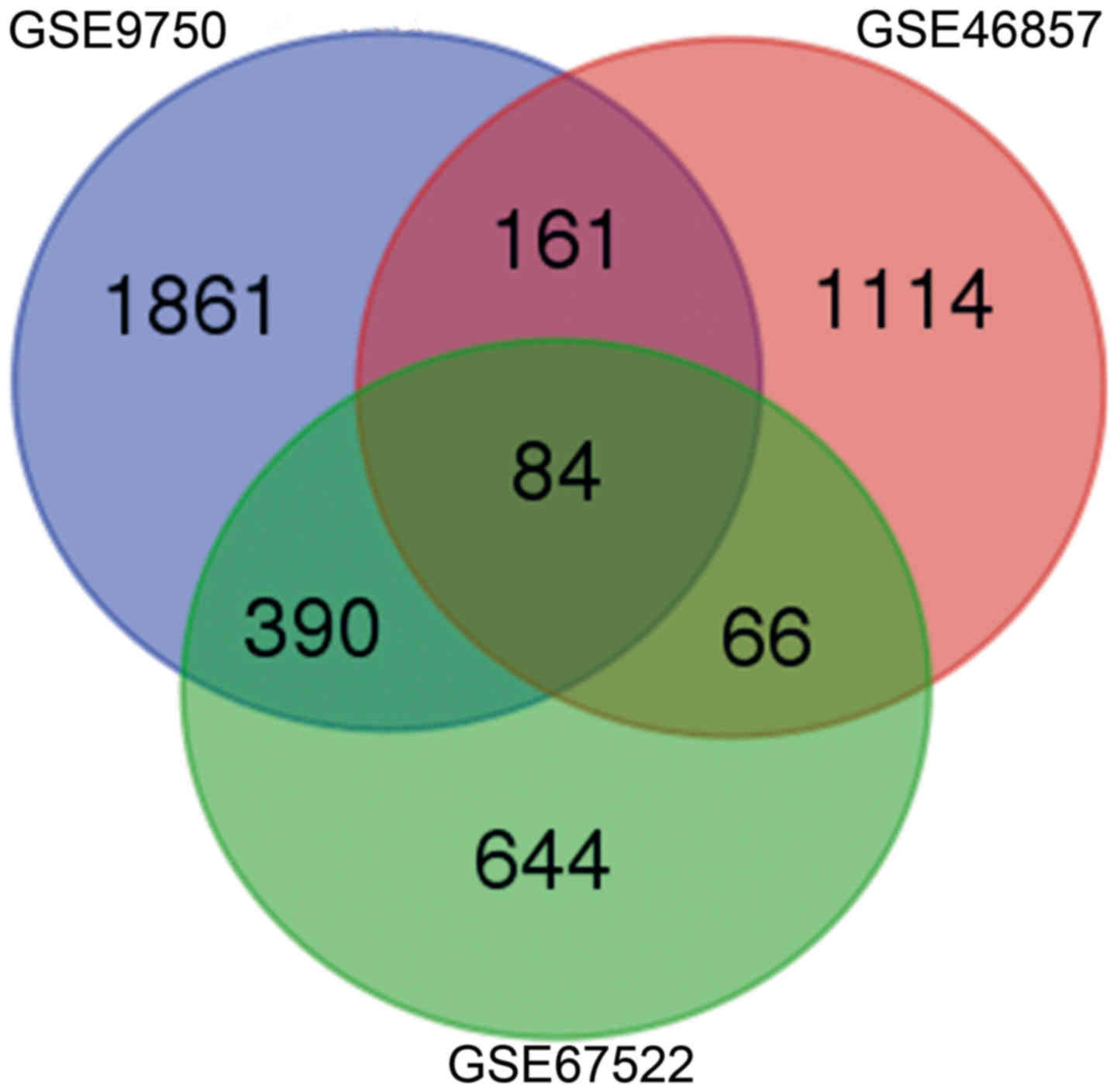
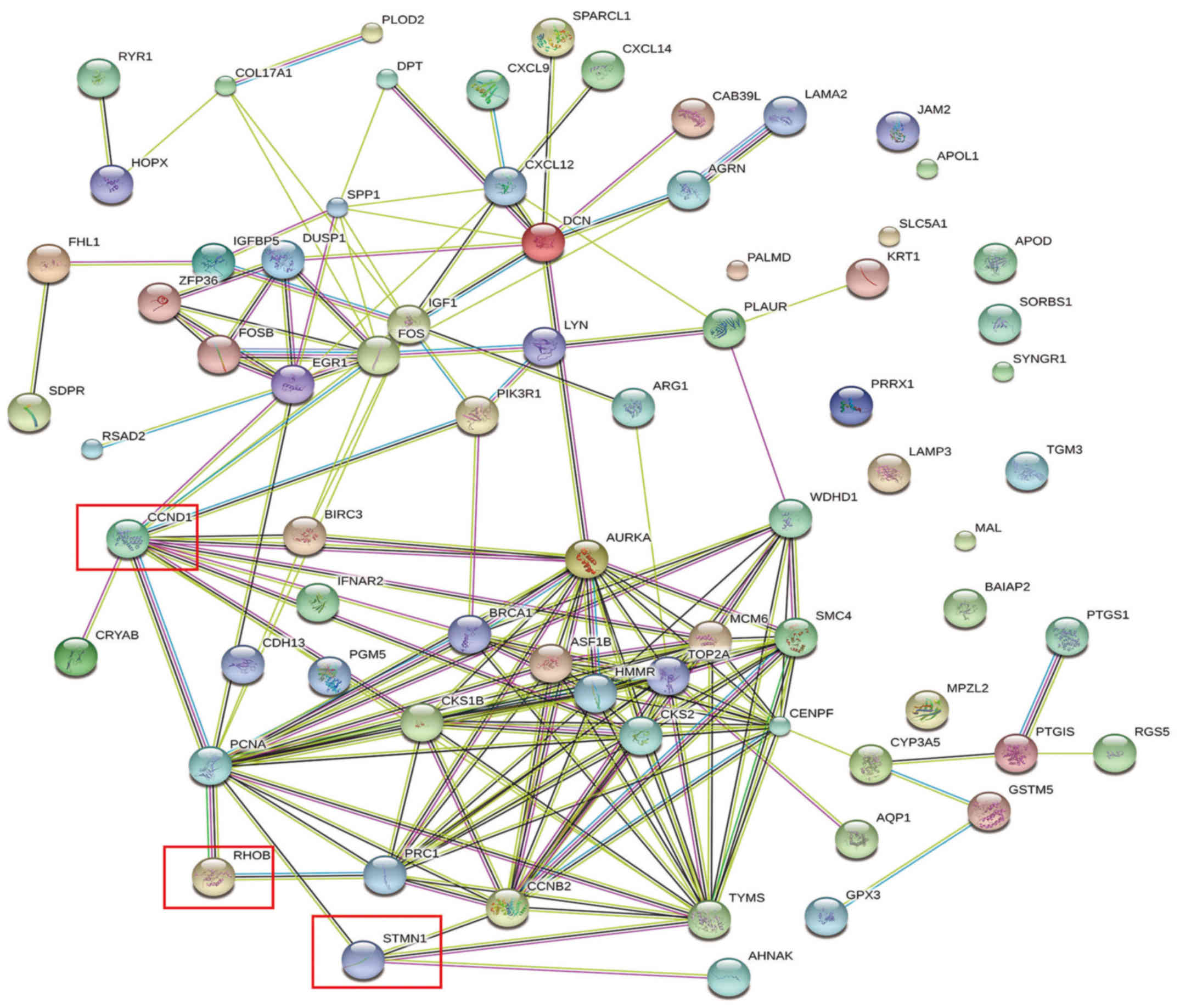
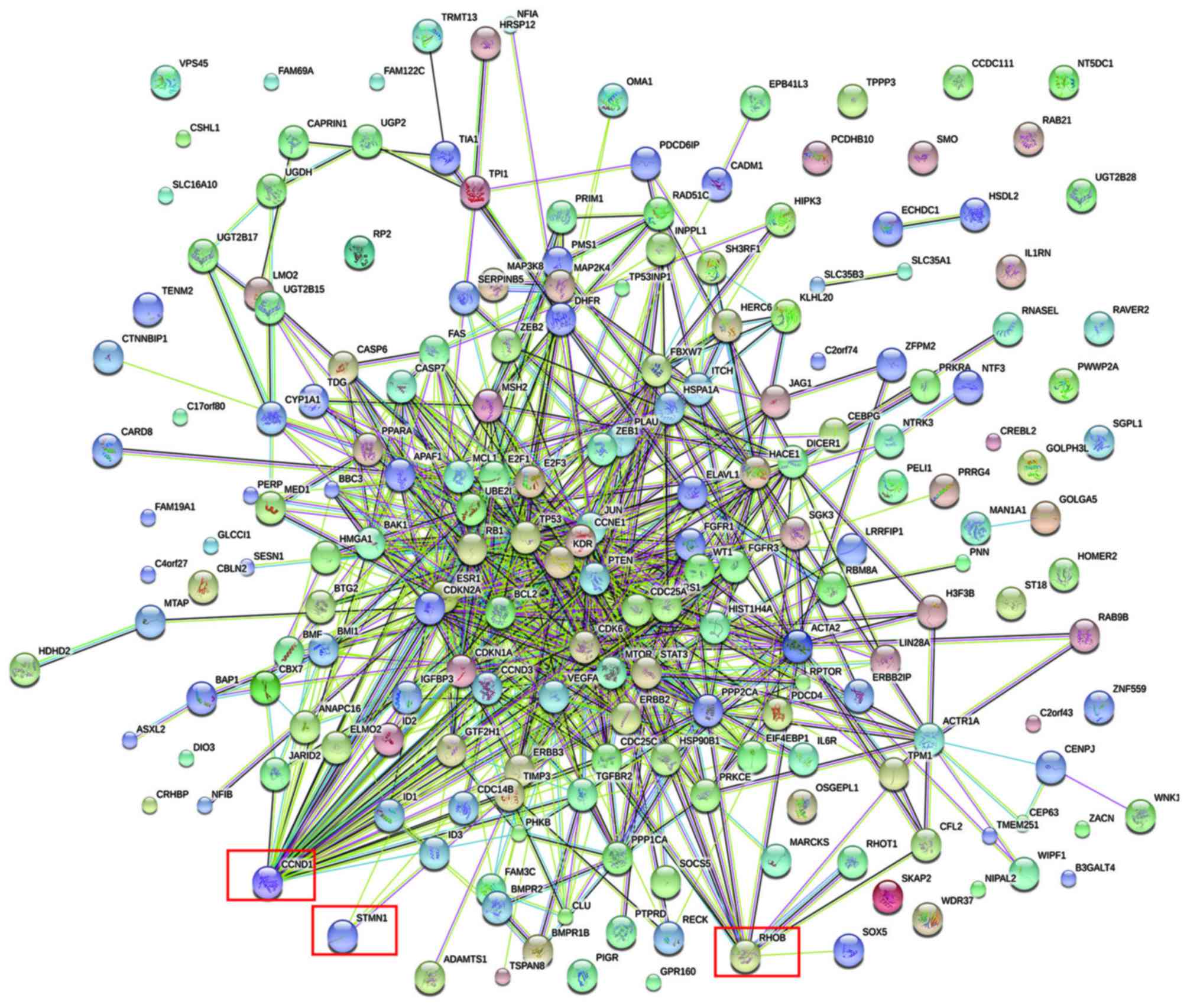
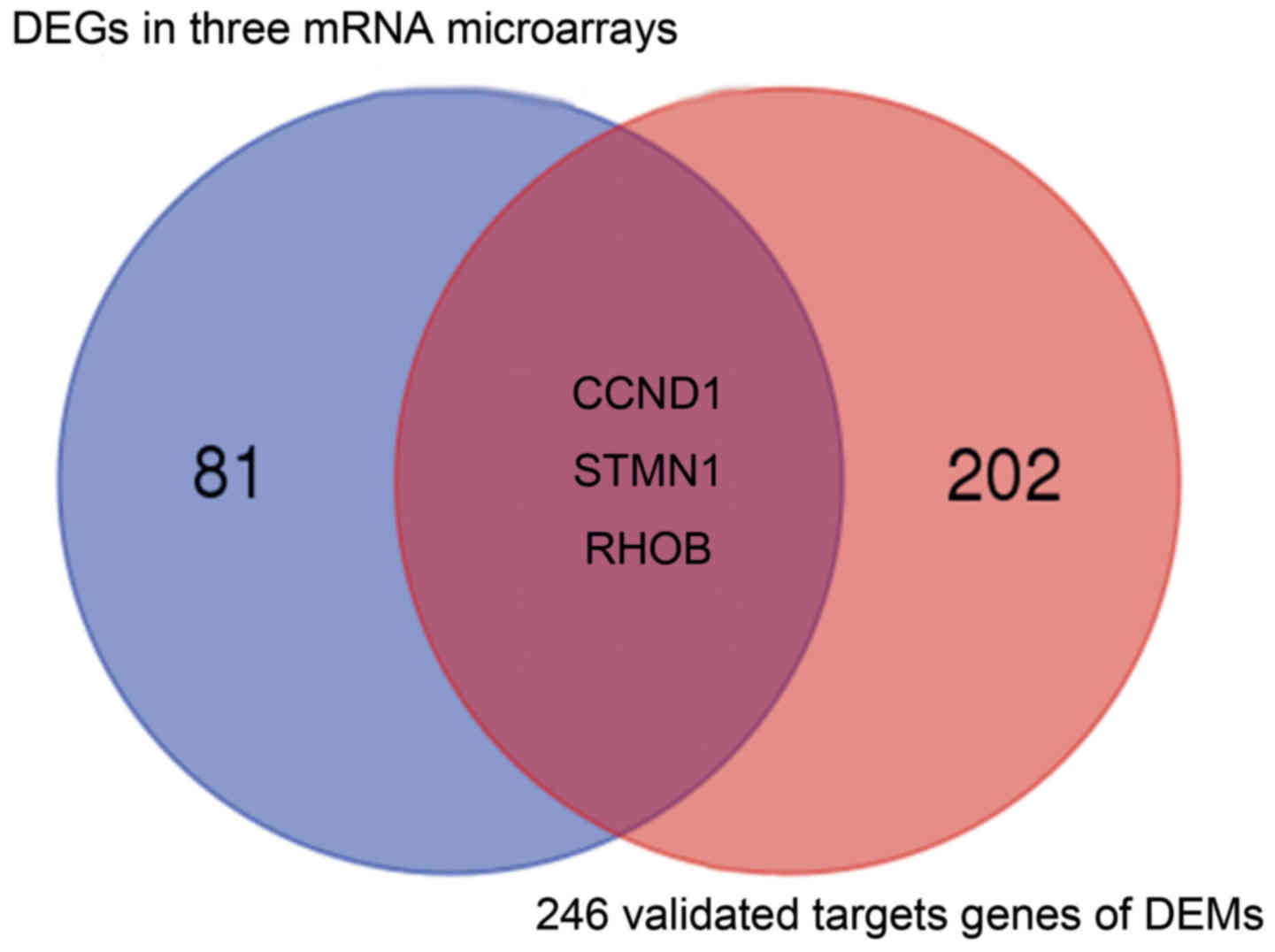
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Angelis R, Sant M, Coleman MP,
Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H,
Ardanaz E, et al: Cancer survival in Europe 1999–2007 by country
and age: Results of EUROCARE-5-a population-based study. Lancet
Oncol. 15:23–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vaccarella S, Laversanne M, Ferlay J and
Bray F: Cervical cancer in Africa, Latin America and the Caribbean,
and Asia: Regional inequalities and changing trends. Int J Cancer.
141:1997–2001. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Williams SP and McDermott U: The pursuit
