msi1 binds with the 3′-Untranslated Region (UTR) of mRNA and
regulates mRNA stability and the translation of proteins involved
in essential oncogenic signaling pathways, and it is implicated in
maintaining cancer stem cell populations and promoting diverse
types of cancer (11–15).
The present study identified a novel splice
transcript of Msi1, namely Msi1 variant 2, which consists of 13
exons. Exon 3 and 4 skipping led to the acquisition of a new
in-frame TGA stop codon in exon 5, predicting a truncated product
lacking two RNA recognition motif (RRM) domains. This exon-skipping
event was confirmed using minigene plasmid, and could be detected
as a major RNA splice form of Msi1 compared with its canonical
transcript Msi1 in various cell lines. The present study further
investigated the preliminary function of exon 3 and 4 skipping of
Msi1 pre-mRNA in mediating hypoxia-induced cellular resistance to
cisplatin.
Materials and methods
Cell culture
NSCLC cell lines, including H460, H1792, H1975,
A549/DDP (16) and H1299, and one
established primary NSCLC cell strain, Lac521 (17), were cultured in RPMI-1640 medium
(Wisent Biotechnology, Nanjing, China) supplemented with 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA). The NSCLC A549 and SW1573 cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM) (Wisent Biotechnology)
supplemented with 10% FBS. Human colorectal carcinoma (CRC) cell
lines, including SW620, SW480, HT29, HCT116, Lovo and Caco-2, were
cultured in DMEM supplemented with 10% FBS. Human leukemia cell
lines, including Jurkat, HL-60, U937 [ATCC® CRL1593.2™;
American Type Culture Collection (ATCC), Manassas, VA, USA],
CCRF-CEM, K562 and THP-1, were cultured in RPMI-1640 medium
supplemented with 10% FBS. Human hepatocellular carcinoma (HCC)
cell lines, including SMMC7721 and BEL-7402, were cultured in DMEM
supplemented with 10% FBS. The human breast carcinoma MDA-MB-231
cell line was cultured in DMEM supplemented with 10% FBS. The human
cervical squamous carcinoma HeLa cell line and the human
osteosarcoma MG63 cell line were cultured in DMEM supplemented with
10% FBS. As non-cancerous cell lines, human normal bronchial
epithelial cell lines, including BEAS-2B and HBE1, were cultured in
RPMI-1640 medium supplemented with 10% FBS. The human normal liver
cell line HL-7702 (18) was cultured
in RPMI-1640 medium supplemented with 10% FBS. The cell line 293T
(used for transfection and expression analysis) was cultured in
DMEM supplemented with 10% FBS. The human normal pancreatic ductal
epithelial HPDE6-C7 cell line (19)
was cultured in RPMI-1640 medium supplemented with 10% FBS. The
A549/DDP, HL-7702 and HPDE6-C7 cell lines were purchased from
Shanghai Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences (Shanghai, China), as mentioned in previous
studies (16,18,19). The
remaining cell lines were obtained from the ATCC and were supplied
by its corporate agent, Zhongyuan Ltd. (Beijing, China).
Cell lines
In the present study, the Human non-small cell lung
carcinoma (NSCLC) H1792 cell line was used to amplify the
full-length cDNA of Msi1 variants while the 293T cell line was used
to Non-cancerous cells including BEAS-2B, HBE1, HPDE6-C7 and
HL-7702 and cancerous cells including NSCLC, CRC, HCC and leukemia
cell lines, in addition to Lac521 cells, were used to detect the
endogenous skipping of exons 3 and 4 of Msi1 pre-mRNA. The 293T
cell line was used to verify the expression of Msi1 constructs and
to confirm the existence of exon 3 and 4 skipping by a minigene
assay. The H460 cell line was used to investigate the role of
musashi-1 in mediating chemoresistance under hypoxia condition.
Antibodies
A rabbit polyclonal antibody against Msi1 (cat. no.
GTX101540) was purchased from GeneTex, Inc. (Irvine, CA, USA).
Mouse monoclonal anti-GAPDH immunoglobulin G (IgG) (cat. no.
sc-47724) and horseradish peroxidase (HRP)-conjugated secondary
IgGs, including goat-anti-rabbit IgG (cat. no. sc-2004) and
goat-anti-mouse IgG (cat. no. sc-2005), were purchased from Santa
Cruz Biotechnology, Inc. (Dallas, TX, USA).
Construction of Msi1 minigene
plasmid
Genomic DNA was extracted from the H1792 cell line
using DNAiso Reagent (Takara, Japan). Briefly, cells were scraped
and homogenized using 1 ml DNAiso Reagent. The supernatant was
removed into a new tube following centrifugation (12,000 × g for 10
min at 4°C) and 500-µl 1/2 volumes of ethanol was added. The DNA
pellet was collected into a new tube and washed with 75% ethanol
twice. The exon 2-intron 2-exon 3-intron 3-exon 4-intron 4-exon 5
(E2-I2-E3-I3-E4-I4-E5) region of Msi1 pre-mRNA was amplified using
he LA Taq polymerase (Takara, Japan) following amplification
protocol: 94°C for 1 min, 60°C, for 1 min, 72°C for 5 min followed
by post-extension for 15 min. Prior to amplification, the DNA
template was pre-denatured at 98°C for 10 sec. The amplified
fragment was digested with BamHI and HindIII and then
ligated into the mammalian expression vector pRK5 with a FLAG tag
sequence (The backbone plasmid was kindly provided by professor
Zichun Hua in School of Life Sciences in Nanjing University).
