Introduction
Although it has a low morbidity rate, pancreatic
cancer (PC) is one of the most fatal malignant tumor types, with
the highest mortality rate worldwide (1). Nearly all patients with PC succumb to
the disease within 1–2 years (2).
Following pancreaticoduodenectomy (Whipple procedure), the 5-year
survival rate is 25–30% for node-negative (3) and 10% for node-positive (4) diseases. The majority of patients are
diagnosed at an advanced stage, owing to a lack of effective
diagnostic techniques for PC diagnosis at the early stages of
disease (5). The traditional tumor
serum markers, carcinoembryonic antigen and carbohydrate antigen
19-9, are neither sensitive nor specific for screening patients
with PC (6,7). A number of environmental factors,
including alcohol consumption, smoking history, body mass index,
diabetes history and family history of PC have been demonstrated to
be high-risk factors for PC (8,9). However,
certain individuals exposed to these risk factors do not develop
PC, which suggests that genetic factors may also influence cancer
progression.
The xeroderma pigmentosum group C (XPC) gene is
located at chromosome 3p25 (10); it
contains 16 exons and 15 introns, and encodes a protein of 940
amino acids (11). The encoded
protein is an indispensable component in the early stages of global
genome nucleotide excision repair (NER), particularly in the damage
recognition and initiation of NER (12); it is involved in initiating protein
complex formation and repair of these complexes (13,14). There
are >687 single nucleotide polymorphisms (SNPs) in the XPC gene,
with >100 SNPs in the coding regions (http://www.ncbi.nlm.nih.gov/projects/SNP). However, to
date, only a small number of correlation analysis studies have
focused on XPC polymorphisms and PC risk. There are three
polymorphisms most frequently detected in the XPC gene: poly AT
insertion/deletion on intron 9 (PAT), A to C substitution in exon
15 (Lys939Gln, rs2228001) and C to T substitution in exon 9
(Ala499Val, rs2228000). The PAT polymorphism has been demonstrated
to confer an increased risk of PC (15,16);
however, an association with PC risk has not been observed for
rs2228001 and rs2228000 (17,18).
The use of tag-SNPs markedly improves the
effectiveness of candidate gene and disease correlation analyses
(19). Therefore, in the present
study, to understand the association between XPC polymorphisms and
PC susceptibility, XPC gene tag-SNPs and functional SNPs were
investigated in patients with pathologically proven PC.
Materials and methods
Study subjects
A total of 205 patients with PC, with an age range
between 24 and 87 years (mean age, 63.69±11.40 years), who were
treated at The Affiliated Cancer Hospital and The First Affiliated
Hospital of Xinjiang Medical University (Urumqi, Xinjiang, China)
between December 2007 and December 2015 were enrolled in the
present study. All patients had pathologically proven PC. Among
these patients, 131 cases underwent pancreaticoduodenectomy, 19
cases underwent iodine-125 seed implantation and palliative surgery
(biopsy obtained during surgery), 51 cases underwent distal
(combined with the spleen) pancreatectomy and 4 cases underwent
fine-needle aspiration biopsy under computed tomography scan
guidance.
In addition, a total of 230 non-cancer subjects,
with an age range between 26 and 88 years (mean age, 63.69±11.86
years), who were admitted to The First Affiliated Hospital of
Xinjiang Medical University during the same period, were recruited
as a control group. These subjects had no previous history of
pancreatic disease and had not been diagnosed with any malignant
cancer.
All subjects recruited to this study signed informed
consent forms and the study protocol was approved by the Ethical
Committee of The First Affiliated Hospital of Xinjiang Medical
University.
‘Drinking’ was defined as consuming alcohol more
than once a week, continuously over a 6-month period in a lifetime.
‘Smoking’ was defined as accumulative smoking of >100 cigarettes
in a lifetime. The accumulative smoking amount (packets/year)
indicated the smoking status according to the following formula:
Accumulative smoking amount (packets/year)=mean number of
cigarettes per day/20 times the number of years of smoking. The
median of the accumulative smoking amount was used as the cut-off
point to define mild and heavy smokers (20).
Blood collection and DNA
extraction
Peripheral blood (3 ml) was collected from each
participant, placed in an EDTA tube and stored at −80°C within 30
min. Genomic DNA was extracted from blood samples using a DNA blood
extraction kit (BioTeke, Beijing, China) according to the
manufacturer's protocol.
SNP selection and genotyping
SNPs were selected from the HapMap database
(https://www.genome.gov/10001688/international-hapmap-project/
HapMap Data Rel 24/Phase II, Nov08, on NCBI B36 assembly, dbSNP
b126), which provided the genotype data collected from Han Chinese
individuals living in Beijing. SNPs in the XPC gene were selected
by combined analysis of functional SNPs and tag-SNPs from the dbSNP
(http://www.ncbi.nlm.nih.gov/SNP/) and
HapMap databases. The minor allele frequencies were >5%, and the
linkage disequilibrium (LD) coefficient r2 values were
>0.8. A total of 7 tag-SNPs were located, 2 of which were in the
5′-untranslated region (5′UTR) (rs2607775 and rs3731055), 3 of
which were in introns (intron 5, rs3729587; intron 6, rs3731114;
and intron 12, rs2470353) and another 2 of which were in exons
(exon 9, rs2228000; and exon 15, rs2228001).
