Introduction
The incidence of pancreatic cancer is increasing
globally (1). The latest data from
the American Cancer Society for the USA reveals that the incidence
of pancreatic cancer is 11 per 100,000 people, ranking it tenth of
the known malignant tumors; the mortality rate has been ranked
fourth out of the known malignant tumors (2). In addition, epidemiological data from
China demonstrates an increasing incidence in pancreatic cancer
(3–6).
Pancreatic cancer stem cells (CSCs) are considered
to influence the early metastasis of pancreatic cancer, are
insensitive to radiation and chemotherapy and are the main cause of
rapid disease progression (7). CSCs
cannot be eradicated, and as such drive invasion, metastasis and
recurrence, and are the source radiation and chemotherapy
resistance (8). In 2007, Lee et
al (9) using flow cytometry to
isolate cluster of differentiation
(CD)44+CD24+, epithelial surface
antigen+ pancreatic cancer cells from surgically removed
specimens. These cells have the abilities of self-renewal and
multi-directional differentiation, and may adapt to alterations in
circumstances. Lee et al (9)
hypothesized that this group of pancreatic cancer cells were
pancreatic CSCs. In recent years, one study has validated that the
Hedgehog (Hh) signaling pathway exhibits a function in the process
of embryonic development (10). The
abnormal activation of growth and development is associated with
the development of tumors (1,2). In 2003, Berman et al (11) and Thayer et al (12) demonstrated an association between the
Hh signaling pathway and pancreatic cancer. Thayer et al
(12) used 26 cancer cell lines,
including pancreatic cancer (either from a human primary source or
a metastatic tumor), to screen the expression of members of the Hh
signaling pathway. Thayer et al (12) demonstrated that expression of members
of the Hh signaling pathway, smoothened (Smo), zinc finger protein
Gli1 (GLI-3) and Patch-1, was observed in each pancreatic cell
line. The occurrence of Hh signaling pathway activation in
pancreatic cancer serves an important role in its development
(11,12). Smo a typical G protein coupled
receptor, is a positive regulator of cellular proliferation and
differentiation in insects and vertebrates (13,14).
Abnormal activation of Smo leads to development of a number of
types of cancer, which makes Smo an attractive therapeutic target.
At present, vismodegib, an inhibitor of Smo, is approved by the
food and drug administration for the treatment of cancer; however,
cancer cells may acquire resistance (15,16). A
number of resistance mechanisms have been demonstrated, including
mutations in Smo, the negative control receiver, suppressor of
fused homolog (Sufu) and the mitogen-activated protein kinase
signaling pathway (13). Therefore,
an improved understanding of the Smo regulating mechanism to
develop effective therapies to treat cancer caused by Smo mutations
is required.
Small interfering RNA (siRNA) is an RNA molecule
that can efficiently and specifically degrade target gene mRNA to
inhibit the excessive expression of genes. In addition, they may be
used as a treatment in patients and are a useful tool for studying
gene function in the future. The occurrence and development of
pancreatic cancer is a multi-factor and multi-stage process
(17,18). In the present study, experimental
siRNA stability infection in pancreatic cancer cells of
CD44+ CD24+ cells was used to determine
whether or not RNA and protein expression was affected.
Patients and methods
Patients
A total of 19 cases (11 males, 8 females; mean age,
59.89 years; range, between 40 and 78 years) of pancreatic cancer
were included in the present study. There were 9 tumors of the head
of the pancreas, 9 tumors of the body and tail of the pancreas, and
1 pancreatic neck tumor. Pathological results according to the
pathology classification system of the Cancer Hospital Affiliated
to Xinjiang Medical University (Urumqi, China), included the
following: 2 cases of highly differentiated carcinoma, 4 cases of
moderately/highly differentiated carcinoma, 8 cases of moderately
differentiated carcinoma, 4 cases of moderately/poorly
differentiated carcinoma and 1 case of poorly differentiated
carcinoma. Local lymph node metastasis was identified in 7 cases.
The patients were all treated at the Cancer Hospital Affiliated to
Xinjiang Medical University between January 2014 and December 2015.
The Ethics committee of the Affiliated Cancer Hospital to Xinjiang
Medical University approved the present study and all patients
provided written informed consent.
Materials
SDS-PAGE gel preparation kit, dNTPs and Taq DNA
polymerase, 2 kb plus DNA Marker and PCR primers were obtained from
Shanghai Shenggong Biology Engineering Technology Service, Ltd.
(Shanghai, China). The PCR reagent, primers and dsDNA oligos were
obtained from Shanghai Jikai Gene Chemical Technology Co., Ltd.
(Shanghai, China). Taq polymerase was obtained from Clontech
Laboratories, Inc. (Mountainview, CA, USA) and the Qiagen plasmid
Maxi kit was obtained from Qiagen, Inc. (Valencia, CA, USA). Bovine
serum albumin (BSA) was obtained from Shanghai JIEBEISI Gene
Technology Co., Ltd. (Shanghai, China). Lysogeny broth (LB), super
optimal broth (SOB) and super optimal broth with catabolite
repression (SOC) were obtained from American Type Culture
Collection (Manassas, VA, USA). T4 DNA ligase, T4 DNA ligase
buffer, and the restriction enzymes AgeI, EcoRI,
HpaI and XhoI were obtained from New England BioLabs,
Inc. (Ipswich, MA, USA). MgSO4 was obtained from Wuhua
Chemical Industry Co., Ltd. (Shanghai, China). Agarose was obtained
from Saibaisheng Biochemical Co., Ltd. (Beijing, China). The 250 bp
DNA ladder marker was obtained from Beijing Huada Jierui
Biotechnology Co., Ltd. (Beijing, China). Gene sequencing for the
positive clone was performed by Shanghai Genechem Co., Ltd.
(Shanghai, China).
The intermediate clone vector pUC57,
pLenti6/V5-D-TOPO recombinant lentiviral vector,
ViraPower™ Packaging Mix, viral quantitative primers and
the TaKaRaMiniBEST Viral RNA/DNA Extraction kit were obtained from
Baiao Maike Bio-Technology Co., Ltd. (Nantong, China).
Lipofectamine® 2000 transfection reagent kit was
obtained from Invitrogen (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). TOP10 chemical competent cells and 293T virus packaging
cells were obtained from the Cell Bank of Type Culture Collection
of Chinese Academy of Sciences (Shanghai, China). Dulbecco's
modified Eagle's medium (DMEM), fetal bovine serum (FBS), trypsin,
and OptiMEM medium were obtained from Gibco; Thermo Fisher
Scientific, Inc.
Sorting of
CD44+CD24+ cells
BXPC-3, PANC-1 and SW1990 (all from Shanghai
Genechem Co., Ltd., Shanghai, China) pancreatic cancer cell lines
were cultured at 37°C in an atmosphere containing 5% CO2
in RMPI-1640 (Hyclone; Thermo Fisher Scientific, Inc.) or DMEM,
supplemented with 10% FBS. Subsequently,
CD44+CD24+ cells were sorted from the three
cell lines using flow cytometry as previously described (19,20).