Small interfering RNA (siRNA)
transfections
For transient knockdown studies, 1×106
H460 cells were transfected with NC control or Msi1-specific siRNAs
supplied by Shanghai GenePharma Co., Ltd, (Shanghai, China). (100
nM) using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
The sequences of the designed siRNAs were as follows: Sense, Msi1-1
5′-GUGGAGAAAGUGUGUGAAAUU-3′ and Msi1-2 5′-GGGUUACCCAGGUUUCCAAGC-3′.
To examine the effect of hypoxia on chemoresistance to cisplatin,
H460 cells were transfected with Msi1-1 and Msi1-2 siRNA and then
cultured under normoxia (95% O2, 5% CO2) or
hypoxia (1% O2, 5% CO2, 94% N2).
After 24 h of siRNA-mediated knockdown, cells were treated with 10
µg/ml cisplatin purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany) and maintained under the same conditions for an
additional 24 h.
Cell apoptosis assay
Cell apoptosis was measured by Annexin V-fluorescein
isothiocyanate/propidium iodide double staining, according to the
manufacturer's protocols (BioVision, Inc., Milpitas, CA, USA). A
total of 10,000 cells were analyzed by flow cytometry. Data were
analyzed using FlowJo 7.6.1 software (FlowJo LLC, Ashland, OR,
USA).
Western blot analysis
Cells were lysed in ice-cold cell lysis buffer [50
mM Tris-HCl (pH 7.4), 250 mM NaCl, 50 mM NaF, 5 mM EDTA, 5 mM
β-glycerophosphate, 1 mM sodium vanadate, 1% Nonidet P-40 and
complete protease inhibitor cocktail (Roche Diagnostics,
Indianapolis, IN, USA). Upon centrifugation (12,000 × g for 10 min
at 4°C), the protein concentration was determined using the
Bradford method. Protein samples (40–60 µg) were separated by 10%
SDS-PAGE, and then transferred onto polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA), which were then
blocked with 5% w/v non-fat powdered milk in TBS Tween-20 (TBST)
for 1 h at room temperature (0.5 M NaCl, 20 mM Tris-HCl and 0.05%
v/v Tween-20; pH 7.4). The membranes were subsequently incubated
with primary antibodies, including anti-Msi1 IgG (1:1,000 dilution)
or anti-GAPDH IgG (1:1,000 dilution), at 4°C overnight. Following
three washes with TBS for 5 min each, the membranes were incubated
with HRP-conjugated secondary antibodies (1:500) at room
temperature for 2 h. The blots were visualized upon incubation with
the Amersham ECL start western blotting detection reagent (GE
Healthcare Life Sciences, Little Chalfont, UK).
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNAs were isolated with TRIzol Reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
following the manufacturer's protocol. RT reactions were performed
at 42°C for 1 h with oligo (dT) as a primer using the RevertAid RT
Reverse Transcription kit (Fermentas; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The sequences of the primers used for
amplifying the full length of Msi1 (primer set A) are
5′-TTGGATCCATGGAGACTGACGCGCCCCAG-3′ (forward) and
5′-TTAAGCTTTCAGTGGTACCCATTGGTGAAGGCT-3′ (reverse). The PCR product
was electrophoresed through a 0.8% agarose gel. The sequences of
the primers used for detecting the exon 3-exon 4 skipping event
(primer set B) are 5′-CAAGATGTTCATCGGGGGACTCAG-3′ (forward) and
5′-CTTGGGCTGTGCTCGCCGAGGGAAG-3′ (reverse). The sequences of primers
for the reference control GAPDH are 5′-TGTCCCCACTGCCAACGTGTCA-3′
(forward) and 5′-AGCGTCAAAGGTGGAGGAGTGGGT-3′ (reverse). To confirm
the existence of Msi1 variant 2, the FLAG tag was used as the
forward primer, alongside the downstream reverse primer in primer
set B, in order to detect the exogenous exon 2-exon 5 skipping
event using a minigene plasmid. The PCR protocol was as follows:
94°C for 30 sec, 60°C, for 45 sec, 72°C for 30 sec followed by
post-extension for 5 min. PCR products were electrophoresed through
a 2% agarose gel. Each gel was stained with ethidium bromide for
visualization under ultraviolet light.
Statistical analysis
Data are presented as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference. Multiple comparisons were performed with two-way
analysis of variance followed by Bonferroni's post hoc test. For
data analysis, GraphPad Prism 5.0 software for Windows was used
(GraphPad Software, Inc., La Jolla, CA, USA).
Results
Amplification of RNA variants of
Msi1
The Msi1 RT-PCR amplification products in the NSCLC
H1792 cell line were separated on a 0.8% agarose gel for analysis,
and two bands of Msi1 were observed (Fig.
1A). We speculated that at least two variants of Msi1 could
exist, and thus, each amplification product was subsequently cloned
and sequenced. Unexpectedly, three additional variants besides the
canonical Msi1 (1,089 bp) were observed. These new variants were
sequenced, and the 1,056-, 889- and 1,064-bp products were
identified as new variants of Msi1, termed variant 1, 2 and 3,
respectively (Fig. 1B).