The 7 SNPs were detected using a SNaPshot assay
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) with PCR primers designed with Primer 5 software (21) The primers of the SNPs were as follows:
rs2228001 forward, 5′-CTGTAGTGGGGCAGCAGCAACT-3′ and reverse,
5′-AGAGGAGGGGACCAGCTCTCAA-3′; rs2470353 forward,
5′-TGCTGGGCAGGAAGAGGTACAC-3 and reverse,
5′-GACCTGGGCCTGTTTGGCTACT-3′; rs2228000 forward,
5′-CCCACTTTTCCTCCTGCTCACA-3′ and reverse,
5′-AGGACAAAGGCTGGGTCCAAGA-3′; rs3731114 forward,
5′-ACCCGCCTGCCTCTGTCCTA-3′ and reverse, 5′-TGCCAGACTGGTGGGGAGAC-3′;
rs3729587 forward, 5′-GAAACTTGCCATGGCCACAGAG-3′ and reverse,
5′-AAGGGGTCCATGAGGACACACA-3′; rs2607775 forward,
5′-GTTTCCGAGCCATGTTGCTTGT-3′ and reverse,
5′-CTTTCCTGCTTCCCGCAGTTTT-3′; and rs3731055 forward,
5′-TCCGGAGATTGACGTTGCTCTT-3′ and reverse
5′-CTCAGGGCCTACGGCAAAATTC-3′. Results were analyzed using
GeneMapper 4.0 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.). For quality control, genotyping was performed in
a double-blinded manner with 5% randomly duplicated samples. Hence,
reproducibility was 100%.
Bioinformatics analysis
The functions of the XPC SNPs were predicted using
the SNPinfo Web Server (https://snpinfo.niehs.nih.gov/). XPC expression and
survival analysis in PC was evaluated using The Cancer Genome Atlas
data by the online analysis tool UALCAN (http://ualcan.path.uab.edu/analysis.html) (22).
Statistical analysis
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) was used
for statistical analysis. A goodness-of-fit χ2 test was
used to assess the Hardy-Weinberg equilibrium. Allele frequencies
were assessed by χ2 test. Using unconditional logistic
regression with adjustment for age and sex, odds ratios (ORs) and
95% confidence intervals (CIs) were calculated to estimate the
relative risks of PC associated with SNP genotypes. HaploView
version 4.2 (Broad Institute, Cambridge, MA, USA) was used to
generate the LD plot and to assess the association between
haplotypes and PC. Comparisons of all variables between cases and
control subjects were performed using the online tool on the
website http://ualcan.path.uab.edu/.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics
The demographic characteristics of subjects and
related risk factors are presented in Table I. According to χ2 tests, no
significant differences in age, sex, drinking status, body mass
index, diabetes history, smoking history or family history of
cancer were identified between the case and control groups
(P>0.05). Heavy smokers (tobacco ≥25 packets/year) in the case
group accounted for 32.0%, which was a significantly higher value
compared with that in the control group (17.5%) (P=0.019).
 | Table I.General characteristics of the
pancreatic cancer cases (n=205) and controls (n=230). |
Table I.
General characteristics of the
pancreatic cancer cases (n=205) and controls (n=230).
|
Characteristics | Cases, n (%) | Controls, n
(%) | χ2 | P-value |
|---|
| Age, years |
|
| 0.600 | 0.896 |
|
≤49 | 31 (15.1) | 36 (15.7) |
|
|
|
50–59 | 33 (16.1) | 36 (15.7) |
|
|
|
60–69 | 65 (31.7) | 66 (28.6) |
|
|
|
≥70 | 76 (37.1) | 92 (40.0) |
|
|
| Sex |
|
| 0.003 | 0.953 |
|
Male | 126 (61.5) | 142 (61.7) |
|
|
|
Female | 79 (38.5) | 88 (38.3) |
|
|
| Diabetes |
|
| 2.330 | 0.127 |
| No | 135 (65.9) | 167 (72.6) |
|
|
|
Yes | 70 (34.1) | 63 (27.4) |
|
|
| BMI,
kg/m2 |
|
| 6.026 | 0.110 |
|
<18.5 | 84 (41.0) | 69 (30.0) |
|
|
|
18.5–23.9 | 71 (34.6) | 93 (40.4) |
|
|
|
24-27.9 | 33 (16.1) | 48 (20.9) |
|
|
|
≥28 | 17 (8.3) | 20 (8.7) |
|
|
| Smoking |
|
| 1.909 | 0.167 |
|
Non-smoker | 105 (51.2) | 133 (57.8) |
|
|
|
Smoker | 100 (48.8) | 97 (42.2) |
|
|
| Packets/year
smokeda |
|
| 5.520 | 0.019 |
|
<25 | 68 (68.0) | 80 (82.5) |
|
|
|
≥25 | 32 (32.0) | 17 (17.5) |
|
|
| Drinking |
|
| 0.011 | 0.917 |
|
Seldom | 156 (76.1) | 176 (76.5) |
|
|
|
Often | 49 (23.9) | 54 (23.5) |
|
|
| Family history of
cancer |
|
| 2.812 | 0.094 |
| No | 179 (87.3) | 212 (92.2) |
|
|
|
Yes | 26 (12.7) | 18 (7.8) |
|
|
Association analysis of PC
susceptibility
The distribution of allelic gene frequency in the 7
tag-SNP loci conformed to the Hardy-Weinberg equilibrium in the
case and control groups (P>0.05; Table II). The C allelic gene frequency of
rs2470353 in patients with pathologically proven PC was
significantly increased compared with that in the control group
(P=0.003). Compared with the GG gene type, PC risk was
significantly increased in subjects with the variant allele C (GC
and GC+CC; P=0.012 and P=0.006, respectively). The G allelic gene
frequency of rs2607775 was significantly increased in patients with
PC compared with that in the control group (P=0.003). Compared with
the CC gene type, PC risk was significantly increased in subjects
with the variant allele G (CG and CG+GG; P=0.013 and P=0.005;
Table III). The distribution of
gene type and allelic gene frequency in the other 5 tag-SNP loci
were not significantly different between the case and control
groups (P>0.05; Tables II and
III).
 | Table II.Characteristics of the 7 tag-SNPs in
the XPC gene. |
Table II.