RT-PCR
TRIzol® (Thermo Fisher Scientific, Inc.)
was used to extract total RNA from SW1990 cellsof each experimental
group, according to the manufacturers protocol, and the RNA
concentration and purity were detected using a spectrophotometer.
The RNA was reverse transcribed into complementary DNA according to
the manufacturers protocol of the two-step M-MuLV First Strand cDNA
Synthesis kit (cat no. B532435; Sangon Biotech Co., Ltd., Shanghai,
China) according to the manufacturers protocol. For the PCR
reaction, sonic hedgehog protein (Shh), Smo, protein patched
homolog 1 (Ptch1), zinc finger protein Gli1 (Gli1), zinc finger
protein Gli2 (Gli2), Sufu and apoptosis regulator Bcl-2 (Bcl-2)
primers (Table I) were designed and
synthesized according to the sequences in GeneBank, as previously
described (21). Primer sequences and
their annealing temperatures, including for the reference gene
β-actin, are provided in Table I. PCR
was performed with the Bio-Rad MyiQ™2 two-color
real-time PCR detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) using SYBR Green I as the fluorophore. The
components and their volumes in the polymerase chain reaction are
shown in Table II. The reaction
conditions used were: 95°C for 5 min, followed by 40 cycles of 95°C
for 10 sec, 60°C for 30 sec. Quantitative analysis of Shh, Smo,
Ptch1, Gli1 and Gli2, Sufu and Bcl2 mRNA expression was performed
with the 2−∆∆Cq method (22). Data analysis was performed using
Microsoft Excel 2016.
 | Table I.Primers used for reverse
transcription-polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-polymerase chain reaction.
| Gene | Primer
sequences | Amplification
fragment size, bp | Annealing
temperature, °C |
|---|
| Sonic hedgehog | Forward:
5′-GTCTCCTCGCTGCTGGTATG-3′ | 150 | 56 |
|
| Reverse:
5′-TTGGGGATAAACTGCTTGTAGG-3′ |
|
|
| Protein patched
homolog 1 | Forward:
5′-CTCCTTTGCGGTGGACAA-3′ | 109 | 54 |
|
| Reverse:
5′-CCTCAGCCTTATTCAGCATTTC-3′ |
|
|
| Smoothened | Forward:
5′-CTCCTACTTCCACCTGCTCAC-3′ | 105 | 57 |
|
| Reverse:
5′-CAAAACAAATCCCACTCACAGA-3′ |
|
|
| Zinc finger protein
GLI1 | Forward:
5′-ATCCTTACCTCCCAACCTCTGT-3′ | 84 | 55 |
|
| Reverse:
5′-AACTTCTGGCTCTTCCTGTAGC-3′ |
|
|
| Zinc finger protein
GLI2 | Forward:
5′-GCGGAATTCGCAACGGAATG-3′ | 472 | 55 |
|
| Reverse:
5′-GCTGGATCCTTAGTCACA-3′ |
|
|
| Suppressor of
fused | Forward:
5′-CGGACCCACCAGAAGCGG-3′ | 398 | 52 |
| homolog | Reverse:
5′-GGAGGCGTCCTTCCGAC-3′ |
|
|
| B-cell
lymphoma | Forward:
5′-ACCTTAGCCCCATGCATTCTG-3′ | 287 | 54 |
|
| Reverse:
5′-CTAATCGGCTAGCTTCGAAAT-3′ |
|
|
| β-actin | Forward:
5′-GGGACCTGACTGACTACCTC-3′ | 543 | 56 |
|
| Reverse:
5′-CGTCATACTCCTGCTTGCTG-3′ |
|
|
 | Table II.Polymerase chain reaction components
and volumes. |
Table II.
Polymerase chain reaction components
and volumes.
| Reagent | Volume per
reaction/µl |
|---|
| 2X ES Tap Master
Mix | 12.5 |
| Upstream primer (10
µmol/l) | 1 |
| Downstream primer
(10 µmol/l) | 1 |
| RNase-free
water | 8.5 |
| Complementary DNA
template | 2 |
| Total volume | 25 |
Western blot analysis
A total protein extraction kit (Trizma®
base Vetec reagent grade ≥99%; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) was used to extract the total protein of the
cells and the quantity of protein was determined using a BCA
protein concentration detection kit. A total of 125 µg of protein
was used for 12% SDS-PAGE electrophoresis. Proteins were
transferred to polyvinylidene fluoride membranes and blocked with
5% bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.) at
room temperature for 2 h. The membranes were washed three times
with TBS-Tween-20 (TBST) and were incubated with the following
primary antibodies overnight at 4°C with gentle agitation: Bcl-2
(1:1,000; cat no. ab32124), Gli1 (1:500; cat no. ab92611), SMO
(1:1,000; cat no. ab38686), PTCH1 (1:500; cat no. ab53715), Gli2
(1:1,000; cat no. ab26056) and SHH (1:2,000; cat no. ab53281;
Abcam, Cambridge, MA, USA). The membranes were washed three times
with TBST prior incubation with the alkaline phosphatase-conjugated
anti-rabbit secondary antibody (1:2,000; cat no. WB7105;
WesternBreeze; Thermo Fisher Scientific, Inc.) for 30 min at room
temperature. Membranes were washed three times with TBST and the
blots were developed using enhanced chemiluminescence kit (Western
Breeze Chemiluminescence Reagent kit; cat. no. WB7105; Thermo
Fisher Scientific, Inc.). Using β-actin as a reference, the
relative protein expression was calculated. The Gel Doc XR+ system
(Bio-Rad Laboratories, Inc.) was used to capture and analyze
results of gels.
Construction of the lentiviral
expression vector expressing Smo siRNA
For the design and synthesis of Smo siRNA fragments,
the information and the full-length nucleotide sequence of the
human Smo gene were obtained by searching the NCBI GenBank database
(https://www.ncbi.nlm.nih.gov/genbank/; no. NM_005631)
and used to design three siRNA targets (Table III). Three siRNAs were used in the
current study: siRNA1 (TGATGGACACAGAACTCAT); siRNA2
(GGAGAAGATCAACCTGTTT) and siRNA3 (TGACTGTGAGATCAAGAAT) (Shanghai
Genechem CO., Ltd.).
 | Table III.siRNA targets designed using the NCBI
GenBank database. |
Table III.
siRNA targets designed using the NCBI
GenBank database.
| siRNA target | Target sequence
information | GC content, % | Start position |
|---|
|
SMO-RNAi(37303–1)a |
TGATGGACACAGAACTCAT | 42.1 | 2604 |
|
SMO-RNAi(37304–1)b |
GGAGAAGATCAACCTGTTT | 42.1 | 1831 |
|
SMO-RNAi(37305–1)c |
TGACTGTGAGATCAAGAAT | 36.8 | 1795 |
293T cells were cultured in DMEM, containing 10% FBS
and seeded into 24-well culture plates (3.5×10−4
cells/ml) one day prior to transfection using carrier GV plasmid
(20 µg;), PHelper1.0 carrier plasmid (15 µg) and PHelper 2.0
carrier plasmid, 10 µg) (Shanghai Genechem, Co., Ltd.), with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) After 24 h, the expression of the intracellular
fluorescent-labeled gene (GFP) was observed under a fluorescence
microscope, and then the cells were collected for western blot
analysis 36 h following transfection. The viral titer was
determined by Virus drops degree detection; fluorescence/absolute
quantitative method (23).