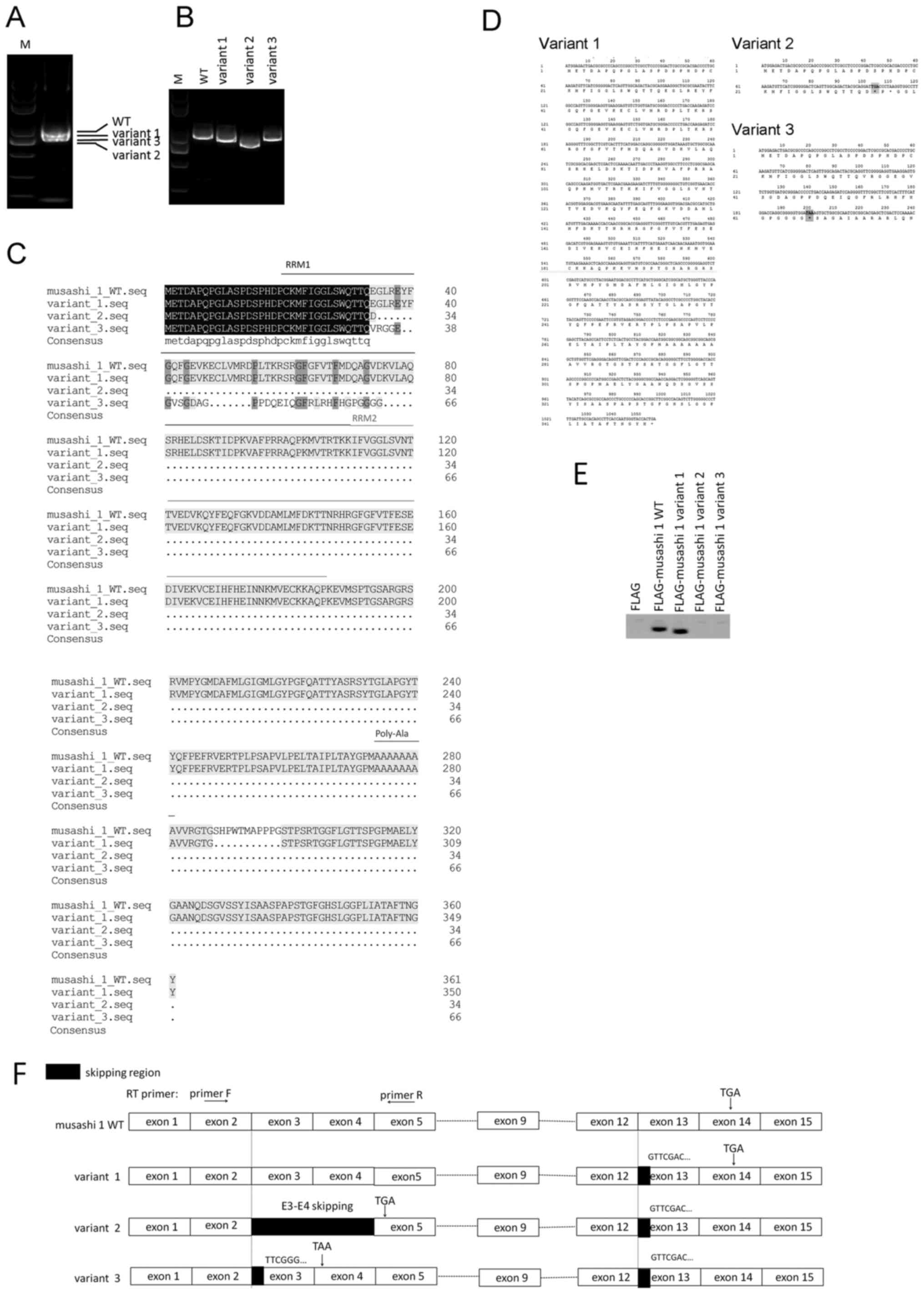 | Figure 1.Identification and translational
analysis of Msi1 variants. (A) Amplification of Msi1 transcripts.
(B) Cloning and separation of Msi1 variants. RT-PCR assays in (A)
and (B) were performed using primer set A, as described in the
Materials and methods. Bands were separated using 0.8% agarose gel
electrophoresis. (C) Comparison of the amino acid alignment of the
Msi1 canonical transcript and its variants. The predicted amino
acid sequences of three Msi1 variants are aligned with that of the
canonical transcript of Msi1. (D) Novel in-frame stop codons in
Msi1 variants 2 and 3. (E) Protein translation analysis of Msi1
variants. All Msi1 variants were cloned into the FLAG-tagged
mammalian expression vector pRK5 and transfected into 293T cells.
Western blotting was performed following 8% SDS-PAGE. (F) Schematic
representation of exon organization in Msi1 splice variants. The
Msi1 transcript contains 15 exons, while variant 2 exhibits a
skipping event of exons 3 and 4. Frameshift was observed in
variants 2 and 3 with the existence of new in-frame stop codons
(namely, TGA in variant 2 and TAA in variant 3). The arrows
indicate the position of the RT-PCR primers used to detect the exon
3 and 4 skipping event. Msi1, musashi-1; M, marker; WT, wild-type;
RT-PCR, reverse transcription-polymerase chain reaction; RRM, RNA
recognition motif; F, forward; R, reverse; E, exon. |
Protein translation analysis of novel
Msi1 transcripts
Alignment of the amino acid sequences of these novel
Msi1 transcripts demonstrated the presence of the premature stop
codons TGA in exon 5 for variant 2 and TAA in exon 4 for variant 3
due to ribosomal frameshift, predicting two truncated open reading
frames (ORFs) without the intact RRM domains (Fig. 1C, D). The three Msi1 variants and the
canonical form were cloned into the mammalian expression vector
pRK5, containing a FLAG tag. Then these plasmids were transfected
into 293T cells for 24 h. The exogenous FLAG was used as the
negative control. As demonstrated by western blotting in Fig. 1E, a band shift was observed between
Msi1 variant 1 and its wild-type version due to the lack of 11
amino acids in the C-terminus of variant 1 compared with the
canonical Msi1. However, Msi1 variants 2 and 3 could not be
detected by western blotting due to the acquisition of the
premature stop codons (Fig. 1E).