Characteristics of the 7 tag-SNPs in
the XPC gene.
|
|
|
|
| MAF | HWE P-value |
|
|---|
|
|
|
|
|
|
|
|
|---|
| SNP | Chromosome
position | Location | Alleles | Case | Control | Case | Control | P-value |
|---|
| rs2228001 | 14187449 | Extron 15 | A/C | 0.351 | 0.365 | 0.600 | 0.633 | 0.667 |
| rs2470353 | 14190268 | Intron 12 | G/C | 0.124 | 0.065 | 0.595 | 0.981 | 0.003 |
| rs2228000 | 14199887 | Extron 9 | C/T | 0.305 | 0.300 | 0.728 | 0.300 | 0.876 |
| rs3731114 | 14206622 | Intron 6 | C/G | 0.205 | 0.217 | 0.146 | 0.409 | 0.652 |
| rs3729587 | 14208625 | Intron 5 | G/C | 0.334 | 0.302 | 0.780 | 0.754 | 0.312 |
| rs2607775 | 14220095 | 5′UTR | C/G | 0.163 | 0.096 | 0.197 | 0.149 | 0.003 |
| rs3731055 | 14220439 | 5′UTR | G/A | 0.232 | 0.241 | 0.697 | 0.348 | 0.740 |
 | Table III.Association between polymorphisms of
XPC genes and pancreatic cancer. |
Table III.
Association between polymorphisms of
XPC genes and pancreatic cancer.
|
|
|
|
| χ2
test | Logistic
regression |
|---|
|
|
|
|
|
|
|
|---|
| SNP | Genotype | Case, n | Control, n | OR (95% CI) | P-value | OR (95%
CI)a |
P-valuea |
|---|
| rs2228001 | A/A | 88 | 91 | 1.000 |
| 1.000 |
|
|
| A/C | 90 | 110 | 0.846
(0.565–1.268) | 0.418 | 0.824
(0.547–1.239) | 0.352 |
|
| C/C | 27 | 29 | 0.963
(0.528–1.755) | 0.901 | 1.009
(0.548–1.857) | 0.977 |
|
| A/C+C/C | 117 | 139 | 0.870
(0.594–1.276) | 0.477 | 0.860
(0.585–1.265) | 0.444 |
| rs2470353 | G/G | 158 | 201 | 1.000 |
| 1.000 |
|
|
| G/C | 43 | 28 | 1.954
(1.162–3.285) | 0.011 | 1.942
(1.154–3.267) | 0.012b |
|
| C/C | 4 | 1 | 5.089
(0.563–45.980) | 0.108 | 5.253
(0.577–47.822) | 0.141 |
|
| G/C+C/C | 47 | 29 | 2.062
(1.241–3.425) | 0.005 | 2.053
(1.235–3.412) | 0.006b |
| rs2228000 | C/C | 98 | 116 | 1.000 |
| 1.000 |
|
|
| C/T | 89 | 90 | 1.171
(0.786–1.742) | 0.438 | 1.164
(0.780–1.737) | 0.457 |
|
| T/T | 18 | 24 | 0.888
(0.455–1.731) | 0.727 | 0.914
(0.463–1.803) | 0.795 |
|
| C/T+T/T | 107 | 114 | 1.111
(0.762–1.619) | 0.584 | 1.113
(0.762–1.626) | 0.579 |
| rs3731114 | C/C | 133 | 143 | 1.000 |
| 1.000 |
|
|
| C/G | 60 | 74 | 0.872
(0.576–1.319) | 0.516 | 0.848
(0.557–1.291) | 0.442 |
|
| G/G | 12 | 13 | 0.992
(0.437–2.252) | 0.986 | 1.006
(0.441–2.292) | 0.989 |
|
| C/G+G/G | 72 | 87 | 0.890
(0.602–1.316) | 0.559 | 0.885
(0.596–1.313) | 0.543 |
| rs3729587 | G/G | 90 | 111 | 1.000 |
| 1.000 |
|
|
| G/C | 93 | 99 | 1.159
(0.779–1.723) | 0.467 | 1.132
(0.759–1.689) | 0.543 |
|
| C/C | 22 | 20 | 1.357
(0.697–2.641) | 0.369 | 1.392
(0.711–2.726) | 0.335 |
|
| G/C+C/C | 115 | 119 | 1.192
(0.817–1.740) | 0.363 | 1.184
(0.810–1.732) | 0.383 |
| rs2607775 | C/C | 146 | 190 | 1.000 |
| 1.000 |
|
|
| C/G | 51 | 36 | 1.844
(1.143–2.974) | 0.011 | 1.839
(1.139–2.970) | 0.013b |
|
| G/G | 8 | 4 | 2.603
(0.769–8.811) | 0.112 | 2.500
(0.733–8.522) | 0.143 |
|
| C/G+G/G | 59 | 40 | 1.920
(1.217–3.028) | 0.005 | 1.914
(1.212–3.024) | 0.005b |
| rs3731055 | G/G | 122 | 135 | 1.000 |
| 1.000 |
|
|
| G/A | 71 | 79 | 0.995
(0.664–1.489) | 0.979 | 0.981
(0.655–1.471) | 0.928 |
|
| A/A | 12 | 16 | 0.830
(0.378–1.824) | 0.642 | 0.830
(0.377–1.829) | 0.644 |
|
| G/A+A/A | 83 | 95 | 0.967
(0.659–1.418) | 0.863 | 0.965
(0.657–1.417) | 0.856 |
Function prediction and expression
analysis
The XPC SNP functions were predicted using the
SNPinfo Web Server (https://snpinfo.niehs.nih.gov/). rs2470353 was
identified to be located in the region of intron 12, but was not
predicted to be a functional SNP. rs2607775 was located in the
transcription factor binding site (TFBS) of the 5′UTR of XPC.