The CD44+CD24+ cells that were
sorted as previously mentioned were cultured to the logarithmic
growth phase and then digested into single cell suspension with
trypsin. 293T cells were cultured for 24 h to 90% confluence,
washed three times, then incubated in a 5% CO2 incubator
at 37°C for 20 min. Cells were transfected with the
Lipofectamine® 2000 transfection kit, according to the
manufacturer's protocol. Following incubation at 37°C for 72 h, the
SMO gene expression in the cells of each group was determined by
the RT-PCR method, the Smo siRNA with the highest transfection
efficiency was screened for the follow-up study.
Statistical analysis
Analysis of expression of the genes was performed
using the SASJMP10.0 software (SAS Institute, Cary, NC, USA). Data
are expressed as the mean ± standard deviation. Differences among
>2 groups were assessed using one-way analysis of variance with
Bonferroni post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
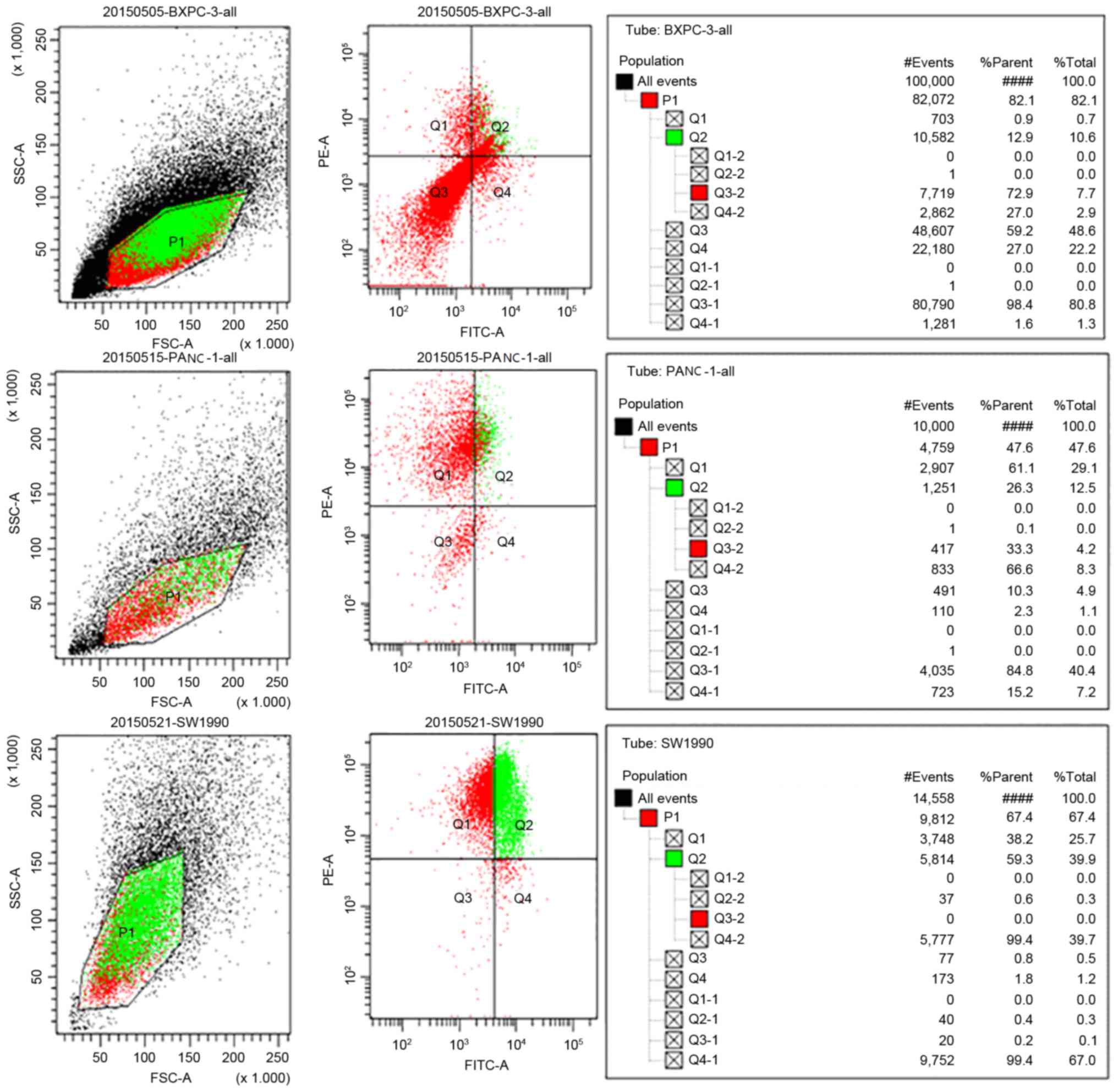
Sorting of
CD44+CD24+ cells
Three lines of
CD44+CD24+pancreatic cancer cells were
isolated using flow cytometry (BXPC-3, 10.6%
CD44+CD24+ cells; PANC-1, 12.5%
CD44+CD24+ cells; SW1990, 39.9%
CD44+CD24+ cells; Fig. 1). CD44+CD24+
SW1990 pancreatic cancer cells were selected for further
experiments.