Exons 3 and 4 skipping in Msi1 variant
2 leads to the premature termination of Msi1 translation
In order to verify the site where the frameshift of
the Msi1 transcript occurs, an incomplete exon 3 in Msi1 variant 3
and a skipping event of two exons in variant 2 were identified,
which led to the existence of the premature stop codons TGA in
variant 2 and TAA in variant 3 (Fig.
1F). Msi1 variant 2 contains exons 2 and 5, but lacks exons 3
and 4, and the acquisition of the new in-frame stop codon TGA
results in a truncated ORF lacking the complete RRM1 and the
following domains (Fig. 2A). Next, a
minigene plasmid comprising E2-I2-E3-I3-E4-I4-E5 was constructed
and transfected into 293T cells. As revealed by RT-PCR analysis,
exogenous transcripts E2-E3-E4-E5 and E2-E5 were observed,
confirming the existence of exon 3 and 4 skipping of Msi1 pre-mRNA
in human cells (Fig. 2B).
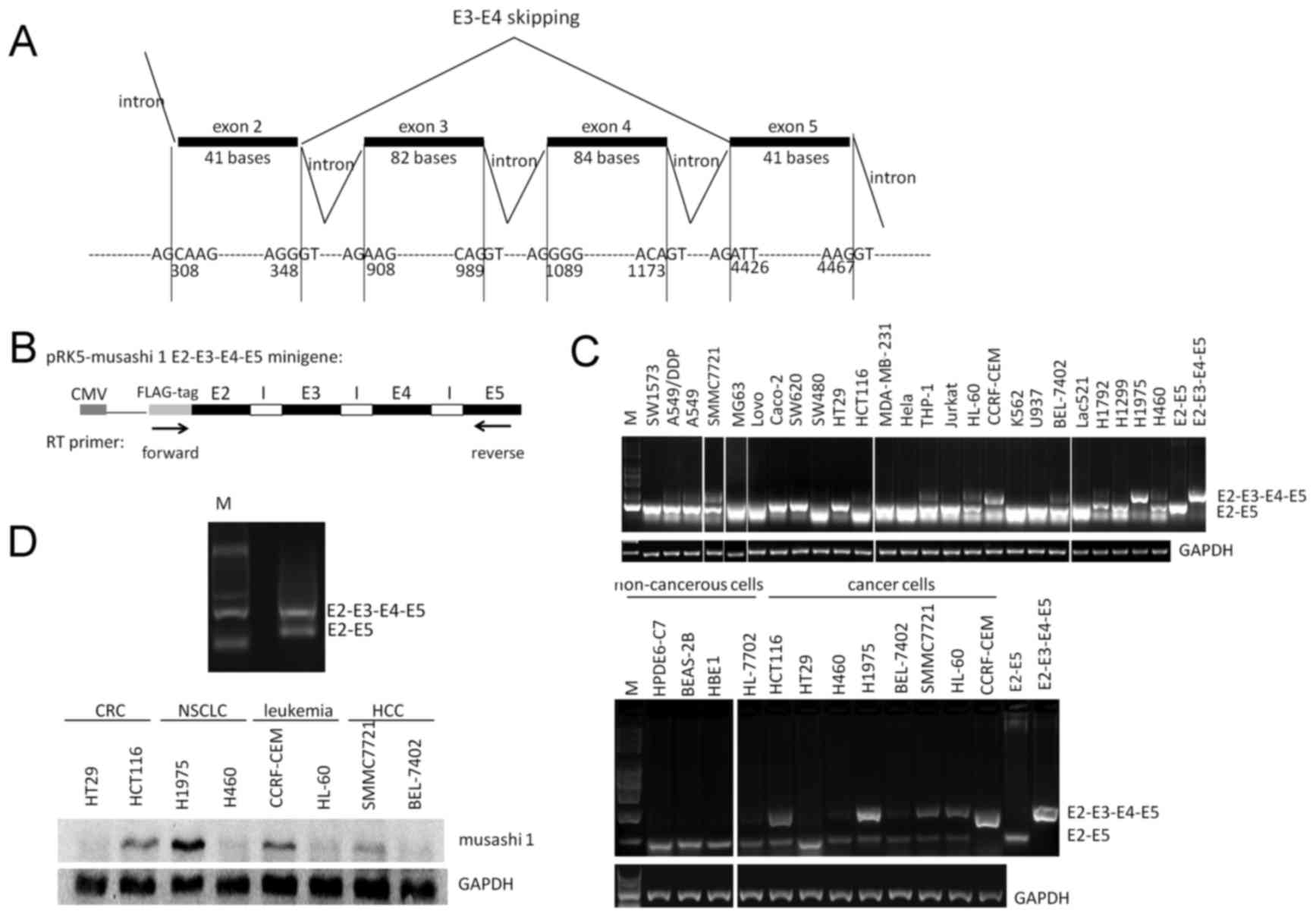 | Figure 2.Detection of exon 3 and 4 skipping of
Msi1 pre-mRNA. (A) Structure of the Msi1 spliceosome from exon 2 to
exon 5. Exon 3 and 4 skipping of Msi1 pre-mRNA results in a
sequence depletion of 166 nucleotides in Msi1 transcripts. (B)
Detection of exogenous exon 3 and 4 skipping of Msi1 pre-mRNA using
Msi1 E2-I2-E3-I3-E4-I4-E5 minigene plasmid. The arrows indicate the
position of the RT-PCR primers used to detect minigene splicing.