Therefore, rs2607775 was predicted to influence XPC expression.
Since only blood samples were collected in the
present study, XPC expression in pancreatic adenocarcinoma (PAAD)
was analyzed using the online tool UALCAN (http://ualcan.path.uab.edu/analysis.html) (22). XPC expression was decreased in PAAD
patients (P>0.05; Fig. 1A)
irrespective of sex (Fig. 1B), age
(Fig. 1C), cancer stage (Fig. 1D), drinking habit (Fig. 1E), chronic pancreatitis status
(Fig. 1F), diabetes status (Fig. 1G) and ethnicity (Fig. 1H), with few exceptions. Notably, XPC
expression was demonstrated to be significantly decreased in Asian
patients with PAAD (P=0.02; Fig. 1H).
Although a general trend of decreased XPC expression was observed
in patients with PAAD, the majority of these differences were not
statistically significant, potentially due to the small number of
control samples (n=4) and the inevitably large individual
differences in expression.
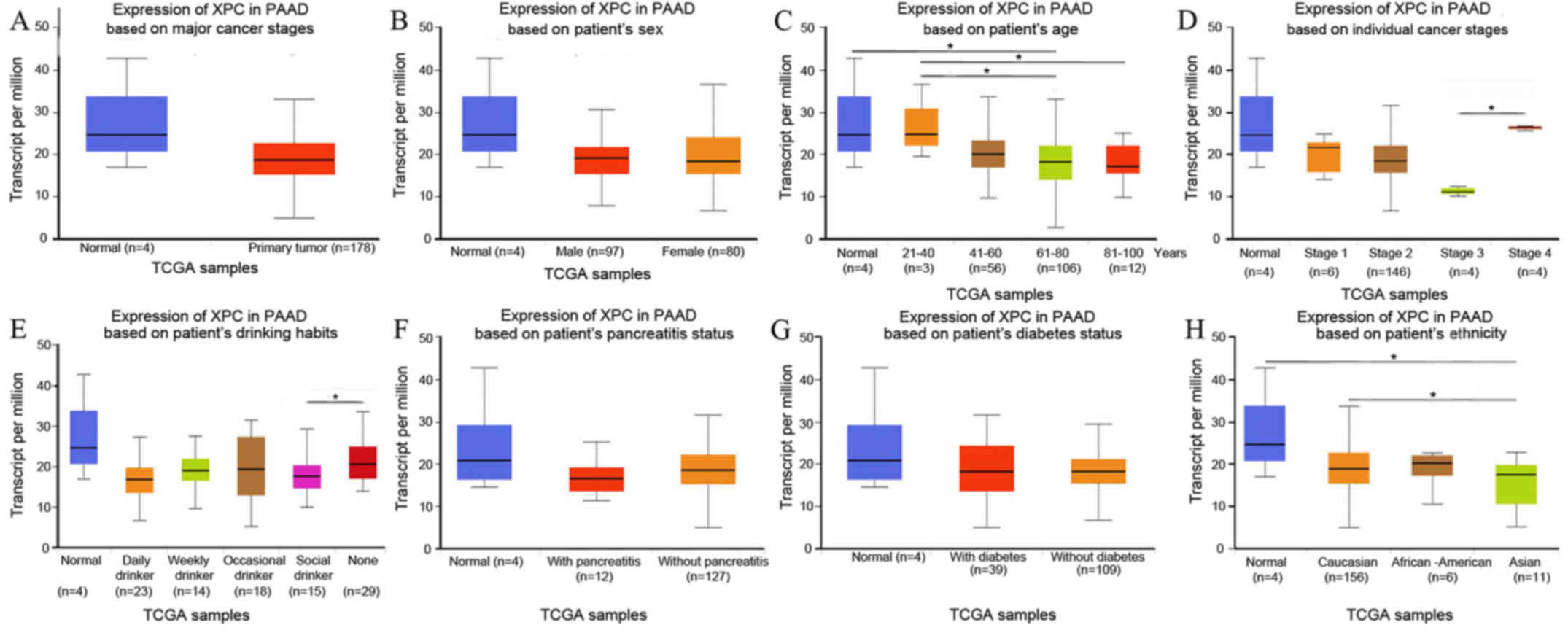 | Figure 1.(A) XPC expression in PAAD patients,
(B) sex, (C) age, (D) cancer stage, (E) drinking habit, (F) chronic
pancreatitis status, (G) diabetes status, and (H) patient ethnicity
was analyzed using the online tool UALCAN (18). *P<0.05. XPC, xeroderma pigmentosum
group C; PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome
Atlas. |
LD and haplotype association
analysis
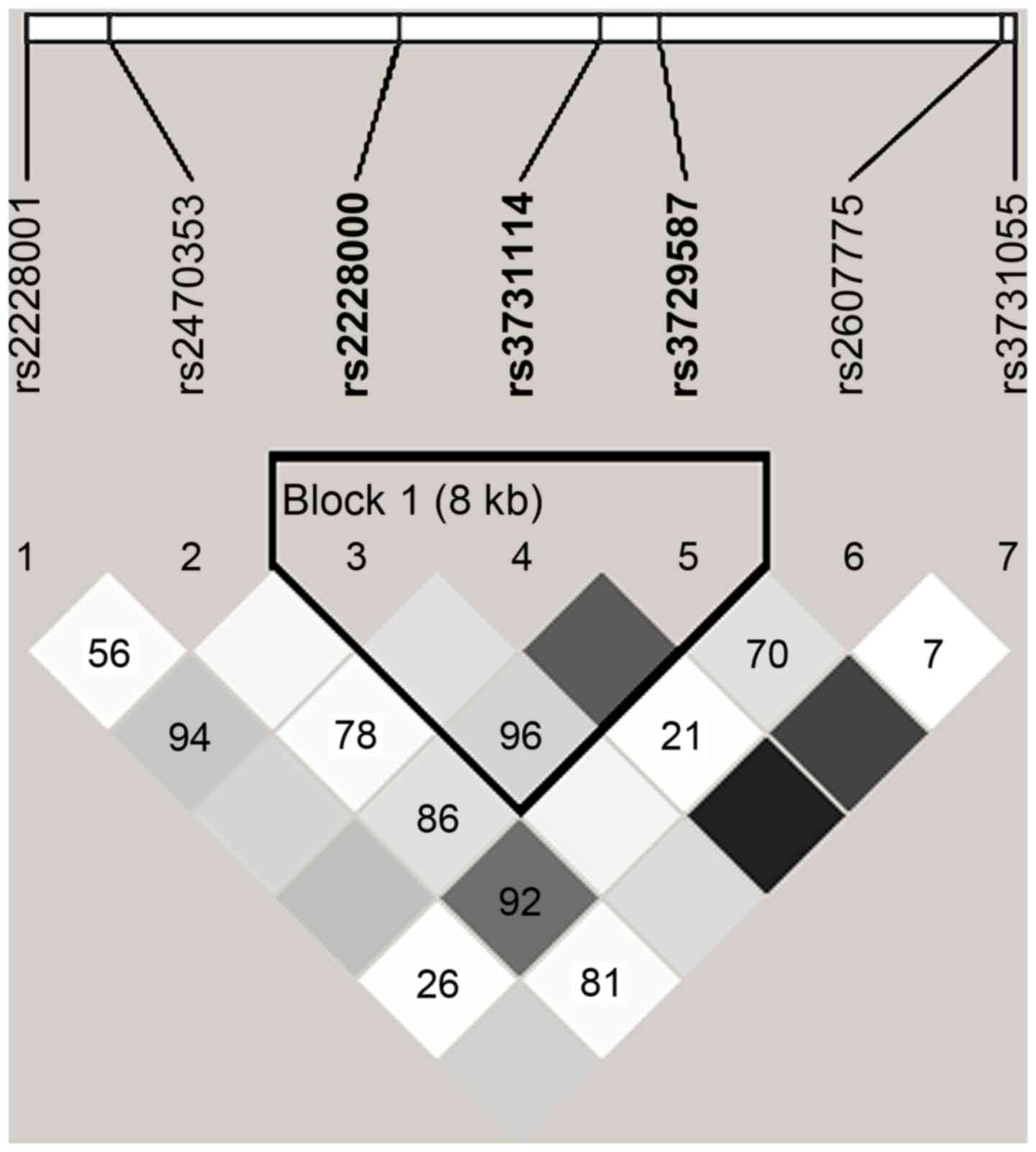
In Fig. 2, the range
of the area surrounded by black lines indicates that 3 tag-SNPs of
the XPC gene were contained in a haplotype and in a state of
linkage disequilibrium. Block 1 comprised rs2228000, rs3731114 and
rs3729587 (Fig. 2). The frequencies