Expression of Hedgehog (Hh) signaling
pathway members in pancreatic cancer cells
The expression of the Hh signaling pathway members
Shh, Smo, Ptch1, Gli1, Gli2, Sufu and Bcl2 (24) was investigated in unsorted SW1990
pancreatic cancer cells, sorted CD44+CD24+
cells, sorted CD44−CD24− cells and normal
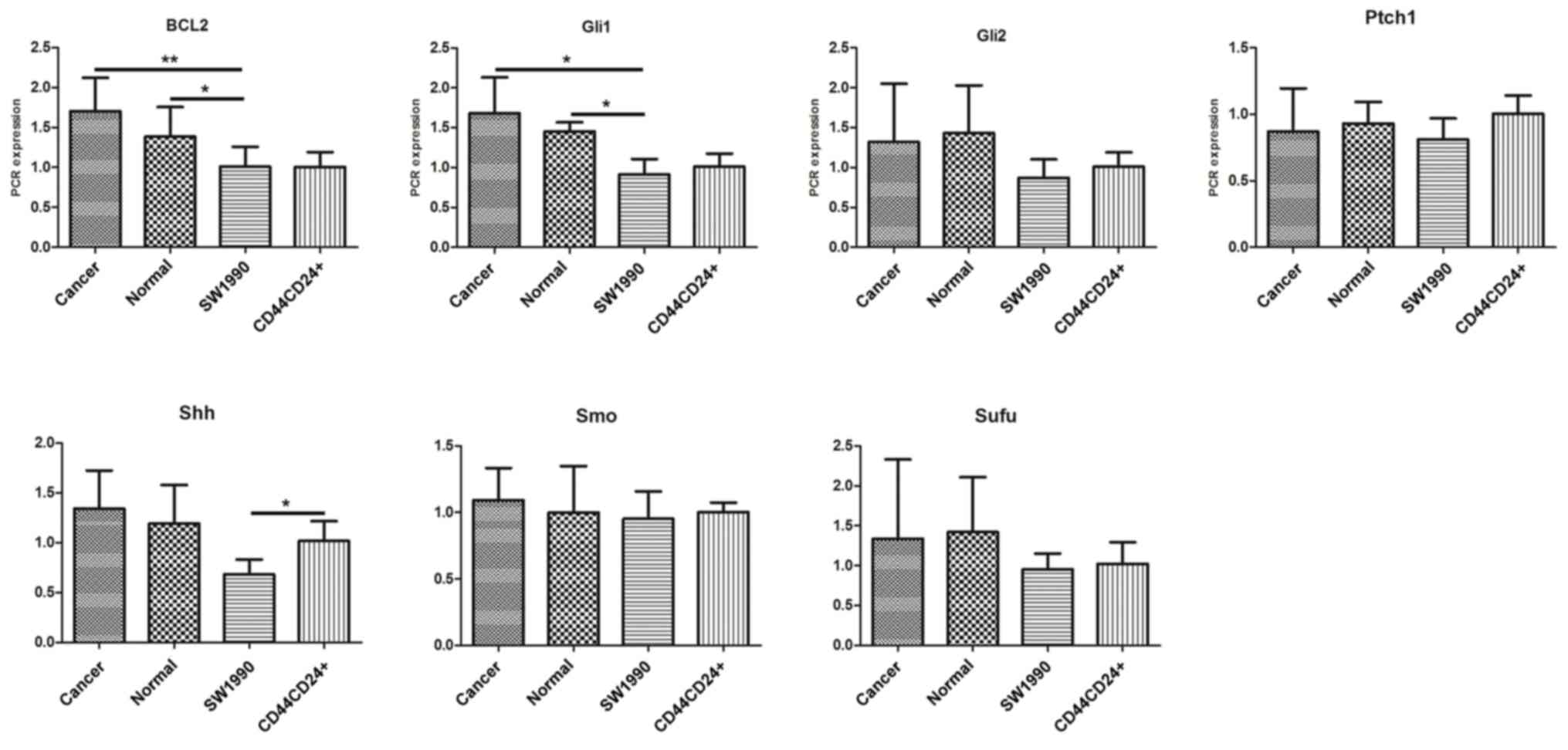
pancreatic duct cells obtained from patients using RT-PCR (Fig. 2) and western blot analysis (Fig. 3). The expression of Gli1 and Bcl2 in
pancreatic ductal cells and cancer cells was increased compared
with that in SW1990 pancreatic cancer cells (Fig. 4); however, no significant differences
were observed between the expression of the other proteins in
pancreatic cancer cells compared with that in the pancreatic duct
cells. The expression of Shh and Smo in
CD44+CD24+ cells was markedly increased
compared with that in pancreatic cancer cells; however, no
significant differences were observed between the expression of the
other proteins in the two groups. In addition, no significant
differences in expression were observed between the
CD44−CD24− cells and pancreatic cancer
cells.
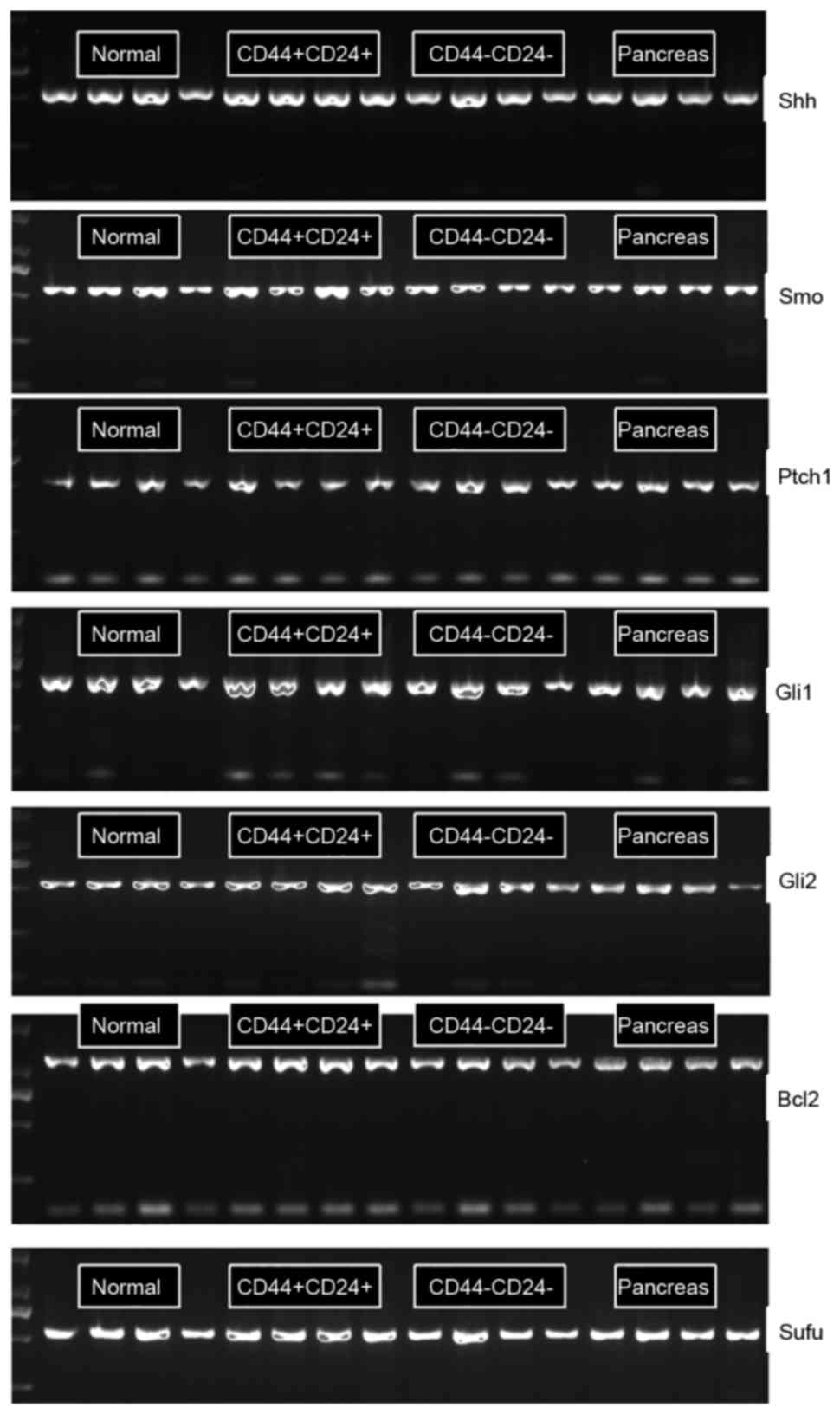 | Figure 2.The electrophoresis maps of reverse
transcription-polymerase chain reaction products of members in the
Hedgehog signaling pathway in pancreatic cancer tissue, CD44+CD24+
SW1990 cells, CD44-CD24- SW1990 cells, normal pancreatic tissue
adjacent to carcinoma cell search group. Shh, sonic hedgehog; Smo,
smoothened; Ptch1, protein patched homolog 1; Gli1, zinc finger