The exogenous exon skipping event of the Msi1 minigene plasmid was
detected using a forward primer specific for the FLAG tag and a
reverse primer from primer set B. (C) Detection of endogenous exon
3 and 4 skipping of Msi1 transcript in various cancer cells,
including primary Lac521 cells, and NSCLC, CRC, leukemia, HCC and
non-cancerous cell lines. Detection of exon 3 and 4 skipping was
performed via RT-PCR assays using primer set B, as described in the
Materials and methods. (D) Detection of Msi1 protein in various
cancer cell lines. Msi1, musashi-1; E, exon; I, intron; CMV,
cytomegalovirus; RT-PCR, reverse transcription-polymerase chain
reaction; CRC, colorectal carcinoma; NSCLC, non-small cell lung
carcinoma; HCC, hepatocellular carcinoma; M, marker. |
Msi1 variant 2 transcript is
preferentially expressed in non-cancerous cells
The exon 3 and 4 skipping event in Msi1 variant 2
could be distinguished from wild-type Msi1 using RT-PCR primers
specifically designed to flank the E2-E3-E4-E5 coding region of
Msi1. The mRNA level of Msi1 variant 2 was observed to be
universally expressed in various non-cancerous and cancerous cells,
including primary NSCLC Lac521 cells (Fig. 2C). Thus, Msi1 variant 2 could be a
major form of its splicing transcripts. While wild-type Msi1 was
predominantly expressed in cancerous cell lines, including NSCLC,
HCC, CRC and leukemia, Msi1 variant 2 was preferentially expressed
in non-cancerous cell lines (Fig.
2C). In addition, the expression level of Msi1 variant 2
transcript inversely associated with the protein level of wild-type
Msi1 in cancer cell lines (Fig.
2D).
Hypoxia-induced retention of exons 3
and 4 in Msi1 increases cisplatin resistance
Considering that Msi1 wild-type was preferentially
expressed in cancer cells, the present study evaluated the
contributing factors to exon 3 and 4 skipping of Msi1 pre-mRNA in
the tumor microenvironment. As a salient feature of solid tumors
(20), hypoxia (1% O2, 5%
CO2 and 94% N2) was observed to suppress
exogenous exon 3 and 4 skipping of Msi1 pre-mRNA (Fig. 3A). Consistent with the aforementioned
observation, endogenous detection of exon 3 and 4 skipping of Msi1
revealed that hypoxia induced the retention of exons 3 and 4 of
Msi1, and upregulated Msi1 protein levels (Fig. 3B). Hypoxic tumors are often resistant
to conventional cancer therapies. Thus, the present study examined
the effect of hypoxia on the chemoresistance to cisplatin. With the
upregulation of Msi1 protein level, hypoxia significantly increased
cisplatin resistance in H460 cells, which was compromised in the
absence of Msi1 (Fig. 3C). The
results of western blotting confirmed the knockdown effect of Msi1
using the RNAi method (Fig. 3D).
These observations indicate that hypoxia-induced cisplatin
resistance requires downregulation of exons 3 and 4 skipping of
Msi1 pre-mRNA.
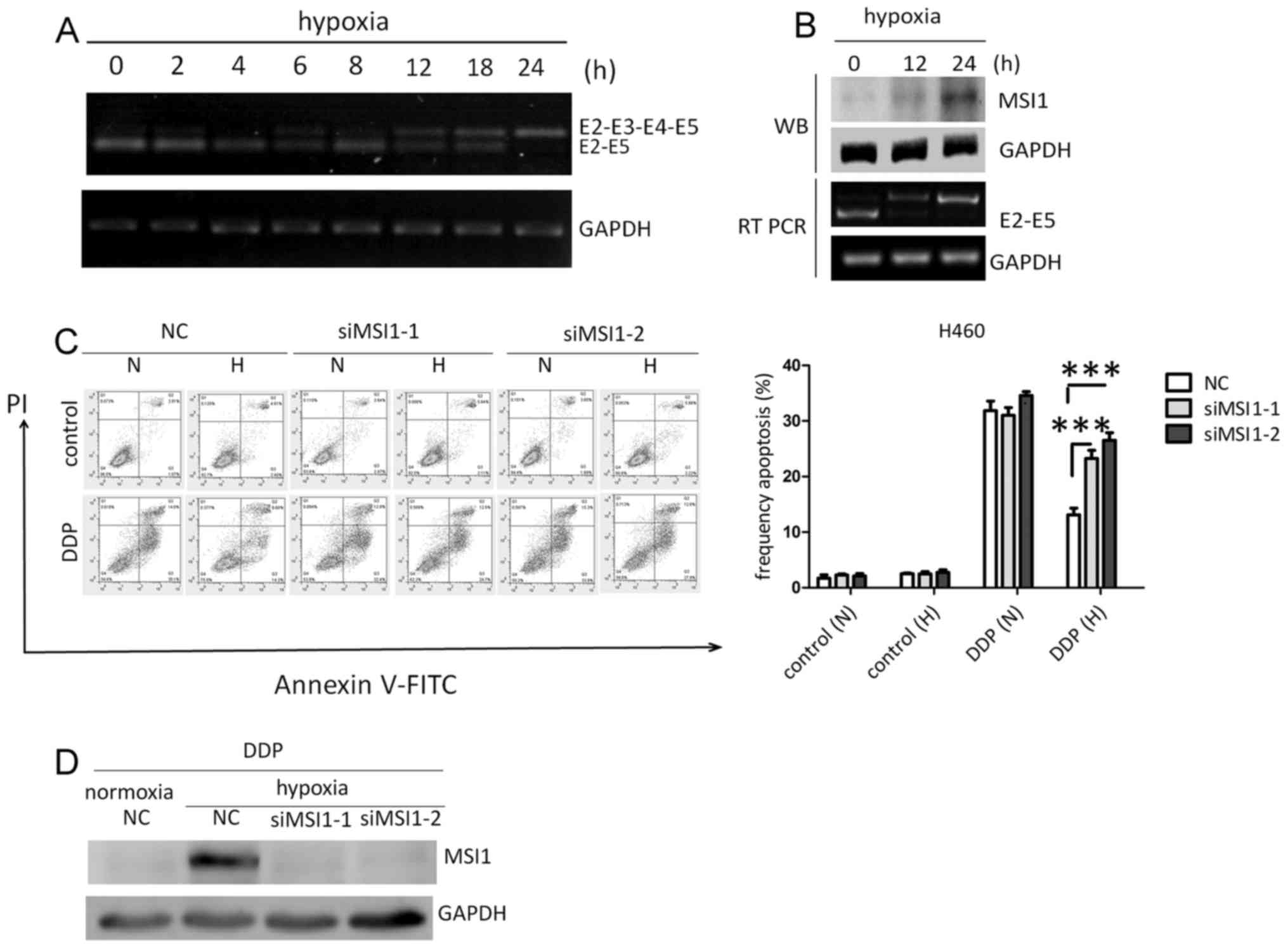 | Figure 3.Hypoxia increases cisplatin resistance
by promoting the retention of exons 3 and 4 of Msi1 pre-mRNA. (A)
Effect of hypoxia on the exogenous exon 3 and 4 skipping event of
Msi1 using Msi1 E2-I2-E3-I3-E4-I4-E5 minigene plasmid. (B) Effect
of hypoxia on the exogenous exon 3 and 4 skipping event of Msi1 and
its protein level. (C) Cisplatin-induced apoptosis in H460 cells
under normoxia or hypoxia, in the presence or absence of Msi1
protein. (D) Knockdown efficiency of Msi1 protein. Data from
triplicates are presented as the mean ± standard deviation.
***P<0.001 (two-way analysis of variance). Msi1, musashi-1; E,
exon; I, intron; WB, western blotting; RT-PCR, reverse