of the haplotypes CCC and TCG were higher in patients with cancer
compared with those in the non-cancer controls, and the CCC
haplotype for Block 1 significantly increased the risk of PC (OR,
1.610; 95% CI, 1.035–2.481; P=0.034; Table IV).
 | Table IV.XPC haplotype of rs2228000, rs3731114
and rs3729587 frequencies and associations with pancreatic cancer
risk. |
Table IV.
XPC haplotype of rs2228000, rs3731114
and rs3729587 frequencies and associations with pancreatic cancer
risk.
| Haplotype | Freq | Cases (freq) | Controls
(freq) | OR (95% CI) | P-value |
|---|
| CGC | 0.208 | 0.201 | 0.217 | 0.928
(0.667–1.285) | 0.646 |
| CCC | 0.106 | 0.129 | 0.082 | 1.610
(1.035–2.481) | 0.034a |
| TCG | 0.299 | 0.301 | 0.297 | 1.021
(0.765–1.367) | 0.887 |
| CCG | 0.384 | 0.365 | 0.401 | 0.863
(0.651–1.127) | 0.276 |
Interaction analysis of smoking and
XPC gene polymorphism
Compared with non-smoking subjects with wild-type GG
gene in the rs2470353 locus, PC risk did not significantly increase
in smoking subjects with the GC gene type (P>0.05). PC risk
increased by 3.505-fold in heavy smokers (tobacco ≥25 packets/year)
with the variant allele C (OR=4.505, 95% CI=1.418–15.007, P=0.008;
Table V). Compared with non-smoking
subjects with wild gene type CC in the rs2607775 locus, PC risk did
not increase in smoking subjects with the CC gene type (P>0.05).
PC risk increased by 3.950-fold in heavy smokers (tobacco ≥25
packets/year) with the variant allele G (CG+GG) (OR, 4.950; 95% CI,
1.758–13.924; P=0.001; Table
VI).
 | Table V.Risk of XPC genotypes at rs2470353
with pancreatic cancer by smoking status. |
Table V.