protein GLI1; Gli2, zinc finger protein GLI2; Bcl-2, B-cell
lymphoma 2; Sufu, suppressor of fused homolog; CD, cluster of
differentiation. |
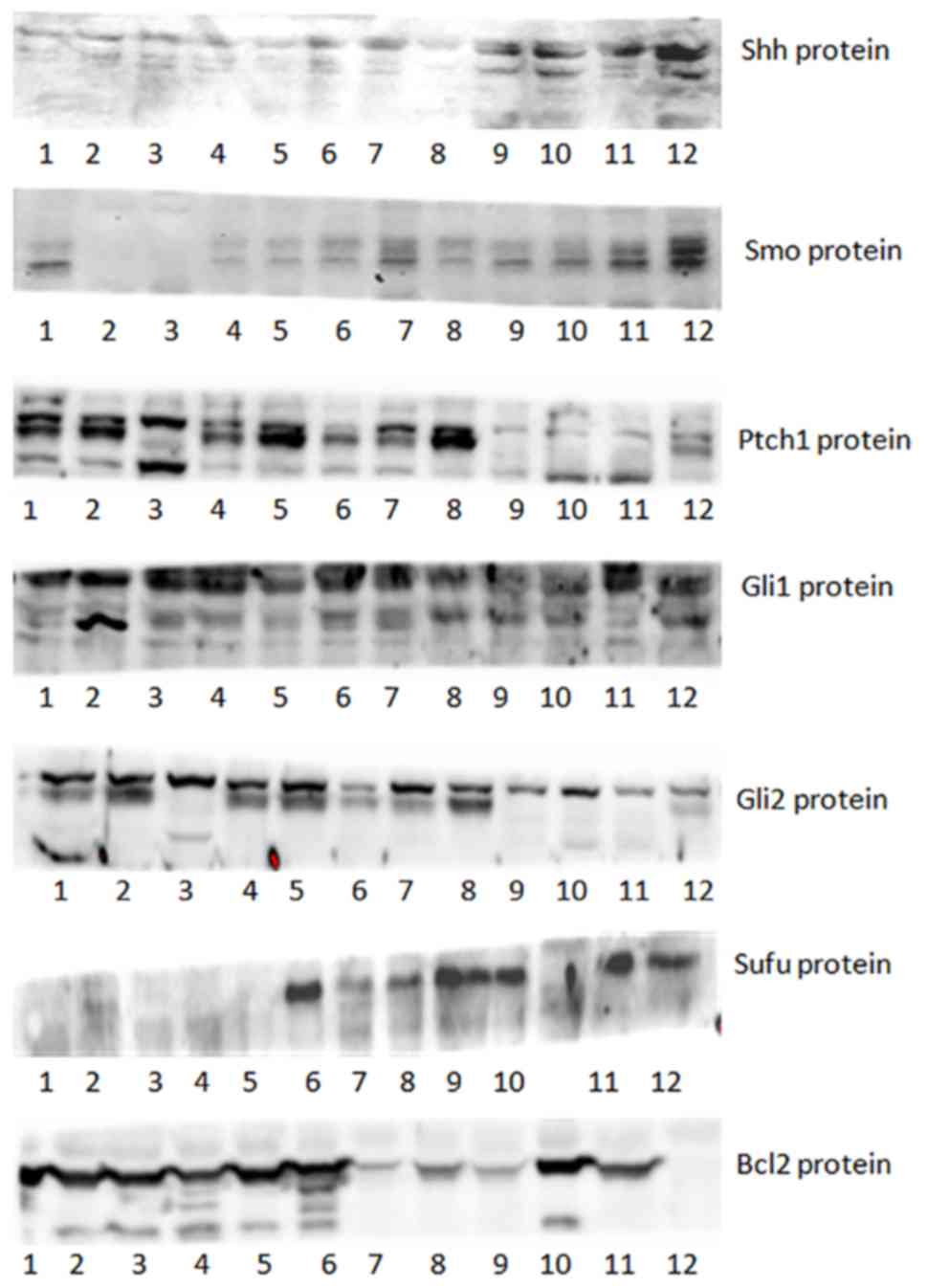 | Figure 3.Western blot analysis of members of
the Hedgehog signaling pathway in pancreatic cancer tissue cells,
CD44+CD24+ SW1990 cells, CD44-CD24- SW1990 cells, normal pancreatic
tissue adjacent to carcinoma cells. Shh, sonic hedgehog; Smo,
smoothened; Ptch1, protein patched homolog 1; Gli1, zinc finger
protein GLI1; Gli2, zinc finger protein GLI2; Bcl-2, B-cell
lymphoma 2; Sufu, suppressor of fused homolog. |
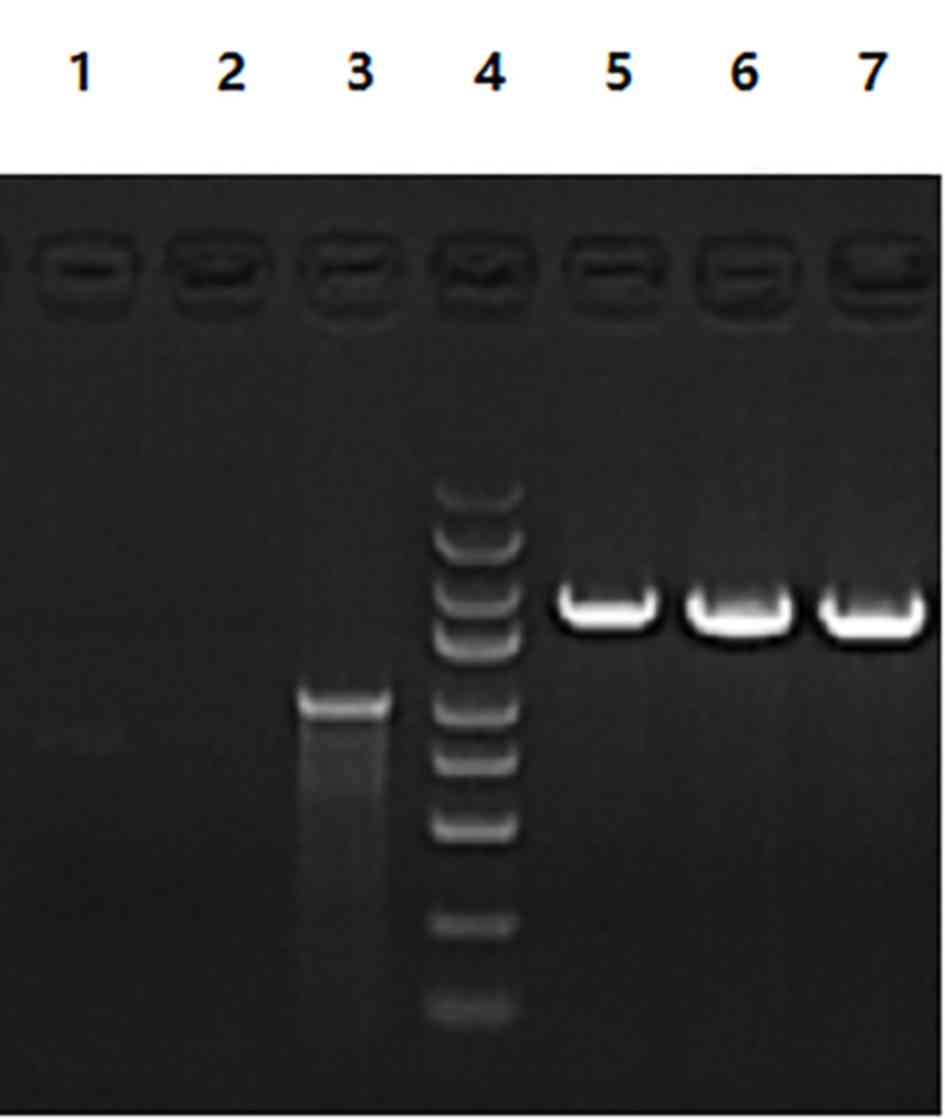
Construction of the siRNA expression
vector carrying Smo
Sequencing of the KL15570-Smo siRNA sequencing
demonstrated that the sequence was consistent with the designed
fragment (data not shown). The electrophoresis map of the PCR
products of the KL15570-Smo siRNA vector digested by SacII B
restriction endonucleases confirmed that the fragment had been
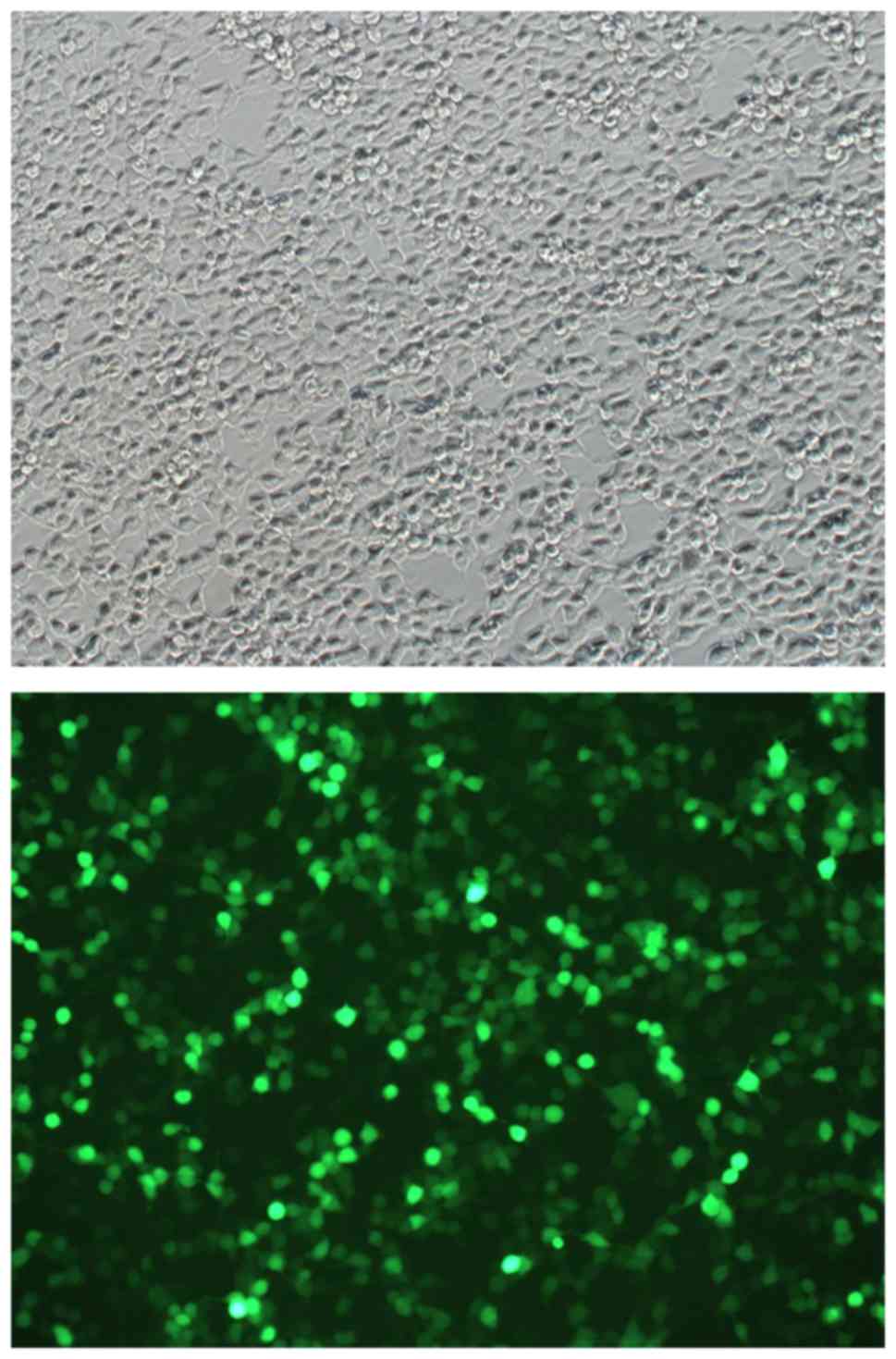
inserted (Fig. 5). Fluorescence
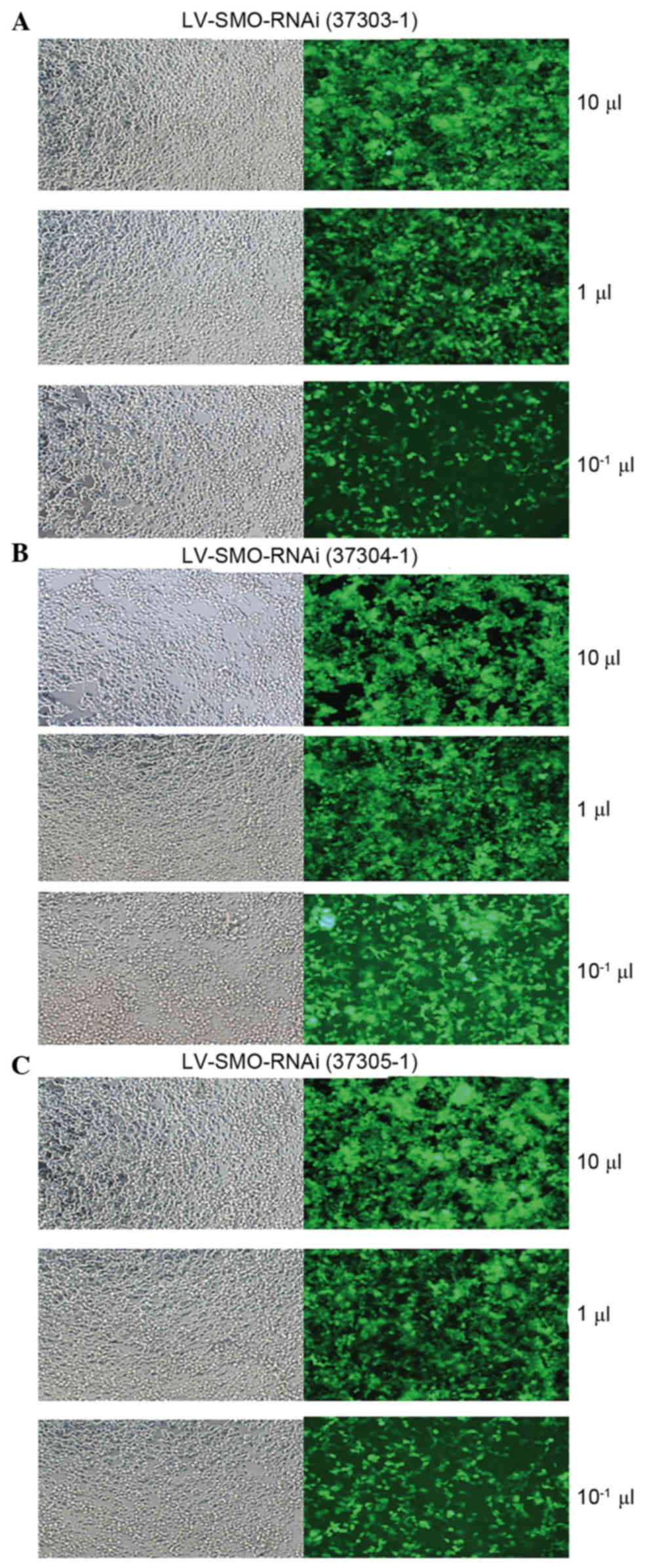
microscopy confirmed that Three groups of interfering lentivirus
cells had been successfully transfected with the plasmid (Fig. 6). The viral titers for the siRNA1,
siRNA2 and siRNA3 plasmids were 5×108, 6×108
and 5×108 viral TU/ml, respectively.
Detection of Smo gene expression in
CD44+CD24+ cells infected with three Smo-
siRNA lentiviral expression vectors
RT-PCR analysis demonstrated that Smo expression was