transcription-polymerase chain reaction; NC, negative control; si,
small interfering; PI, propidium iodide; FITC, fluorescein
isothiocyanate; N, normoxia; H, hypoxia; DDP, cisplatin. |
Discussion
The present study identified a novel splice variant
of Msi1, termed Msi1 variant 2, consisting of 13 exons. Msi1 has
been identified as a marker of progenitor stem cells (21). Notably, Msi1 was also identified as a
marker of CSCs (22,23). The present study demonstrated that the
skipping of exons 3 and 4 of Msi1 variant 2 led to the acquisition
of a premature stop codon in exon 5 of the Msi1 transcript,
resulting in the absence of complete RRM domains on the truncated
Msi1 transcripts. Thus, this exon skipping event of Msi1 could be a
way of negatively regulating cancer stemness stemness.
The biochemical mechanism by which Msi1 positively
regulates cancer stemness or tumorigenesis is currently under
investigation. Previous studies revealed that Msi1 suppresses genes
such as Numb and p21 (Waf1) by competing with eIF4G (9,10,24). These mRNAs are implicated in cell
proliferation and protein modification, which are involved in
tumorigenesis (25–27). However, the role of Msi1 in the
progenitor stem cell compartment has been described only in a
number of systems (12,28). In the present study, the expression of
Msi1 variant 2 was observed in various cancerous cell lines,
including NSCLC, breast, colon and leukemia cell lines, as well as
in several non-cancerous cell lines, suggesting that Msi1 variant 2
may be a broad-spectrum splice variant compared with its wild-type
variant, which was preferentially expressed in malignant cells.
In addition, the Msi1 variant 2 does not contain two
complete RRM domains, which are required for binding RNAs (29,30).
Although the present study has not demonstrated whether Msi1 and
its variant 2 differ in their ability to interact with RNA, we
anticipate that in the absence of RRM domains the function of Msi1
variant 2 may not be associated with the repression of
translational initiation by competitively binding to
Musashi-binding-element in the 3′-untranslated region of target
transcripts. Thus, it is likely that Msi1 and its variant 2 may
have different spectra of biological activity. The existence of
distinct isoforms of Msi1 suggests that this gene may serve a
complex role in regulating tumorigenesis, and elucidating the
precise functions of its variants may provide further insights into
the regulation of tumorigenesis and stemness.
An important strength of the current study is the
association between the exon 3 and 4 skipping event of Msi1 and
hypoxia-induced chemoresistance in NSCLC. Although solid tumors
typically develop hostile microenvironments characterized by stress
conditions, including irregular vascularization, and poor oxygen
and nutrient supply, the cancer cells in these tumors exhibit
unregulated growth under such hostile conditions through the
activation of a series of survival pathways (31,32). Under
hypoxia, chemoresistance has been reported to be upregulated in
various types of cancer (33,34). Notably, Msi1 has been identified as an
oncogene in several malignancies (15,35). In
the present study, hypoxia-induced cisplatin resistance in H460
cells required the retention of exons 3 and 4 in the Msi1
transcript, which frequently occurred in cancer cells compared with
non-cancerous cells. Thus, the generation of Msi1 variant 2 by
alternative splicing serves a tumor-suppressing role, and reversal
of this exon skipping event of Msi1 under stress conditions may be
responsible for tumorigenesis and malignant progression with
increased chemoresistance.