Risk of XPC genotypes at rs2470353
with pancreatic cancer by smoking status.
| Genotype | Smoking status,
pack-years | Cases, n | Controls, n | OR (95% CI) | P-value |
|---|
| GG | Non-smoker | 83 | 115 | 1.000 |
|
|
| <25 | 56 | 73 | 1.041
(0.598–1.635) | 0.724 |
|
| ≥25 | 19 | 13 | 2.071
(0.967–4.431) | 0.082 |
| GC+CC | Non-smoker | 22 | 18 | 1.683
(0.846–3.345) | 0.096 |
|
| <25 | 12 | 7 | 2.366
(0.891–6.278) | 0.076 |
|
| ≥25 | 13 | 4 | 4.505
(1.418–15.007) | 0.008a |
 | Table VI.Risk of XPC genotypes at rs2607775
with pancreatic cancer by smoking status. |
Table VI.
Risk of XPC genotypes at rs2607775
with pancreatic cancer by smoking status.
| Genotype | Smoking status,
pack-years | Cases, n | Controls, n | OR (95% CI) | P-value |
|---|
| CC | Non-smoker | 77 | 106 | 1.000 |
|
|
| <25 | 55 | 72 | 1.047
(0.661–1.658) | 0.831 |
|
| ≥25 | 14 | 12 | 1.503
(0.668–3.376) | 0.288 |
| CG+GG | Non-smoker | 28 | 27 | 1.414
(0.771–2.595) | 0.270 |
|
| <25 | 13 | 8 | 2.251
(0.891–5.759) | 0.089 |
|
| ≥25 | 18 | 5 | 4.950
(1.758–13.924) | 0.001a |
Discussion
Previous studies have reported XPC polymorphisms to
be associated with cancer risk. There are three polymorphisms most
frequently detected in the XPC gene: Poly AT insertion/deletion on
intron 9 (PAT), A to C substitution in exon 15 (Lys939Gln,
rs2228001) and C to T substitution in exon 9 (Ala499Val, rs2228000)
(23). Epidemiological studies have
demonstrated that the PAT+/+ genotype results in a
1.85-fold increase in the risk of squamous cell carcinoma of the
head and neck (24) and a 1.6-fold
increase in the risk of lung cancer (25). Meta-analysis revealed that the exon 15
Lys939Gln (rs2228001 A>C) C/C gene type is associated with
increased risk of lung cancer and esophageal cancer (26,27). The
XPC Ala499Val (rs2228000, C>T) polymorphism is associated with
the risk of endometrial, colorectal and liver cancer, as well as
other malignant cancer types (28–30).
However, the association between other XPC polymorphisms and
bladder cancer remains controversial (31–33).
In the present study, the associations between
genetic polymorphisms of XPC and PC risk were investigated using a
tag-SNP method. The results revealed that variant alleles at two
loci were associated with increased PC risk, even though the
rs2470353 locus was located in the intron area and the rs2607775
locus was located in the 5′UTR (P<0.05). The other five tag-SNP
loci, including in exon 9 (rs2228000) and exon 15 (rs222800), did
not exhibit significant differences in the distribution of gene
type or allelic gene frequency between the case and control groups
(P>0.05; Tables III and IV).
G/C polymorphisms at or near the exonic boundaries
in intron 12 of the XPC gene may affect mRNA translation through
exon skipping and/or aberrant mRNA folding (34–37). SNPs
in the 5′UTR may affect XPC gene expression via promoter modulation
(34), resulting in reduced DNA
repair capacity (DRC) and increased risk of PC. Unfortunately,
although rs2470353 is located in the region of intron 12, it is not
a predicted functional SNP. rs2607775 is located on the TFBS of the
5′UTR of XPC, and is therefore predicted to influence XPC
expression (38,39). Furthermore, XPC expression in PAAD was
analyzed using UALCAN (22) in the
present study. XPC expression was identified to be decreased in
patients with PAAD irrespective of sex, age, cancer stage, drinking
habits, chronic pancreatitis status, diabetes status, or race, with
few exceptions. The most notable finding was that XPC expression
was decreased significantly in Asian patients with PAAD. However,
due to the small number of normal control samples, the majority of
these differences were not statistically significant.
SNPs in coding regions, as well as non-coding
regions of the XPC gene, that are in LD with each other as part of
a given haplotype may act in a collective manner to influence the
phenotype. The results of the present study revealed that the CCC
haplotype of rs2228000, rs3731114 and rs3729587 exhibited a higher
frequency in patients with PC, compared with that in the control
group, indicating that the CCC haplotype may result in an increased
risk of PC. However, the exact mechanism of this remains
unclear.
Smoking is recognized as a traditional risk factor
for PC (9,40). The present study also revealed that
smoking in the XPC rs2470353 (GC+CC) and/or rs2607775 (CG+GG)
subjects significantly increased PC risk. In subjects with an
accumulative smoking amount of ≥25 packets/year, PC risk increased
3.505-fold with rs2470353 (GC+CC) (OR, 4.505; 95% CI, 1.418–15.007;
P=0.008) and 3.950-fold with rs2607775 (CG+GG) (OR, 4.950; 95% CI,
1.758–13.924; P=0.001), indicating that the combination of
mutations and smoking may serve an important role in PC
progression. Smoking causes genetic damage and/or cell mutations
(41) that may not be repaired by the
NER pathway (13,14), since genetic damage may not be
recognized by XPC with the rs2470353 (GC+CC) and/or rs2607775
(CG+GG) mutant gene type.