markedly decreased in CD44+CD24+ cells
following infection with the constructed Smo siRNA lentiviral
expression vectors (Fig. 7). The
blocking efficiency of the three siRNAs were 54.293, 32.188 and
62.531%, respectively (data not shown). The efficiency of siRNA3
was the highest; therefore the lentiviral expression vector
carrying Smo-siRNA3 was used in the follow-up study to block the Hh
signaling pathway.
Discussion
The Hh signaling pathway serves key roles in the
proliferation of epidermal stem cells, the development of embryonic
cells with stem cell characteristics, and the development of mouse
cerebral cortex cells following birth (25). Its abnormal activation is associated
with the occurrence and development of a number of types of
malignant tumor, multiple drug resistance and other
characteristics. The presence of CD44+ CD24−
cells was detected by Tao et al (8) using the RT-PCR method; the results
demonstrated that the expression of Smo and GLI-1 genes was
significantly increased between CD44+ CD24−
cells and non CD44+ CD24− cells, and
indicated that associated factors of the Hh signal pathway were
overexpressed in breast cancer stem cells (CSCs). The Hh signal
pathway serves important roles in the proliferation,
differentiation and maintenance of malignant biological
characteristics of CSCs (26).
Therefore, multiple studies have focused on the association between
the Hh signaling pathway and the development of pancreatic cancer.
Onishi et al (27) confirmed
in vivo and in vitro that cyclopamine, a specific