of therapeutic biomarkers with high-throughput cancer cell drug
screens. Cell Chem Biol. 24:1066–1074. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun HH, Vaynblat A and Pass HI: Diagnosis
and prognosis-review of biomarkers for mesothelioma. Ann Transl
Med. 5:2442017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abdollah F, Dalela D, Haffner MC, Culig Z
and Schalken J: The role of biomarkers and genetics in the
diagnosis of prostate cancer. Eur Urol Focus. 1:99–108. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel SA and DeMichele A: Adding adjuvant
systemic treatment after neoadjuvant therapy in breast cancer:
Review of the data. Curr Oncol Rep. 19:562017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Verdaguer H, Saurí T and Macarulla T:
Predictive and prognostic biomarkers in personalized
gastrointestinal cancer treatment. J Gastrointest Oncol. 8:405–417.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Herrera-Marcos LV, Lou-Bonafonte JM, Arnal
C, Navarro MA and Osada J: Transcriptomics and the mediterranean
diet: A systematic review. Nutrients. 9:E4722017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong HJ, Koom WS and Koh WG: Cell
microarray technologies for high-throughput cell-based biosensors.
Sensors (Basel). 17:E12932017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimizu D, Kanda M and Kodera Y: Review of
recent molecular landscape knowledge of gastric cancer. Histol
Histopathol. 33:11–26. 2018.PubMed/NCBI
|
|
13
|
Syed P, Gidwani K, Kekki H, Leivo J,
Pettersson K and Lamminmäki U: Role of lectin microarrays in cancer
diagnosis. Proteomics. 16:1257–1265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomas A, Mahantshetty U, Kannan S,
Deodhar K, Shrivastava SK, Kumar-Sinha C and Mulherkar R:
Expression profiling of cervical cancers in Indian women at
different stages to identify gene signatures during progression of
the disease. Cancer Med. 2:836–848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sharma S, Mandal P, Sadhukhan T, Roy
Chowdhury R, Ranjan Mondal N, Chakravarty B, Chatterjee T, Roy S
and Sengupta S: Bridging links between long noncoding RNA HOTAIR
and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci Rep.