The present study has initially indicated that the
XPC gene rs2470353 and rs2607775 loci polymorphisms are associated
with PC risk. The haplotype CCC of rs2228000, rs3731114 and
rs3729587 was also identified to be associated with increased PC
risk. However, additional SNP loci have been continuously selected
for future study, particularly the functional loci of the exon
regions and loci at splicing regions. In addition, the chromosome
hereditary variation of the XPC gene and its association with PC
could be further verified by transcription analysis, in order to
evaluate the effects on regulation and splicing of the XPC
gene.
Acknowledgements
Not applicable.
Funding
This study was supported by research grants from the
National Natural Science Foundation of China (grant no.
30960433).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request. The XPC expression and survival analysis in PC was
evaluated using the online analysis tool UALCAN (http://ualcan.path.uab.edu/analysis.html) (21).
Authors' contributions
XHL, DY and XYW conceived and designed the
experiments. DY, WD and XYW participated in clinical data
collection. XHL and DYA performed the experiments. XHL, DY and JXZ
analyzed the data. XHL, DY and JXZ contributed to the
interpretation of results obtained and manuscript construction. DY
and XYW analyzed and interpreted the patients' data regarding the
clinical characteristics. XHL and DY wrote the paper. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All samples were obtained with the informed consent
of the participants prior to their inclusion in the study,
according to Helsinki Declaration principles and with approval of
the Ethical Committee of The First Affiliated Hospital of Xinjiang
Medical University.
Patient consent for publication
All patients provided informed consent for the
publication of any associated data.
Competing interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chakraborty S, Baine MJ, Sasson AR and
Batra SK: Current status of molecular markers for early detection
of sporadic pancreatic cancer. Biochim Biophys Acta. 1815:44–64.
2011.PubMed/NCBI
|
|
3
|
Trede M, Schwall G and Saeger HD: Survival
after pancreatoduodenectomy. 118 consecutive resections without an
operative mortality. Ann Surg. 211:447–458. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang MJ, Jang JY, Chang YR, Kwon W, Jung W
and Kim SW: Revisiting the concept of lymph node metastases of
pancreatic head cancer: Number of metastatic lymph nodes and lymph
node ratio according to N stage. Ann Surg Oncol. 21:1545–1551.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Eng J Med. 371:1039–1049. 2014.
View Article : Google Scholar
|
|
6
|
DiMagno EP, Reber HA and Tempero MA: AGA
technical review on the epidemiology, diagnosis, and treatment of
pancreatic ductal adenocarcinoma. American Gastroenterological
Association. Gastroenterol. 117:1464–1484. 1999. View Article : Google Scholar
|
|
7
|
Lamerz R: Role of tumour markers,
cytogenetics. Ann Oncol 10 Suppl. 4:145–149. 1999. View Article : Google Scholar
|
|
8
|
Pandol S, Gukovskaya A, Edderkaoui M,
Dawson D, Eibl G and Lugea A: Epidemiology, risk factors, and the
promotion of pancreatic cancer: Role of the stellate cell. J
Gastroenterol Hepatol. 27 Suppl 2:S127–S134. 2012. View Article : Google Scholar
|
|
9
|
Krejs GJ: Pancreatic cancer: Epidemiology
and risk factors. Dig Dis. 28:355–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dou K, Xu Q and Han X: The association
between XPC Lys939Gln gene polymorphism and urinary bladder cancer
susceptibility: A systematic review and meta-analysis. Diagn
Pathol. 8:1122013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li L, Peterson C and Legerski R: Sequence
of the mouse XPC cDNA and genomic structure of the human XPC gene.
Nucleic Acids Res. 24:1026–1028. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Melis JP, Luijten M, Mullenders LH and van
Steeg H: The role of XPC: Implications in cancer and oxidative DNA
damage. Mutat Res. 728:107–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khan SG, Muniz-Medina V, Shahlavi T, Baker
CC, Inui H, Ueda T, Emmert S, Schneider TD and Kraemer KH: The
human XPC DNA repair gene: Arrangement, splice site information
content and influence of a single nucleotide polymorphism in a
splice acceptor site on alternative splicing and function. Nucleic
Acids Res. 30:3624–3631. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sugasawa K, Ng JM, Masutani C, Iwai S, Van
der Spek PJ, Eker AP, Hanaoka F, Bootsma D and Hoeijmakers JH:
Xeroderma pigmentosum group C protein complex is the initiator of
global genome nucleotide excision repair. Mol Cell. 2:223–232.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duell EJ, Bracci PM, Moore JH, Burk RD,
Kelsey KT and Holly EA: Detecting pathway-based gene-gene and
gene-environment interactions in pancreatic cancer. Cancer
Epidemiol Biomarkers Prev. 17:1470–1479. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Lin DX, Lu XH, Miao XP and Li H:
Polymorphisms of the DNA repair genes XRCC1 and XPC: Relationship
to pancreatic cancer risk. Wei Sheng Yan Jiu. 35:534–536. 2006.(In
Chinese). PubMed/NCBI
|
|
17
|
Zhao F, Shang Y, Zeng C, Gao D and Li K:
Association of single nucleotide polymorphisms of DNA repair genes
in NER pathway and susceptibility to pancreatic cancer. Int J Clin
Exp Pathol. 8:11579–11586. 2015.PubMed/NCBI
|
|
18
|
McWilliams RR, Bamlet WR, Cunningham JM,
Goode EL, de Andrade M, Boardman LA and Petersen GM: Polymorphisms
in DNA repair genes, smoking, and pancreatic adenocarcinoma risk.