blocking agent for the Hh signaling pathway, can significantly
improve the sensitivity of pancreatic cancer cells to
chemotherapeutic medicines, including fluorouracil and gemcitabine.
Hao et al (28) demonstrated
that the invasion and metastasis of pancreatic cancer cells could
be significantly inhibited by blocking the Hh signaling pathway. In
addition, gene expression studies have demonstrated that expression
of Hh signaling pathway-associated molecules are significantly
higher in pancreatic cancer tissues compared with normal tissues
(29).
Gene therapy has been developed using DNA
recombination technology to correct mutated or defective genes by
introducing normal genes or therapeutic genes into cells, including
tumor and immune cells, in order to modify the biological behavior
of tumor cells (30).
MicroRNAs are endogenous non-coding small RNA
molecules, which serve important roles in tissue inflammation, cell
proliferation and apoptosis, tissue differentiation, and malignant
tumor occurrence and development (31). siRNA viral vector transfection is a
primary method used for studying signal transduction pathways at
present. In addition, it is possible to use these molecules in
clinical applications for tumor-targeted therapy due to low cost
and high efficiency (32).
The tumorous characteristics, including cell
morphology and growth characteristics, of three pancreatic cancer
cell lines (BXPC-3, PANC-1 and SW1990) and the percentage of
CD44+CD24+/CD44−CD24−
cell subsets of pancreatic cancer cells isolated by flow
cytometry.