Collectively, the present study achieved the
following: i) Identified a novel untranslated isoform of Msi1
generated by skipping of exons 3 and 4; ii) evaluated its
expression in various cell lines; and iii) preliminarily described
the role of this exon skipping event in mediating chemoresistance.
The aforementioned results indicate that the occurrence of the exon
3 and 4 skipping event in Msi1 could represent an important tool
for malignancy diagnosis and therapy. With regard to therapeutic
implications, targeting exon 3 and 4 skipping in Msi1 pre-mRNA
could represent a valuable strategy to repress Msi1 signaling in
tumors overexpressing this protein. However, there are several
limitations affecting the present study. First, small molecules
that could promote exon 3 and 4 skipping of Msi1 could not be
identified due to the limited number of drugs tested. Second, the
effect of exon 3 and 4 skipping of Msi1 on cancer stemness should
be further investigated. In addition, future studies are required
in order to clarify the exact role of Msi1 variant 2 in the
development of malignancies.
Acknowledgements
The authors would like to thank Professor Zichun Hua
(State Key Laboratory of Pharmaceutical Biotechnology, College of
Life Sciences, Nanjing University) for providing the plasmid
pRK5-FLAG.
Funding
The present study was funded by the Natural Science
Foundation of China (grant nos. 31071250, 81673462, 81473293,
91540119 and J1103521) and the Fundamental Research Funds for
Central Universities via the Six Talent Peaks Project of Jiangsu
(grant no. YY-012). The funders had no role in the study design,
data collection, data analysis, decision to publish or preparation
of the present manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY, LM and HM designed the project. LM, YS, KY
performed the experiments. HM, IE and MN analyzed the experimental
data and offered discussion and suggestion. LM, YS, KY, IE and MN
wrote the manuscript. WY and HM provided the financial support.
Ethics approval and consent to
participate
The present article does not report any studies on
human participants performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Rosenthal N: Regulation of gene
expression. N Engl J Med. 331:931–933. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krijger PH and De Laat W: Regulation of
disease-associated gene expression in the 3D genome. Nat Rev Mol
Cell Biol. 17:771–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopez AJ: Alternative splicing of
pre-mRNA: Developmental consequences and mechanisms of regulation.
Annu Rev Genet. 32:279–305. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hong H, Braunschweig U,
Gonatopoulos-Pournatzis T, Weatheritt RJ, Hirsch CL, Ha KCH,
Radovani E, Nabeel-Shah S, Sterne-Weiler T, Wang J, et al:
Multilayered control of alternative splicing regulatory networks by
transcription factors. Mol Cell. 65:539–553.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Talavera D, Orozco M and de la Cruz X:
Alternative splicing of transcription factors' genes: Beyond the
increase of proteome diversity. Comp Funct Genomics. 2009:Article
ID 9058942009. View Article : Google Scholar
|
|
6
|
Singh B and Eyras E: The role of
alternative splicing in cancer. Transcription. 8:91–98. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim YJ and Kim HS: Alternative splicing
and its impact as a cancer diagnostic marker. Genomics Inform.
10:74–80, 2017. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hofstetter G, Berger AH, Slade N, Zorić A,
Holzer B, Schuster E, Schuster E, Mobus VJ, Reimer D, Daxenbichler
G, et al: Alternative splicing of p53 and p73: The novel p53 splice
variant p53delta is an independent prognostic marker in ovarian
cancer. Oncogene. 29:1997–2004. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hironori K, Takao I, Hiroaki I, Masafumi
T, Ken M and Hideyuki O: Neural RNA-binding protein Musashi1
inhibits translation initiation by competing with eIF4G for PABP. J
Cell Biol. 181:639–653. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Imai T, Tokunaga A, Yoshida T, Hashimoto
M, Mikoshiba K, Weinmaster G, Nakafuku M and Okano H: The neural
RNA-binding protein Musashi1 translationally regulates mammalian
numb gene expression by interacting with its mRNA. Mol Cell Biol.
21:3888–3900. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen HY, Lin LT, Wang ML, Lee SH, Tsai ML,
Tsai CC, Liu WH, Chen TC, Yang YP, Lee YY, et al: Musashi-1
regulates AKT-derived IL-6 autocrinal/paracrinal malignancy and
chemoresistance in glioblastoma. Oncotarget. 7:42485–42501.
2016.PubMed/NCBI
|
|
12
|
Rezza A, Skah S, Roche C, Nadjar J,
Samarut J and Plateroti M: The overexpression of the putative gut
stem cell marker Musashi-1 induces tumorigenesis through Wnt and
Notch activation. J Cell Sci. 123:3256–3265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Toda M, Iizuka Y, Yu W, Imai T, Ikeda E,
Yoshida K, Kawase T, Kawakami Y, Okano H and Uyemura K: Expression
of the neural RNA-binding protein Musashi1 in human gliomas. Glia.