Cancer Res. 68:4928–4935. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tabor HK, Risch NJ and Myers RM:
Candidate-gene approaches for studying complex genetic traits:
Practical considerations. Nat Rev Genet. 3:391–397. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hirooka N, Kadowaki T, Sekikawa A, Ueshima
H, Choo J, Miura K, Okamura T, Fujiyoshi A, Kadowaki S, Kadota A,
et al: Influence of cigarette smoking on coronary artery and aortic
calcium among random samples from populations of middle-aged
Japanese and Korean men. J Epidemiol Community Health. 67:119–124.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lalitha S: Primer Premier 5. Biotech
Software Intern Rep. 1:62000.
|
|
22
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Francisco G, Menezes PR, Eluf-Neto J and
Chammas R: XPC polymorphisms play a role in tissue-specific
carcinogenesis: A meta-analysis. Eur J Hum Genet. 16:724–734. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen H, Sturgis EM, Khan SG, Qiao Y,
Shahlavi T, Eicher SA, Xu Y, Wang X, Strom SS, Spitz MR, et al: An
intronic poly (AT) polymorphism of the DNA repair gene XPC and risk
of squamous cell carcinoma of the head and neck: A case-control
study. Cancer Res. 61:3321–3325. 2001.PubMed/NCBI
|
|
25
|
Marín MS, López-Cima MF, García-Castro L,
Pascual T, Marrón MG and Tardón A: Poly (AT) polymorphism in intron
11 of the XPC DNA repair gene enhances the risk of lung cancer.
Cancer Epidemiol Biomarkers Prev. 13:1788–1793. 2004.PubMed/NCBI
|
|
26
|
Jin B, Dong Y, Zhang X, Wang H and Han B:
Association of XPC polymorphisms and lung cancer risk: A
meta-analysis. PLoS One. 9:e939372014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo W, Zhou RM, Wan LL, Wang N, Li Y,
Zhang XJ and Dong XJ: Polymorphisms of the DNA repair gene
xeroderma pigmentosum groups A and C and risk of esophageal
squamous cell carcinoma in a population of high incidence region of
North China. J Cancer Res Clin Oncol. 134:263–270. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Li Z, Liu N and Zhang G:
Association between CCND1 and XPC polymorphisms and bladder cancer
risk: A meta-analysis based on 15 case-control studies. Tumour
Biol. 35:3155–3165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paszkowska-Szczur K, Scott RJ, Górski B,
Cybulski C, Kurzawski G, Dymerska D, Gupta S, van de Wetering T,
Masojć B, Kashyap A, et al: Polymorphisms in nucleotide excision
repair genes and susceptibility to colorectal cancer in the Polish
population. Mol Biol Rep. 42:755–764. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paszkowska-Szczur K, Scott RJ,
Serrano-Fernandez P, Mirecka A, Gapska P, Górski B, Cybulski C,
Maleszka R, Sulikowski M, Nagay L, et al: Xeroderma pigmentosum
genes and melanoma risk. Int J Cancer. 133:1094–1100. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sak SC, Barrett JH, Paul AB, Bishop DT and
Kiltie AE: The polyAT, intronic IVS11-6 and Lys939Gln XPC
polymorphisms are not associated with transitional cell carcinoma
of the bladder. Br J Cancer. 92:2262–2265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sanyal S, Festa F, Sakano S, Zhang Z,
Steineck G, Norming U, Wijkström H, Larsson P, Kumar R and Hemminki
K: Polymorphisms in DNA repair and metabolic genes in bladder
cancer. Carcinogenesis. 25:729–734. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sak SC, Barrett JH, Paul AB, Bishop DT and
Kiltie AE: Comprehensive analysis of 22 XPC polymorphisms and
bladder cancer risk. Cancer Epidemiol Biomarkers Prev.
15:2537–2541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng AJ, Mao YM and Cui RZ: The effect of
gene polymorphism in promoter and intron 1 on human ApoA I
expression. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 23:610–613.
2006.(In Chinese). PubMed/NCBI
|
|
35
|
Duan ZX, Zhu PF, Dong H, Gu W, Yang C, Liu
Q, Wang ZG and Jiang JX: Functional significance of the TLR4/11367
polymorphism identified in Chinese Han population. Shock.
28:160–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kinslow CJ, El-Zein RA, Hill CE, Wickliffe
JK and Abdel-Rahman SZ: Single nucleotide polymorphisms 5′ upstream
the coding region of the NEIL2 gene influence gene transcription
levels and alter levels of genetic damage. Genes Chromosomes
Cancer. 47:923–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Law AJ, Kleinman JE, Weinberger DR and
Weickert CS: Disease-associated intronic variants in the ErbB4 gene
are related to altered ErbB4 splice-variant expression in the brain
in schizophrenia. Hum Mol Genet. 16:129–141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiao B, Ansari AH, Scott GB, Sak SC,
Chambers PA, Elliott F, Teo MT, Bentley J, Churchman M, Hall J, et
al: In vitro functional effects of XPC gene rare variants from
bladder cancer patients. Carcinogenesis. 32:516–521. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bai Y, Xu L, Yang X, Hu Z, Yuan J, Wang F,
Shao M, Yuan W, Qian J, Ma H, et al: Sequence variations in DNA
repair gene XPC is associated with lung cancer risk in a Chinese
population: A case-control study. BMC Cancer. 7:812007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Maisonneuve P and Lowenfels AB:
Epidemiology of pancreatic cancer: An update. Dig Dis. 28:645–656.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee YJ, Shim HS, Kang YA, Hong SJ, Kim HK,
Kim H, Kim SK, Choi SH, Kim JH and Cho BC: Dose effect of cigarette
smoking on frequency and spectrum of epidermal growth factor
receptor gene mutations in Korean patients with non-small cell lung
cancer. J Cancer Res Clin Oncol. 136:1937–1944. 2010. View Article : Google Scholar : PubMed/NCBI
|