5:117242015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wilting SM, Snijders PJ, Verlaat W,
Jaspers A, van de Wiel MA, van Wieringen WN, Meijer GA, Kenter GG,
Yi Y, le Sage C, et al: Altered microRNA expression associated with
chromosomal changes contributes to cervical carcinogenesis.
Oncogene. 32:106–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database Issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37(Database Issue): D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aranda S, Berkley S, Cowal S, Dybul M,
Evans T, Iversen K, Moeti M, Osotimehin B, Peterson S, Piot P, et
al: Ending cervical cancer: A call to action. Int J Gynaecol
Obstet. 138 Suppl 1:S4–S6. 2017. View Article : Google Scholar
|
|
23
|
Huchko MJ, Maloba M, Nakalembe M and Cohen
CR: The time has come to make cervical cancer prevention an
essential part of comprehensive sexual and reproductive health
services for HIV-positive women in low-income countries. J Int AIDS
Soc. 18 Suppl 5:202822015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thumkeo D, Watanabe S and Narumiya S:
Physiological roles of Rho and Rho effectors in mammals. Eur J Cell
Biol. 92:303–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schaefer A, Reinhard NR and Hordijk PL:
Toward understanding RhoGTPase specificity: Structure, function and
local activation. Small GTPases. 5:62014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calvayrac O, Mazières J, Figarol S,
Marty-Detraves C, Raymond-Letron I, Bousquet E, Farella M,
Clermont-Taranchon E, Milia J, Rouquette I, et al: The RAS-related
GTPase RHOB confers resistance to EGFR-tyrosine kinase inhibitors
in non-small-cell lung cancer via an AKT-dependent mechanism. EMBO
Mol Med. 9:238–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Merkus D and Schermuly RT: Farnesylation
of RhoB: The cancer hypothesis of pulmonary hypertension revisited.
Cardiovasc Res. 113:249–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vega FM and Ridley AJ: The RhoB small
GTPase in physiology and disease. Small GTPases. 1–10. 2016.
|
|
29
|
Chen W, Niu S, Ma X, Zhang P, Gao Y, Fan
Y, Pang H, Gong H, Shen D, Gu L, et al: RhoB acts as a tumor
suppressor that inhibits malignancy of clear cell renal cell
carcinoma. PloS One. 11:e01575992016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Delmas A, Cherier J, Pohorecka M,
Medale-Giamarchi C, Meyer N, Casanova A, Sordet O, Lamant L, Savina
A, Pradines A and Favre G: The c-Jun/RHOB/AKT pathway confers
resistance of BRAF-mutant melanoma cells to MAPK inhibitors.
Oncotarget. 6:15250–15264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marlow LA, Bok I, Smallridge RC and
Copland JA: RhoB upregulation leads to either apoptosis or
cytostasis through differential target selection. Endocr Relat
Cancer. 22:777–792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai T, Yokobori T, Altan B, Ide M, Mochiki
E, Yanai M, Kimura A, Kogure N, Yanoma T, Suzuki M, et al: High
STMN1 level is associated with chemo-resistance and poor prognosis
in gastric cancer patients. Br J Cancer. 116:1177–1185. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dou YD, Zhao H, Huang T, Zhao SG, Liu XM,
Yu XC, Ma ZX, Zhang YC, Liu T, Gao X, et al: STMN1 promotes
progesterone production via StAR up-regulation in mouse granulosa
cells. Sci Rep. 6:266912016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang HQ, Guo X, Guo SQ, Wang Q, Chen XQ,
Li XN and Guo LS: STMN1 in colon cancer: Expression and prognosis
in Chinese patients. Eur Rev Med Pharmacol Sci. 20:2038–2044.
2016.PubMed/NCBI
|
|
35
|
Chen Y, Zhang Q, Ding C, Zhang X, Qiu X
and Zhang Z: Stathmin1 overexpression in hypopharyngeal squamous
cell carcinoma: A new promoter in FaDu cell proliferation and
migration. Int J Oncol. 50:31–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Hu GH, Kong FJ, Wu KM, He B, Song K
and Sun WJ: Reduced STMN1 expression induced by RNA interference
inhibits the bioactivity of pancreatic cancer cell line Panc-1.
Neoplasma. 61:144–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li X, Wang L, Li T, You B, Shan Y, Shi S,
Qian L and Cao X: STMN1 overexpression correlates with biological
behavior in human cutaneous squamous cell carcinoma. Pathol Res
Pract. 211:816–823. 2015. View Article : Google Scholar : PubMed/NCBI
|