34:1–7. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishimoto Y and Okano H: New insight into
cancer therapeutics: Induction of differentiation by regulating the
Musashi/Numb/Notch pathway. Cell Res. 20:1083–1085. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo
B, Okano H, Penalva LO and Glazer RI: Musashi1 as a potential
therapeutic target and diagnostic marker for lung cancer.
Oncotarget. 4:739–750. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi H, Pu J, Zhou XL, Ning YY and Bai C:
Silencing long non-coding RNA ROR improves sensitivity of
non-small-cell lung cancer to cisplatin resistance by inhibiting
PI3K/Akt/mTOR signaling pathway. Tumour Biol. 39:2017. View Article : Google Scholar
|
|
17
|
Li X, You M, Liu YJ, Ma L, Jin PP, Zhou R,
Zhang ZX, Hua B, Ji XJ, Cheng XY, et al: Reversal of the apoptotic
resistance of non-small-cell lung carcinoma towards TRAIL by
natural product toosendanin. Sci Rep. 7:427482017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui R, Zhang H, Guo X, Cui Q, Wang J and
Dai J: Proteomic analysis of cell proliferation in a human hepatic
cell line (HL-7702) induced by perfluorooctane sulfonate using
iTRAQ. J Hazard Mater. 299:361–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wan C, Gong C, Ji L, Liu X, Wang Y, Wang
L, Shao M, Yang L, Fan S, Xiao Y, et al: NF45 overexpression is
associated with poor prognosis and enhanced cell proliferation of
pancreatic ductal adenocarcinoma. Mol Cell Biochem. 410:25–35.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Muz B, de la Puente P, Azab F and Azab AK:
The role of hypoxia in cancer progression, angiogenesis,
metastasis, and resistance to therapy. Hypoxia (Auckl). 3:83–92.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okano H, Kawahara H, Toriya M, Nakao K,
Shibata S and Imai T: Function of RNA-binding protein Musashi-1 in
stem cells. Exp Cell Res. 306:349–356. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiou GY, Yang TW, Huang CC, Tang CY, Yen
JY, Tsai MC, Chen HY, Fadhilah N, Lin CC and Jong YJ: Musashi-1
promotes a cancer stem cell lineage and chemoresistance in
colorectal cancer cells. Sci Rep. 7:21722017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao W, Li Y and Zhang X: Stemness-related
markers in cancer. Cancer Transl Med. 3:87–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Battelli C, Nikopoulos GN, Mitchell JG and
Verdi JM: The RNA-binding protein Musashi-1 regulates neural
development through the translational repression of p21WAF-1. Mol
Cell Neurosci. 31:85–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hong J, Liu Z, Zhu H, Zhang X, Liang Y,
Yao S, Wang F, Xie X, Zhang B, Tan T, et al: The tumor suppressive
role of NUMB isoform 1 in esophageal squamous cell carcinoma.
Oncotarget. 5:5602–5614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang Y, Ding M, Tian G, Guo H, Wan Y, Yao
Z, Li B and Lin D: Overexpression of Numb suppresses tumor cell
growth and enhances sensitivity to cisplatin in epithelioid
malignant pleural mesothelioma. Oncol Rep. 30:313–319. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chilosi M, Doglioni C, Magalini A,
Inghirami G, Krampera M, Nadali G, Rahal D, Pedron S, Benedetti A,
Scardoni M, et al: p21/WAF1 cyclin-kinase inhibitor expression in
non-Hodgkin's lymphomas: A potential marker of p53 tumor-suppressor
gene function. Blood. 88:4012–4020. 1996.PubMed/NCBI
|
|
28
|
Wolfe AR, Ernlund A, Mcguinness W, Lehmann
C, Carl K, Balmaceda N and Neufeld KL: Suppression of intestinal
tumorigenesis in Apc mutant mice upon Musashi-1 deletion. J Cell
Sci. 130:805–813. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maris C, Dominguez C and Allain FH: The
RNA recognition motif, a plastic RNA-binding platform to regulate
post-transcriptional gene expression. FEBS J. 272:2118–2131. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pastore C, Topalidou I, Forouhar F, Yan
AC, Levy M and Hunt JF: Crystal structure and RNA binding
properties of the RNA recognition motif (RRM) and AlkB domains in
human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA
hypermodification. J Biol Chem. 287:2130–2143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giampietri C, Petrungaro S, Conti S,
Facchiano A, Filippini A and Ziparo E: Cancer microenvironment and
endoplasmic reticulum stress response. Mediators Inflamm.
2015:4172812015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Urra H, Dufey E, Avril T, Chevet E and
Hetz C: Endoplasmic reticulum stress and the hallmarks of cancer.
Trends Cancer. 2:252–262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sullivan R, Paré GC, Frederiksen LJ,
Semenza GL and Graham CH: Hypoxia-induced resistance to anticancer
drugs is associated with decreased senescence and requires
hypoxia-inducible factor-1 activity. Mol Cancer Ther. 7:1961–1973.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schnitzer SE, Weigert A, Zhou J and Brüne
B: Hypoxia enhances sphingosine kinase 2 activity and provokes
sphingosine-1-phosphate-mediated chemoresistance in A549 lung
cancer cells. Mol Cancer Res. 7:393–401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lan L, Appelman C, Smith AR, Yu J, Larsen
S, Marquez RT, Liu H, Wu X, Gao P, Roy A, et al: Natural product
(−)-gossypol inhibits colon cancer cell growth by targeting
RNA-binding protein Musashi-1. Mol Oncol. 9:1406–1420. 2015.
View Article : Google Scholar : PubMed/NCBI
|

