In the present study, three siRNA fragments were
designed against the Smo gene in the signal pathway, and the
lentiviral expression vectors carrying the three Smo fragments were
successfully constructed. Through the transfection of
CD44+CD24+ cells, the Smo siRNA lentiviral
expression vector that was the most suitable for the follow-up
study was selected according to the inhibition rate of Smo
expression. This not only laid a good experimental basis for the
smooth development of the follow-up study of this research, but
also provided reliable experimental evidence for the gene therapy
of targeting interference of the Hh signaling pathway. The Hh
signaling pathway serves an important role in the maintenance of
pancreatic CSC malignant biological characteristics, but further
studies are required to elucidate the underlying molecular
mechanisms for this association.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81360328).
Availability of data and materials
The datasets used and/or analyzed during the current
study and the and materials are available from the corresponding
author on reasonable request.
Authors contributions
PC was involved in the design of the experiment,
drafted and revised the manuscript. CY was involved in the design
of the experiment, collected and processed the specimens and was
additionally responsible for the collection and analysis of the
data. XYW was involved in the design of the experiment, revised the
important technical and theoretical content and provided final
approval of the version to be published. All the authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
committee of the Affiliated Cancer Hospital to Xinjiang Medical
University and all patients provided written informed consent.
Patient consent for publication
All patients provided written informed consent for
the publication of all the associated data and accompanying images
in the current study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hammerschmidt M, Brook A and McMahon AP:
The world according to hedgehog. Trends Genet. 13:14–21. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee Y, Miller HL, Russell HR, Boyd K,
Curran T and McKinnon PJ: Patched2 modulates tumori-geneis in
patched1 heterozygous mice. Cancer Res. 66:6964–6971. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McMahon AP: More surprises in the Hedgehog
signaling pathway. Cell. 100:185–188. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li GL, Chen WQ, Wang QS, Ma AW and An R:
Analysis of the association between pancreas cancer and diabetes. J
Cancer Control Treat. 23:132–136. 2010.(In Chinese).
|
|
7
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao YJ, Mao J, Zhang QQ and Li LH:
Hedgehog signaling pathway is activated in breast cancer
CD44+ CD24 cells. Shandong Med. 51:33–35. 2011.(In
Chinese).
|
|
9
|
Lee CJ, Dosch J and Simeone DM: Pancreatic
cancer stem cells. J Clin Oncol. 26:2806–2812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mimeault M and Batra SK: Frequent
deregulations in the hedgehog signaling network and cross-talks
with the epidermal growth factor receptor pathway involved in
cancer progression andt argeted therapies. Pharmacol Rev.
62:497–524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berman DM, Karhadkar SS, Maitra A, Montes
De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman
JR, Watkins DN and Beachy PA: Widespread requirement for Hedgehog
ligand stimulation in growth of digestive tract tumours. Nature.
425:846–851. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thayer SP, di Magliano MP, Heiser PW,
Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernández-del
Castillo C, Yajnik V, et al: Hedgehog is an early and late mediator
of pancreatic cancer tumorigenesis. Nature. 425:851–856. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang K and Jia J: Smoothened regulation
in response to Hedgehog stimulation. Front Biol (Beijing).
10:475–486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Xie G, Fan Q and Xie J: Activation
of the hedgehog-signaling pathway in human cancer and the clinical
implications. Oncogene. 29:469–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sekulic A, Migden MR, Oro AE, Dirix L,
Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander
PA, et al: Efficacy and safety of vismodegib in advanced basal-cell
carcinoma. N Engl J Med. 366:2171–2179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Atwood SX, Sarin KY, Whitson RJ, Li JR,
Kim G, Rezaee M, Ally MS, Kim J, Yao C, Chang AL, et al: Smoothened
variants explain the majority of drug resistance in basal cell
carcinoma. Cancer Cell. 27:342–353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Patel R, Ede J, Collins J and David
Willens: Pancreatic cancer presenting as new-onset diabetes. Case
Rep Oncol. 7:171–174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tong GX, Geng QQ, Chai J, Cheng J, Chen
PL, Liang H, Shen XR and Wang DB: Association between pancreatitis
and subsequent risk of pancreatic cancer: A systematic review of
epidemiological studies. Asian Pac J Cancer Prev. 15:5029–5034.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farhana L, Dawson MI, Das JK, Murshed F,
Xia Z, Hadden TJ, Hatfield J and Fontana JA: Adamantyl
retinoid-related molecules induce apoptosis in pancreatic cancer
cells by inhibiting IGF-1R and Wnt/β-catenin pathways. J Oncol.
2012:7967292012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao J, An Y, Qian JJ, Bai DS, Xu ZK and
Miao Y: Hedgehog signaling pathway inhibitor reverts acquired
chemoresistance in human pancreatic cancer SWl990 cell line. Chin J
Pancreatol. 184–188. 2012.
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lizée G, Aerts JL, Gonzales MI, Chinnasamy
N, Morgan RA and Topalian SL: Real-time quantitative reverse
transcriptase-polymerase chain reaction as a method for determining
lentiviral vector titers and measuring transgene expression. Hum
Gene Ther. 14:497–507. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu J, Rodova M, Roy SK, Sharma J, Singh
KP, Srivastava RK and Shankar S: GANT-61 inhibits pancreatic cancer
stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice
xenograft. Cancer Lett. 330:22–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Clement V, Sanchez P, de Tribolet N,
Radovanovic I and Ruiz i Altaba A: HEDEGHOG-GLI1 signaling
regulates human glioma growth, cancer stem cell self-renewal, and
tumorigenicity. Curr Biol. 17:165–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Kong D, Ahmad A, Bao B and Sarkar
FH: Pancreatic cancer stem cells: Emerging target for designing
novel therapy. Cancer Lett. 338:94–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Onishi H, Morifuji Y, Kai M, Suyama K,
Iwasaki H and Katano M: Hedgehog inhibitor decreases
chemosensitivity to 5-fluorouracil and gemcitabine under hypoxic
conditions in pancreatic cancer. Cancer Sci. 103:1272–1279. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao K, Tian XD, Qin CF, Xie XH and Yang
YM: Hedgehog signaling pathway regulates human pancreatic cancer
cell proliferation and metastasis. Oncol Rep. 29:1124–1132. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang H, Li Q, He C, Li F, Sheng H, Shen
X, Zhang X, Zhu S, Chen H, Chen X, et al: Activation of the Wnt
pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J
Cancer Res. 4:537–544. 2014.PubMed/NCBI
|
|
30
|
Fujihira A, Suzuki T, Chang MO, Moriyama
T, Kitajima M and Takaku H: Antitumor effects of
baculovirus-infected dendritic cells against human pancreatic
carcinoma. Gene Ther. 21:849–854. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X,
Li R, Wang W, Hua D and Chen W: A functional variant at miR-132-3p,
miR-212-3p, and miR-361-5p binding site in CD80 gene
alterssusceptibility to gastric cancer in a Chinese Han population.
Med Oncol. 31:602014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tuschl T: Expanding small RNA
interference. Nat Biotechnol. 20:446–448. 2002. View Article : Google Scholar : PubMed/NCBI
|